Antioxidant Effect of Ethyl Acetate Fraction from Kaempferia galanga L.: Integrated Phytochemical Profiling, Network Analysis, and Experimental Validation
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Plant Material and Discovery of Bioactive Fraction in K. galanga
2.3. LC-MS Analysis Conditions
2.4. Target Collection and Potential Target Prediction in KGEA
2.5. Protein–Protein Interaction (PPI) of the Targets
2.6. Functional Annotation and Pathway Analysis
2.7. Component–Target–Pathway Network Construction
2.8. Molecular Docking
2.9. Molecular Dynamics Simulation
2.10. DPPH Radical Scavenging Assay
2.11. ABTS Radical Scavenging Assay
2.12. Cell Culture
2.13. Cell Viability Assay
2.14. ROS Assay
2.15. JC-1 Assay
2.16. Evaluation of Antioxidant Enzyme Activity and Lipid Peroxidation
2.17. Western Blotting Analysis
2.18. Zebrafish Husbandry
2.19. Waterborne Exposure of Zebrafish Embryos to KGEA and H2O2
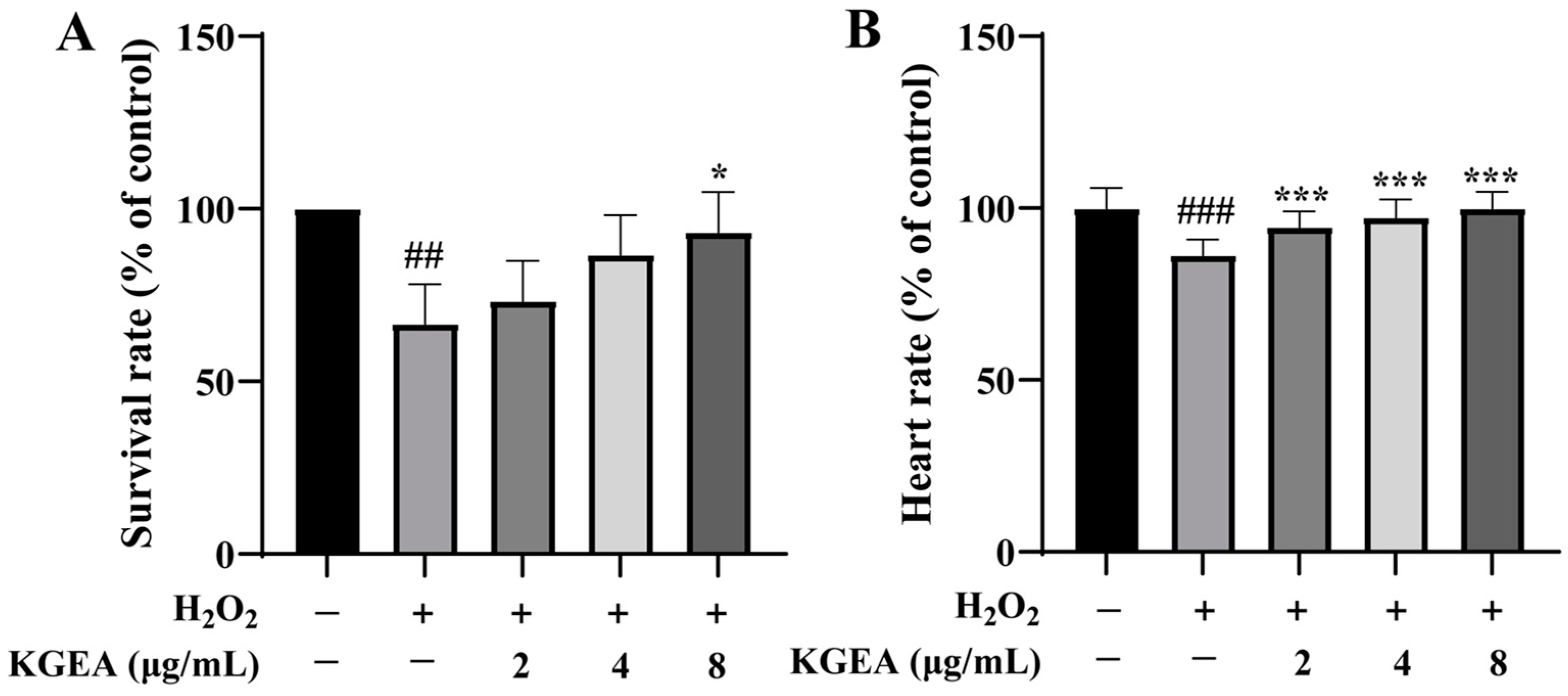
2.20. Heartbeat Rate of Zebrafish Embryos
2.21. Determination of KGEA Against H2O2-Induced Oxidative Stress in Zebrafish Embryos
2.22. Determination of Antioxidant Enzyme Activity and Lipid Peroxidation in Zebrafish Embryos
2.23. Statistical Analysis
3. Results
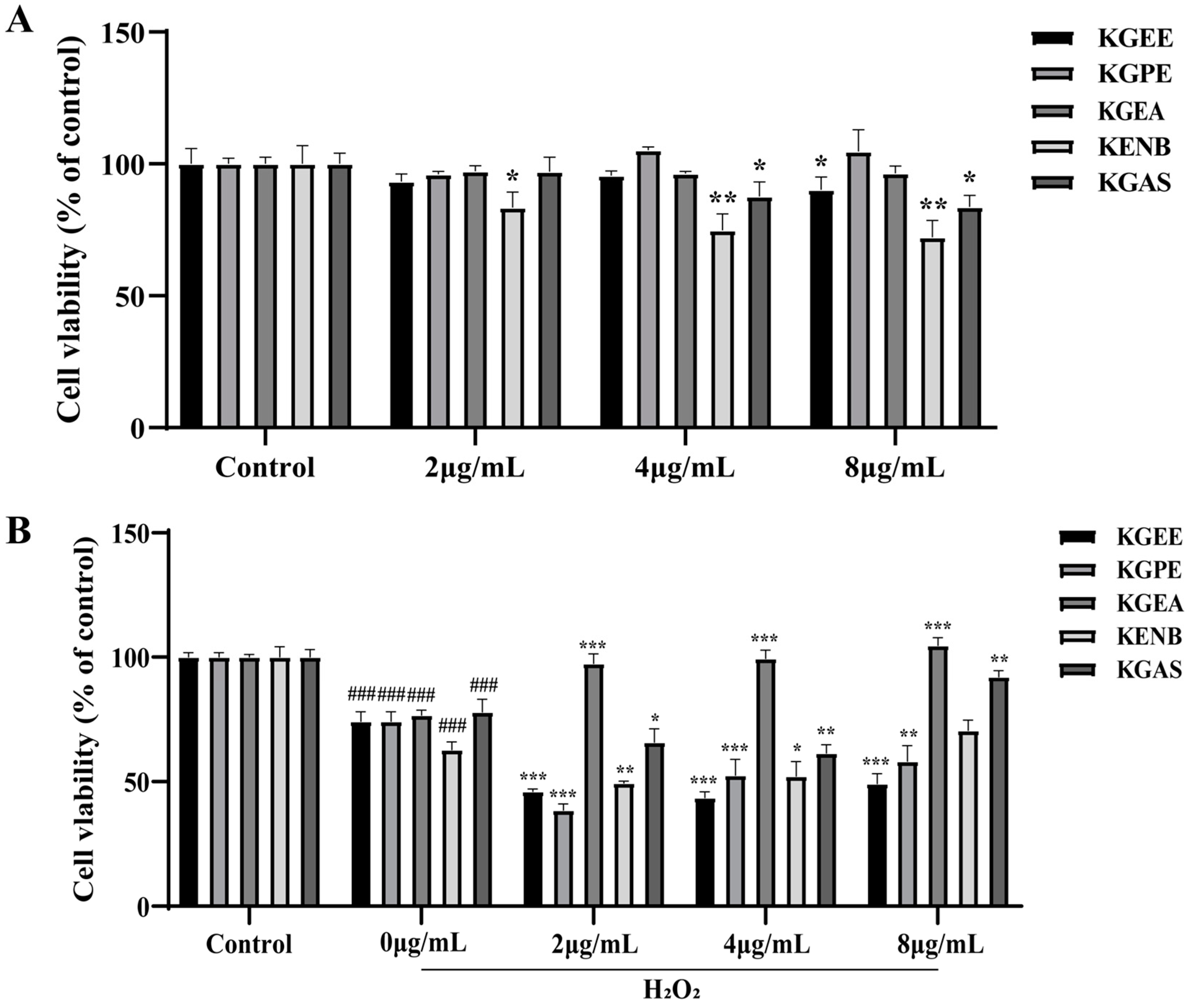
3.1. Discovery of Bioactive Fraction in K. galanga
3.2. LC-MS Analysis of the KGEA
3.3. Potential Targets of KGEA Compounds and Antioxidants
3.4. PPI Network of the Targets
3.5. GO Enrichment and KEGG Pathway Analyses
3.6. Component–Target–Pathway Network Analysis
3.7. Molecular Docking Analysis
3.8. Molecular Dynamics Simulation Analysis
3.9. DPPH Radical Scavenging Activity
3.10. ABTS Radical Scavenging Activity
3.11. KGEA Alleviated Oxidative Stress in H2O2-Induced RAW264.7 Cells
3.12. KGEA Promoted Antioxidant Enzyme Activity and Reduced Lipid Peroxidation
