Proteomic Analyses of Thioredoxins f and m Arabidopsis thaliana Mutants Indicate Specific Functions for These Proteins in Plants
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material and Growth Conditions
2.2. Protein Extraction, Solubilisation
2.3. Isoelectrofocusing, 2-D Electrophoresis, Gel Staining, Image Capture and Analysis
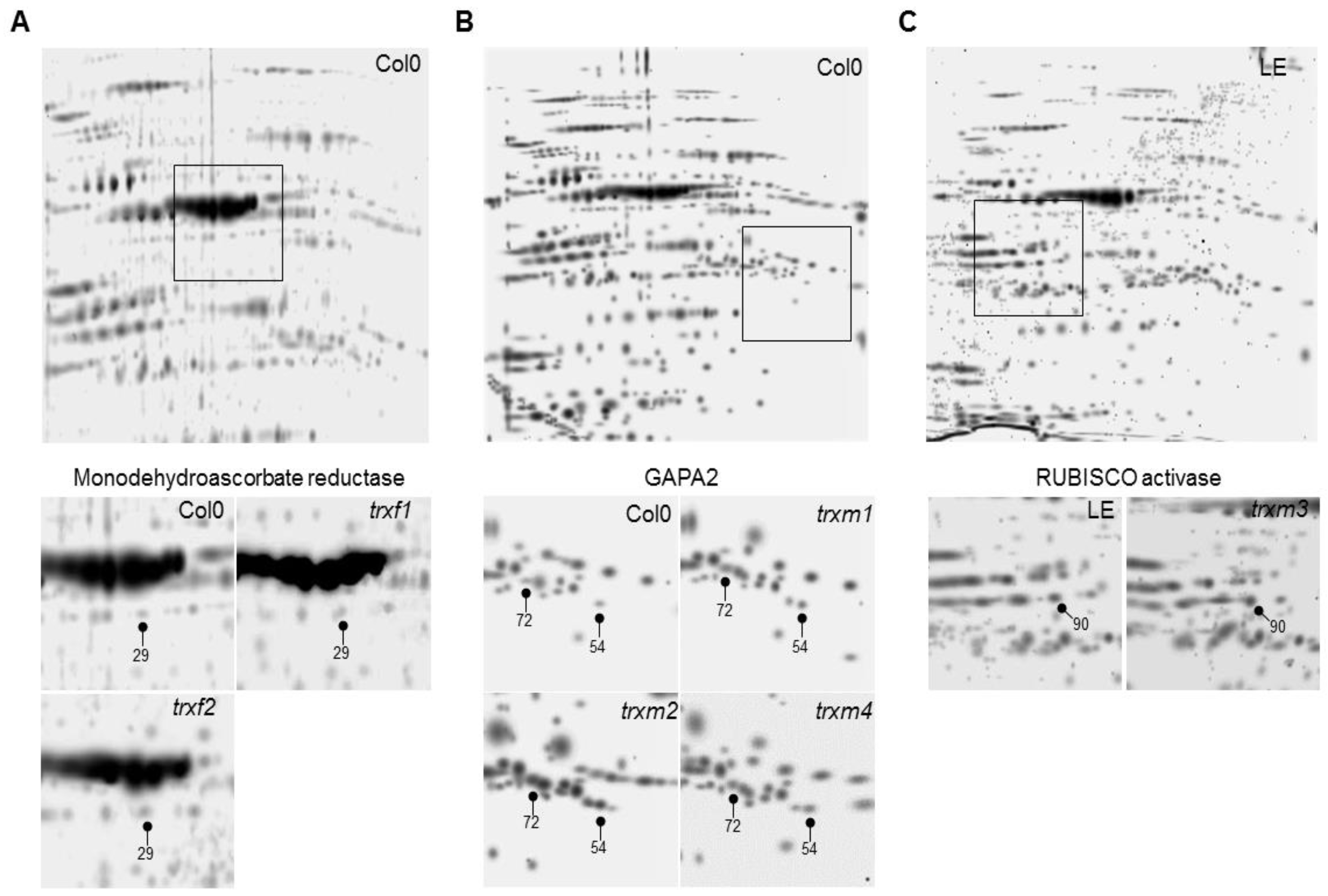
3. Results
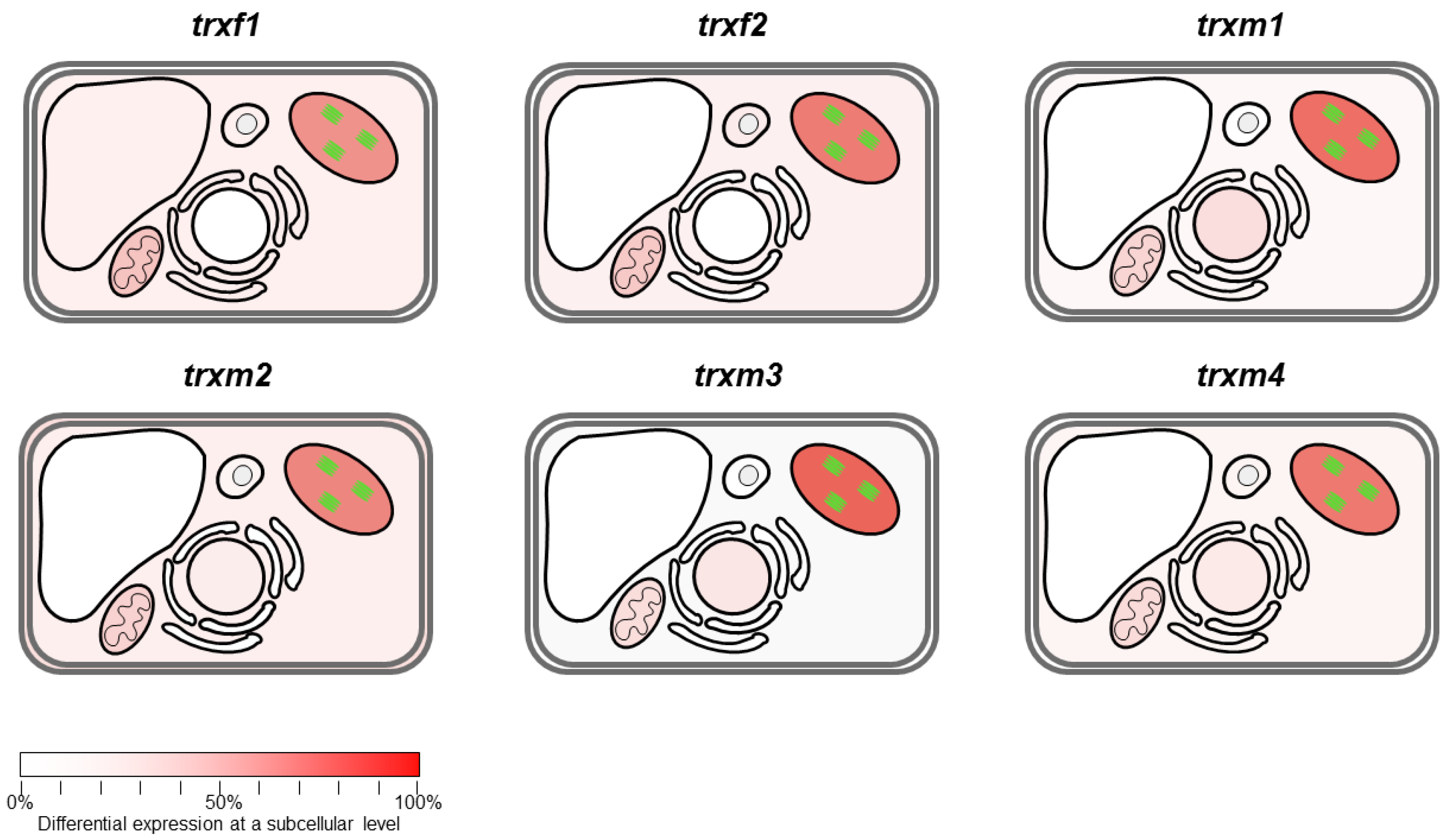
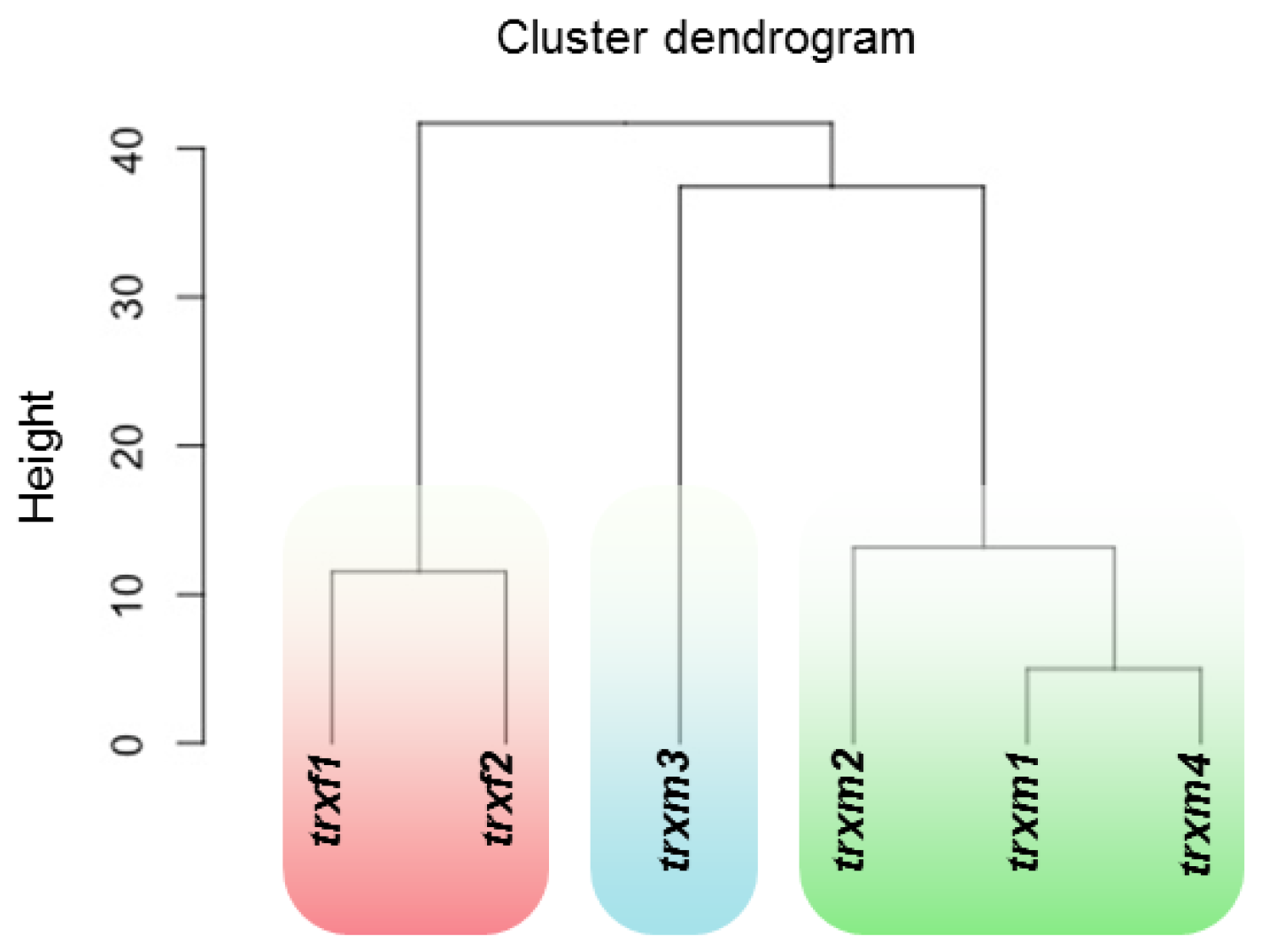
Differentially Regulated Proteins from Rosettes of trxf1, trxf2, trxm1, trxm2, trxm3 and trxm4 Mutants
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Montrichard, F.; Alkhalfioui, F.; Yano, H.; Vensel, W.H.; Hurkman, W.J.; Buchanan, B.B. Thioredoxin targets in plants: The first 30 years. J. Proteom. 2009, 72, 452–474. [Google Scholar] [CrossRef] [PubMed]
- Buchanan, B.B.; Schurmann, P.; Wolosiuk, R.A.; Jacquot, J.P. The ferredoxin/thioredoxin system: From discovery to molecular structures and beyond. Photosynth. Res. 2002, 73, 215–222. [Google Scholar] [CrossRef] [PubMed]
- Barajas-López, J.D.; Serrato, A.J.; Cazalis, R.; Meyer, Y.; Chueca, A.; Reichheld, J.P.; Sahrawy, M. Circadian regulation of chloroplastic f and m thioredoxins through control of the CCA1 transcription factor. J. Exp. Bot. 2011, 62, 2039–2051. [Google Scholar] [CrossRef] [PubMed]
- Chibani, K.; Wingsle, G.; Jacquot, J.P.; Gelhaye, E.; Rouhier, N. Comparative genomic study of the thioredoxin family in photosynthetic organisms with emphasis on Populus trichocarpa. Mol. Plant 2009, 2, 308–322. [Google Scholar] [CrossRef] [PubMed]
- Motohashi, K.; Kondoh, A.; Stumpp, M.T.; Hisabori, T. Comprehensive survey of proteins targeted by chloroplast thioredoxin. Proc. Natl. Acad. Sci. USA 2001, 8, 11224–11229. [Google Scholar] [CrossRef] [PubMed]
- Collin, V.; Issakidis-Bourguet, E.; Marchand, C.; Hirasawa, M.; Lancelin, J.M.; Knaff, D.B.; Miginiac-Maslow, M. The Arabidopsis plastidial thioredoxins: New functions and new insights into specificity. J. Biol. Chem. 2003, 278, 23747–23752. [Google Scholar] [CrossRef] [PubMed]
- Lemaire, S.D.; Michelet, L.; Zaffagnini, M.; Massot, V.; Issakidis-Bourguet, E. Thioredoxins in chloroplasts. Curr. Genet. 2007, 51, 343–365. [Google Scholar] [CrossRef] [PubMed]
- Serrato, A.J.; Fernández-Trijueque, J.; Barajas-López, J.D.; Chueca, A.; Sahrawy, M. Plastid thioredoxins: A “one-for-all” redox-signaling system in plants. Front. Plant Sci. 2013, 4, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Chiadmi, M.; Navaza, A.; Miginiac-Maslow, M.; Jacquot, J.P.; Cherfils, J. Redox signalling in the chloroplast: Structure of oxidized pea fructose-1,6-bisphosphate phosphatase. EMBO J. 1999, 18, 6809–6815. [Google Scholar] [CrossRef] [PubMed]
- Schürmann, P.; Wolosiuk, R.A.; Breazeale, V.D.; Buchanan, B.B. 2 Proteins function in regulation of photosynthetic CO2 assimilation in chloroplasts. Nature 1976, 263, 257–258. [Google Scholar] [CrossRef]
- Buchanan, B.B.; Crawford, N.A.; Wolosiuk, R.A. Ferredoxin-thioredoxin system functions with effectors in activation of NADP glyceraldehyde 3-phosphate dehydrogenase of barley seedlings. Plant Sci. Let. 1978, 12, 257–264. [Google Scholar] [CrossRef]
- Jacquot, J.P.; Vidal, J.; Gadal, P.; Schürmann, P. Evidence for existence of several enzyme-specific thioredoxins in plants. FEBS Let. 1978, 96, 243–246. [Google Scholar] [CrossRef]
- Buchanan, B.B.; Balmer, Y. Redox regulation: A broadening horizon. Ann. Rev. Plant Biol. 2005, 56, 187–220. [Google Scholar] [CrossRef] [PubMed]
- Balmer, Y.; Vensel, W.H.; DuPont, F.M.; Buchanan, B.B.; Hurkman, W.J. Proteome of amyloplasts isolated from developing wheat endosperm presents evidence of broad metabolic capability. J. Exp. Bot. 2006, 57, 1591–1602. [Google Scholar] [CrossRef] [PubMed]
- Barajas-López, J.D.; Serrato, A.J.; Olmedilla, A.; Chueca, A.; Sahrawy, M. Localization in roots and flowers of pea chloroplast thioredoxin f and m proteins reveals new roles in non-photosynthetic organs. Plant Physiol. 2007, 145, 1–15. [Google Scholar]
- Hisabory, T. Thioredoxin affinity chromatography: A useful method for further understanding the thioredoxin network. J. Exp. Bot. 2005, 56, 1463–1468. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, K.; Hisabory, T. Two distinct redox cascades cooperatively regulate chloroplast functions and sustain plant viability. Proc. Natl. Acad. Sci. USA 2016, 113, E3967–E3976. [Google Scholar] [CrossRef] [PubMed]