3.13. Inhibition of KGEA on H2O2-Induced PI3K/AKT and MAPK Pathways
3.14. Protective Effects of KGEA in an H2O2-Induced Zebrafish Model
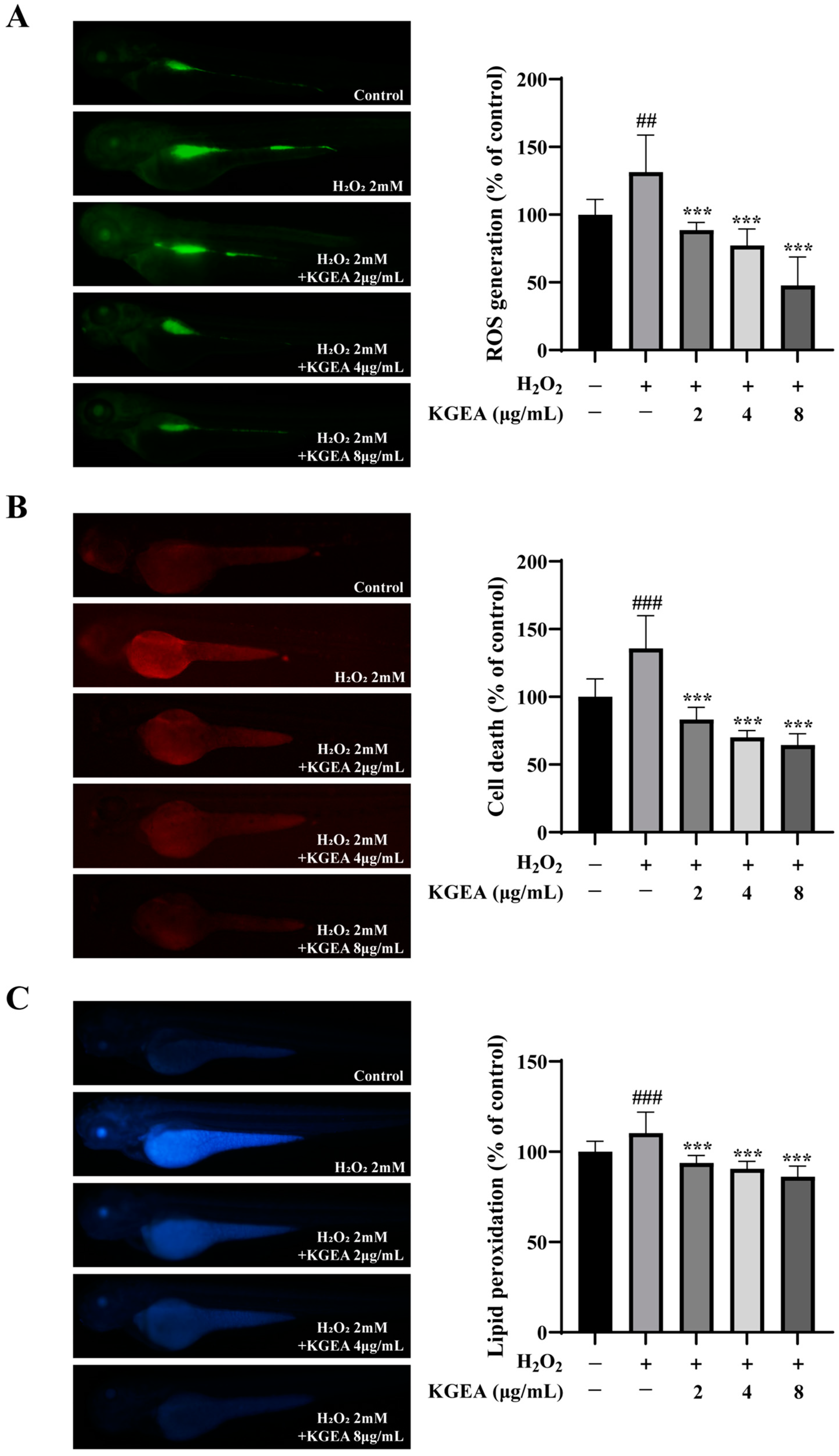
3.15. Antioxidant Effects of the KGEA in an H2O2-Induced Zebrafish Model
3.16. Antioxidant Effects of KGEA on Antioxidant Enzymes Activities and MDA Level in an H2O2-Induced Zebrafish Model
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| KG | Kaempferia galanga L. |
| KGEE | the ethanolic extract from K. galanga |
| KGPE | the petroleum ether fraction of ethanolic extract from K. galanga |
| KGEA | the ethyl acetate fraction of ethanolic extract from K. galanga |
| KGNB | the n-butanol fraction of ethanolic extract from K. galanga |
| KGAS | the remaining aqueous solution fraction of ethanolic extract from K. galanga |
| UV | ultraviolet |
| ROS | reactive oxygen species |
| TCM | traditional Chinese medicine |
| MD | molecular dynamics |
| H2O2 | hydrogen peroxide |
| DPPH | 2,2-diphenyl-1-picrylhydrazyl |
| ABTS | 2,2′-amino-di(2-ethyl benzothiazoline sulfonic acid-6) ammonium salt |
| DCF-DA | 2′,7′-dichlorodihydrofluorescein diacetate |
| DPPP | diphenyl-1-pyrenylphosphine |
| MDA | malondialdehyde |
| SOD | superoxide dismutase |
| CAT | catalase |
| GSH-Px | glutathione peroxidase |
| UHPLC-QTOF-MS | ultra-high-performance liquid chromatography-quadrupole time-of-flight mass spectrometry |
| SMILES | simplified molecular input line entry system |
| ADME | absorption, distribution, metabolism, excretion |
| PPI | protein–protein interactions |
| GO | gene ontology |
| KEGG | Kyoto encyclopedia of genes and genomes |
| RMSD | root-mean-square deviation |
| RMSF | root mean square fluctuation |
| Rg | radius of gyration |
| RMSF | root mean square fluctuation |