- Valot, B.; Negroni, L.; Zivy, M.; Gianinazzi, S.; Dumas-Gaudot, E. A mass spectrometric approach to identify arbuscular mycorrhiza-related proteins in root plasma membrane fractions. Proteom. J. 2006, S145–S155. [Google Scholar] [CrossRef] [PubMed]
- Soto-Suarez, M.; Serrato, A.J.; Rojas-González, J.A.; Bautista, R.; Sahrawy, M. Transcriptomic and proteomic approach to identify differentially expressed genes and proteins in Arabidopsis thaliana mutants lacking chloroplastic 1 and cytosolic FBPases reveals several levels of metabolic regulation. BMC Plant Biol. 2016, 16, 258–274. [Google Scholar] [CrossRef] [PubMed]
- Ramagli, L.S.; Rodríguez, L.V. Quantitation of microgram amounts of protein in two-dimensional polyacrylamide-gel sample buffer electrophoresis electrophoresis. Electrophoresis 1985, 6, 559–563. [Google Scholar] [CrossRef]
- Yan, J.X.; Wait, R.; Berkelman, T.; Harry, R.A.; Westbrook, J.A.; Wheeler, C.H.; Dunn, M.J. A modified silver staining protocol for visualization of proteins compatiblewith matrix-assisted laser desorption/ionization and electrospray ionization massspectrometry. Electrophoresis Int. J. 2000, 21, 3666–3672. [Google Scholar] [CrossRef]
- Castillejo, M.; Fernández-Aparicio, M.; Rubiales, D. Proteomic analysis by twodimensional differential in gel electrophoresis (2D DIGE) of the early response of Pisum sativum to Orobanchecrenata. J. Exp. Bot. 2012, 63, 107–119. [Google Scholar] [CrossRef] [PubMed]
- Courteille, A.; Vesa, S.; Sanz-Barrio, R.; Cazale, A.C.; Becuwe-Linka, N.; Farran, I.; Havaux, M.; Rey, P.; Rumeau, D. Thioredoxin m4 controls photosynthetic alternative electron pathways in Arabidopsis. Plant Physiol. 2013, 161, 508–520. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Liu, J.; Liu, B.; Feng, D.; Da, Q.; Shu, S.; Su, J.; Zhang, Y.; Wang, J.; Wang, H.-B. Evidence for a role of chloroplastic m-type thioredoxins in the biogenesis of photosystem II in Arabidopsis. Plant Physiol. 2013, 163, 1710–1728. [Google Scholar] [CrossRef] [PubMed]
- Benitez-Alfonso, Y.; Cilia, M.; San Roman, A.; Thomas, C.; Maule, A.; Hearn, S.; Jackson, D. Control of Arabidopsis meristem development by thioredoxin-dependent regulation of intercelular transport. Proc. Natl. Acad. Sci. USA 2009, 106, 3615–3620. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, K.; Hara, S.; Hisabori, T. Thioredoxin Selectivity for Thiol-Based Redox Regulation of Target Proteins in Chloroplasts. J. Biol. Chem. 2015, 115, 14278–14288. [Google Scholar] [CrossRef] [PubMed]
- Thormaehlen, I.; Ruber, J.; Von Roepenack-Lahaye, E.; Ehrlich, S.; Massot, V.; Huemmer, C.; Tezycka, J.; Issakidis-Bourguet, E.; Geigenberger, P. Inactivation of thioredoxin f1 leads to decreased light activation of ADP-glucose pyrophosphorylase and altered diurnal starch turnover in leaves of Arabidopsis plants. Plant Cell Environ. 2013, 36, 16–29. [Google Scholar] [CrossRef] [PubMed]
- Stumpp, M.T.; Motohashi, K.; Hisabori, T. Chloroplast thioredoxin mutants without active-site cysteines facilitate the reduction of the regulatory disulphide bridge on the gamma-subunit of chloroplast ATP synthase. Biochem. J. 1999, 341 Pt 1, 157–163. [Google Scholar] [CrossRef]
- Carrillo, L.R.; Froehlich, J.E.; Cruz, J.A.; Savage, L.J.; Kramer, D.M. Multi-level regulation of the chloroplast ATP synthase: The chloroplast NADPH thioredoxin reductase C (NTRC) is required for redox modulation specifically under low irradiance. Plant J. 2016, 87, 654–663. [Google Scholar] [CrossRef] [PubMed]
- Balmer, Y.; Vensel, W.H.; Tanaka, C.K.; Hurkman, W.J.; Gelhay, E.; Rouhier, N.; Jacquot, J.P.; Manieri, W.; Schürmann, P.; Droux, M.; et al. Thioredoxin links redox to the regulation of fundamental processes of plant mitochondria. Proc. Natl. Acad. Sci. USA 2004, 101, 2642–2647. [Google Scholar] [CrossRef] [PubMed]
- Plücken, H.; Müller, B.; Grohmann, D.; Westhoff, P.; Eichacker, L. The HCF136 protein is essential for assembly of the photosystem II reaction center in Arabidopsis thaliana. FEBS Lett. 2002, 532, 85–90. [Google Scholar] [CrossRef]