| ECL | enhanced chemiluminescence |
| AKT | protein kinase B |
| p-AKT | phospho protein kinase B |
| PI3K | phosphoinositide 3-kinase |
| p-PI3K | phospho phosphoinositide 3-kinase |
| ERK1/2 | extracellular signal-regulated kinase 1/2 |
| p-ERK1/2 | phospho extracellular signal regulated kinase 1/2 |
| CCK-8 | cell counting kit-8 (2-(2-methoxy-4-nitrophenyl)-3-(4-nitrophenyl)-5-(2,4-disulfophenyl)-2H-tetrazolium, monosodium salt) |
| JC-1 | 5,5′,6,6′-tetrachloro-1,1′,3,3′-tetraethyl-benzimidazolylcarbocyanine Chloride |
References
- Ungvari, Z.; Bagi, Z.; Feher, A.; Recchia, F.A.; Sonntag, W.E.; Pearson, K.; Cabo, R.D.; Csiszar, A. Resveratrol confers endothelial protection via activation of the antioxidant transcription factor Nrf2. Am. J. Physiol. Heart. Circ. 2010, 299, H18–H24. [Google Scholar] [CrossRef]
- Sahreen, S.; Khan, M.R.; Khan, R.A. Evaluation of antioxidant activities of various solvent extracts of Carissa opaca fruits. Food Chem. 2010, 122, 1205–1211. [Google Scholar] [CrossRef]
- Wu, W.L.; Papagiannakopoulos, T. The pleiotropic role of the Keap1/Nrf2 pathway in cancer. Annu. Rev. Cancer Biol. 2020, 4, 413–435. [Google Scholar] [CrossRef]
- Zhu, Q.; Zeng, J.; Li, J.; Chen, X.M.; Miao, J.X.; Jin, Q.Y.; Chen, H.Y. Effects of compound Centella on oxidative stress and Keap1-Nrf2-ARE pathway expression in diabetic kidney disease rats. Evid.-Based Compl. Alt. 2020, 2020, 9817932. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Mo, Z. Keap1-Nrf2 signaling pathway in angiogenesis and vascular diseases. J. Tissue Eng. Regener. Med. 2020, 14, 869–883. [Google Scholar] [CrossRef] [PubMed]
- Aynur, S.; Bircan, C.T.; Sevil, E.T.; Sevcan, A.; Göksel, K.; Süleyman, K.; Murat, K. Assessment of the Antioxidant Activity of Silybum marianum Seed Extract and Its Protective Efect Against DNA Oxidation, Protein Damage and Lipid Peroxidation. Food Technol. Biotechnol. 2016, 54, 455–461. [Google Scholar]