- Motohashi, K.; Hisabori, T. HCF164 receives reducing equivalents from stromal thioredoxin across the thylakoid membrane and mediates reduction of target proteins in the thylakoid lumen. J. Biol. Chem. 2006, 281, 35039–35047. [Google Scholar] [CrossRef] [PubMed]
- Rintamäki, E.; Martinsuo, P.; Pursisheimo, S.; Aro, E.M. Coorperative regulation of light-harvesting complex II phosphorylation via the plastoquinol and ferredoxin-thioredoxin system in chloroplasts. Proc. Natl. Acad. Sci. USA 2000, 97, 11644–11649. [Google Scholar] [CrossRef] [PubMed]
- Balmer, Y.; Vensel, W.H.; Cai, N.; Manieri, W.; Schürmann, P.; Hurkman, W.J.; Buchanan, B.B. A complete ferredoxin/thioredoxin system regulates fundamental processes in amyloplasts. Proc. Natl. Acad. Sci. USA 2006, 103, 2988–2993. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Trijueque, J.; Barajas-López, J.D.; Chueca, A.; Cazalis, R.; Sahrawy, M.; Serrato, A.J. Plastid thioredoxins f and m are related to the developing and salinity response of post-germinating seeds of Pisum sativum. Plant Sci. 2012, 188–189, 82–88. [Google Scholar] [CrossRef] [PubMed]
- Lambrix, V.; Reichelt, M.; Mitchell-Olds, T.; Kliebenstein, D.J.; Gershenzon, J. The Arabidopsis epithiospecifier protein promotes the hydrolysis of glucosinolates to nitriles and influences Trichoplusiani herbivory. Plant Cell 2001, 13, 2793–2807. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.-Y.; Ober, J.A.; Kliebenstein, D.J. The gene controlling the quantitative trait locus EPITHIOSPECIFIER MODIFIER1 alters glucosinolate hydrolysis and insect resistance in Arabidopsis. Plant Cell 2006, 18, 1524–1536. [Google Scholar] [CrossRef] [PubMed]
- Salvucci, M.E. Association of Rubisco activase with chaperonin-60β: A possible mechanism for protecting photosynthesis during heat stress. J. Exp. Bot. 2008, 59, 1923–1933. [Google Scholar] [CrossRef] [PubMed]



| Functional Category | Protein | Gene ID | Location | TRX Target | Spot | trxf1 | trxf2 |
|---|---|---|---|---|---|---|---|
| Amino acids metabolism | Glutamate-glyoxylate aminotransferase 1 | At1g23310 | Per. | - | 16 | ||
| Aminomethyltransferase | At1g11860 | Mit. | - | 8 | |||
| Glutamate-glyoxylate aminotransferase 2 | At1g70580 | Per. | - | 21 | |||
| 5-methyltetrahydropteroyltrigluta-mate-homocysteine methyltransferase 1 | At5g17920 | Per. | - | 33 | |||
| 5-methyltetrahydropteroyltrigluta-mate-homocysteine methyltransferase 2 | At3g03780 | Cy. | - | 12 | |||
| Calvin-Benson cycle | RUBISCO large subunit | AtCg00490 | Ch. | Yes | 5 | ||
| Transketolase1 | At3g60750 | Ch. | Yes | 20 | |||
| RUBISCO activase | At2g39730 | Ch. | Yes | 27 | |||
| ATP synthesis | ATP synthase subunit ß | AtCg00480 | Ch. | Yes | 15 | ||
| ATP synthase subunit α | AtCg00120 | Ch. | Yes | 4, 24 | |||
| ATP synthase subunit 1 | AtMg01190 | Mit. | Yes | 25 | |||
| Photosynthesis | PSII stability/assembly factor HCF136 | At5g23120 | Ch. | - | 9 | ||
| Ferredoxin-NADP reductase 1 | At5g66190 | Ch. | - | 17 | |||
| ABA signalling | β-D-glucopyranosyl abscisate β-glucosidase | At1g52400 | ER | - | 3 | ||
| Myrosinase 2 | At5g25980 | n.d. | Yes | 22 | |||
| Glycolisis | Triosephosphate isomerase | At3g55440 | Mit. | - | 31 | ||
| Glyceraldehyde-3-phosphate dehydrogenase C2 (GAPC2) | At1g13440 | Cy. | - | 26 | |||
| Stress response | Jacalin-Related lectin | At3g16470 | n.d. | - | 7 | ||
| Monodehydroascorbate reductase | At1g63940 | Ch., Mit. | Yes | 29 | |||
| Protein biosynthesis | Elongation factor Tu | At4g20360 | Ch. | Yes | 6 | ||
| ATP hydrolysis | V-type ATP synthase | At1g78900 | V. | - | 2 | ||
| PSII biogenesis | PSII stability/assembly factor HCF136 | At5g23120 | Ch. | - | 30 | ||
| Refolding activity | Chaperonin 60 subunit ß1 | At1g55490 | Ch. | - | 13 | ||
| Tricarboxylic acid cycle | Malate dehydrogenase 1 | At1g53240 | Mit. | Yes | 35 | ||
| Protein abundance change relative to the control (Col0). |  | ||||||
| Functional Category | Protein | Gene ID | Location | TRX Target | Spot | trxm1 | trxm2 | trxm4 |
|---|---|---|---|---|---|---|---|---|
| Calvin-Benson cycle | Transketolase | At3g60750 | Ch. | Yes | 38 | |||
| RUBISCO large subunit | AtCg00490 | Ch. | Yes | 28 | ||||
| Fructose-bisphosphate aldolase 2 | At4g38970 | Ch. | Yes | 59, 103 | ||||
| RUBISCO activase | At2g39730 | Ch. | Yes | 65 | ||||
| Glyceraldehyde-3-phosphate dehydrogenase B GAPB) | At1g42970 | Ch. | Yes | 68 | ||||