- Ghosh, T.; Basu, A.; Adhikari, D.; Roy, D.; Pal, A.K. Antioxidant activity and structural features of Cinnamomum zeylanicum. 3 Biotech 2015, 5, 939–947. [Google Scholar] [CrossRef]
- Munekata, P.E.S.; Gullon, B.; Pateiro, M.; Tomasevic, I.; Domínguez, R.; Lorenzo, J.M. Natural Antioxidants from Seeds and Their Application in Meat Products. Antioxidants 2020, 9, 815. [Google Scholar] [CrossRef]
- Li, P.; Feng, B.; Jiang, H.; Han, X.; Wu, Z.F.; Wang, Y.Q.; Lin, J.Z.; Zhang, Y.; Yang, M.; Han, L.; et al. A Novel Forming Method of Traditional Chinese Medicine Dispersible Tablets to Achieve Rapid Disintegration Based on the Powder Modifcation Principle. Sci. Rep. 2018, 8, 10319. [Google Scholar]
- Wang, S.Y.; Zhao, H.; Xu, H.T.; Han, X.D.; Wu, Y.S.; Xu, F.F.; Yang, X.B.; Göransson, U.; Liu, B. Kaempferia galanga L.: Progresses in phytochemistry, pharmacology, toxicology and ethnomedicinal uses. Front. Pharmacol. 2021, 12, 675350. [Google Scholar] [CrossRef]
- Munda, S.; Saikia, P.; Lal, M. Chemical composition and biological activity of essential oil of Kaempferia galanga: A review. J. Essent. Oil Res. 2018, 30, 303–308. [Google Scholar] [CrossRef]
- Mustafa, R.A.; Hamid, A.A.; Mohamed, S.; Bakar, F.A. Total phenolic compounds, flavonoids, and radical scavenging activity of 21 selected tropical plants. J. Food Sci. 2010, 75, 28–35. [Google Scholar] [CrossRef] [PubMed]
- Jantan, I.B.; Yassin, M.S.M.; Chin, C.B.; Chen, L.L.; Sim, N.L. Antifungal activity of the essential oils of nine zingiberaceae species. Pharm. Biol. 2003, 41, 392–397. [Google Scholar] [CrossRef]
- Sulaiman, M.R.; Zakaria, Z.A.; Daud, I.A.; Ng, F.N.; Ng, Y.C.; Hidayat, M.T. Antinociceptive and anti-inflammatory activities of the aqueous extract of Kaempferia galanga leaves in animal models. J. Nat. Med. 2008, 62, 221–227. [Google Scholar] [CrossRef] [PubMed]
- Ali, H.; Yesmin, R.; Satter, A.; Habib, R.; Yeasmin, T. Antioxidant and anti-neoplastic activities of methanolic extract of Kaempferia galanga linn. Rhizome against Ehrlich ascites carcinoma cells. J. King Saud Univ. Sci. 2018, 30, 386–392. [Google Scholar] [CrossRef]
- Rahman, I.; Kabir, T.; Islam, N.; Muqaddim, M.; Sharmin, S.; Ullah, M.S.; Uddin, S. Investigation of Antioxidant and Cytotoxic Activities of Kaempferia galanga L. Res. J. Pharm. Technol. 2019, 12, 2189–2194. [Google Scholar] [CrossRef]
- Hopkins, A.L. Network pharmacology: The next paradigm in drug discovery. Nat. Chem. Biol. 2008, 4, 682–690. [Google Scholar] [CrossRef]
- Law, V.; Knox, C.; Djoumbou, Y.; Jewison, T.; Guo, A.C.; Liu, Y.F.; Maciejewski, A.; Arndt, D.; Wilson, M.; Neveu, V.; et al. DrugBank 40: Shedding new light on drug metabolism. Nucleic Acids Res. 2014, 42, 1091–1097. [Google Scholar] [CrossRef]
- Zhang, R.; Zhu, X.; Bai, H.; Ning, K. Network pharmacology databases for traditional Chinese medicine: Review and assessment. Front. Pharmacol. 2019, 10, 123. [Google Scholar] [CrossRef]
- Babgi, B.A.; Alsayari, J.; Alenezi, H.M.; Abdellatif, M.H.; Eltayeb, N.E.; Emwas, A.H.M.; Jaremko, M.; Hussien, M.A. Alteration of Anticancer and Protein-Binding Properties of Gold(I) Alkynyl by Phenolic Schif Bases Moieties. Pharmaceutics 2021, 13, 461. [Google Scholar] [CrossRef]