| Glyceraldehyde-3-phosphate dehydrogenase A2 (GAPA2) | At1g12900 | Ch. | Yes | 54, 72 | ||||
| Amino acids metabolism | Serine hydroxymethyltransferase 1 | At4g37930 | Mit. | Yes | 101 | |||
| Glycine dehydrogenase (decarboxylating) 1 | At4g33010 | Mit. | - | 100 | ||||
| Probable phosphoglycerate mutase 2 | At3g08590 | Mit. | - | 47 | ||||
| Glutamate-glyoxylate aminotransferase 1 | At1g23310 | Per. | Yes | 97 | ||||
| 5-methyltetrahydropteroyltrigluta-mate-homocysteine methyltransferase 1 | At5g17920 | Cy. | Yes | 45, 98 | ||||
| Photosynthesis | Chlorophyll a-b binding protein 2 | At1g29920 | Ch. | Yes | 42 | |||
| Ferredoxin-NADP reductase 1 | At5g66190 | Ch. | - | 43 | ||||
| Chlorophyll a-b binding protein | At2g34420 | Ch. | - | 56 | ||||
| Oxygen-evolving enhancer protein 2-1 | At1g06680 | Ch. | Yes | 57 | ||||
| Oxygen-evolving enhancer protein 1-2 | At3g50820 | Ch. | Yes | 41 | ||||
| Ferredoxin-NADP reductase 2 | At1g20020 | Ch. | - | 104 | ||||
| Stress response | Uncharacterized protein | At2g37660 | Ch. | - | 40 | |||
| Heat shock 70 kDa protein 3 | At3g09440 | N. | - | 46 | ||||
| Monodehydroascorbate reductase 1 | At3g52880 | Per. | Yes | 70 | ||||
| PSII stabilization/repair | Photosystem II stability/assembly factor HCF136 | At5g23120 | Ch. | - | 67 | |||
| Protease Do-like 1 | At3g27925 | Ch. | - | 39 | ||||
| Protein transport | Chaperone protein ClpC1 | At5g50920 | Ch. | - | 37 | |||
| ATP synthesis | ATP synthase subunit beta | AtCg00480 | Ch. | Yes | 93 | |||
| Glycolysis | Glyceraldehyde-3-phosphate dehydrogenase GAPC2 | At1g13440 | Cy. | Yes | 53 | |||
| Carbohydrate metabolism | Chloroplast stem-loop binding protein of 41 kDa b | At1g09340 | Ch. | - | 64 | |||
| Carbon utilization | β carbonic anhydrase 2 | At5g14740 | Ch. | - | 55 | |||
| Lipid degradation | GDSL esterase/lipase ESM1 | At3g14210 | N. | - | 105 | |||
| Protein refolding | Chaperonin 60 subunit beta 2 | At3g13470 | Ch. | Yes | 96 | |||
| Unkown | Polyketide cyclase/dehydrase and lipid transport superfamily protein | At4g14500 | Mit. | - | 61 | |||
| Uncharacterized protein | At2g37660 | Ch. | - | 40 | ||||
| Uncharacterized protein | At5g05113 | Mit. | - | 74 | ||||
| Disease resistance protein (NBS-LRR class) family | At5g40060 | n.d. | - | 75 | ||||
| Protein abundance change relative to the control (Col0). |  | |||||||
| Functional Category | Protein | Gene ID | Location | TRX Target | Spot | trxm3 |
|---|---|---|---|---|---|---|
| Calvin-Benson cycle | RUBISCO activase | At2g39730 | Ch. | Yes | 90 | |
| Fructose-bisphosphate aldolase 2 | At4g38970 | Ch. | Yes | 91 | ||
| ATP synthesis | ATP synthase subunit 1 | AtMG01190 | Mit. | Yes | 82 | |
| ATP synthase γ chain 1 | At4g04640 | Ch. | Yes | 85 | ||
| JA signalling /response | Epithiospecifier protein | At1g54040 | N. | - | 84 | |
| Lipoxygenase 2 | At3g45140 | Ch. | Yes | 76 | ||
| ABA signalling | Myrosinase 2 | At5g25980 | n.d. | Yes | 77 | |
| Carbon utilization | β carbonic anhydrase 2 | At5g14740 | Ch. | Yes | 92 | |
| Refolding activity | Chaperonin 60 subunit β 2 | At3g13470 | Ch. | - | 87 | |
| Stress response | Monodehydroascorbate reductase | At1g63940 | Ch., Mit. | Yes | 83 | |
| Protein abundance change relative to the control (Ler). |  | |||||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fernández-Trijueque, J.; Serrato, A.-J.; Sahrawy, M. Proteomic Analyses of Thioredoxins f and m Arabidopsis thaliana Mutants Indicate Specific Functions for These Proteins in Plants. Antioxidants 2019, 8, 54. https://doi.org/10.3390/antiox8030054
Fernández-Trijueque J, Serrato A-J, Sahrawy M. Proteomic Analyses of Thioredoxins f and m Arabidopsis thaliana Mutants Indicate Specific Functions for These Proteins in Plants. Antioxidants. 2019; 8(3):54. https://doi.org/10.3390/antiox8030054
Chicago/Turabian StyleFernández-Trijueque, Juan, Antonio-Jesús Serrato, and Mariam Sahrawy. 2019. "Proteomic Analyses of Thioredoxins f and m Arabidopsis thaliana Mutants Indicate Specific Functions for These Proteins in Plants" Antioxidants 8, no. 3: 54. https://doi.org/10.3390/antiox8030054
APA StyleFernández-Trijueque, J., Serrato, A.-J., & Sahrawy, M. (2019). Proteomic Analyses of Thioredoxins f and m Arabidopsis thaliana Mutants Indicate Specific Functions for These Proteins in Plants. Antioxidants, 8(3), 54. https://doi.org/10.3390/antiox8030054