- Mu, C.; Sheng, Y.; Wang, Q.; Wang, Q.; Amin, A.; Li, X.G.; Xie, Y.Q. Potential compound from herbal food of Rhizoma polygonati for treatment of COVID-19 analyzed by network pharmacology: Viral and cancer signaling mechanisms. J. Funct. Foods 2021, 77, 104149. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Chen, Z.; Li, W.; Wang, Y.; Li, X.; Yu, H.; Ran, P.; Liu, Z. Systems pharmacology uncovers the mechanisms of anti-asthma herbal medicine intervention (ASHMI) for the prevention of asthma. J. Funct. Foods 2019, 52, 611–619. [Google Scholar] [CrossRef]
- Nie, J.Y.; Li, R.; Jiang, Z.T.; Wang, Y.; Tan, J.; Tang, S.H.; Zhang, Y. Screening and evaluation ofradical scavenging active compounds in the essential oil from Magnolia biondii pamp byelectronic nose coupled with chemical methodology. Ind. Crops Prod. 2020, 144, 112060. [Google Scholar] [CrossRef]
- Tungcharoen, P.; Wattanapiromsakul, C.; Tansakul, P.; Nakamura, S.; Matsuda, H.; Tewtrakul, S. Anti-inflammatory Effect of Isopimarane Diterpenoids from Kaempferia galanga. Phytother. Res. 2020, 34, 612–623. [Google Scholar] [CrossRef] [PubMed]
- Yao, F.; Huang, Y.; Wang, Y.; He, X. Anti-inflammatory Diarylheptanoids and Phenolics from the Rhizomes of Kencur (Kaempferia galanga L.). Ind. Crops Prod. 2018, 125, 454–461. [Google Scholar] [CrossRef]
- Yao, F.Z. Study on the Chemical Constituents of the Rhizome of Kaempferia galanga L. Master’s Thesis, Guangdong Pharmaceutical University, Guangzhou, China, 2018. [Google Scholar]
- Jiao, Z.X.; Xu, W.F.; Zheng, J.S.; Shen, P.; Qin, A.; Zhang, S.Y.; Yang, C. Kaempferide Prevents Titanium Particle Induced Osteolysis by Suppressing JNK Activation During Osteoclast Formation. Sci. Rep. 2017, 7, 16665. [Google Scholar] [CrossRef]
- Wu, H.D. Study on the Chemical Constituents of Rhizoma Kaempferiae. Master’s Thesis, Huazhong University of Science and Technology, Wuhan, China, 2016. [Google Scholar]
- Piao, C.L.; Zhang, Q.; Jin, D.; Wang, L.; Zhang, N.W.; Lian, F.M.; Tong, X. A Study on the Mechanism of Milkvetch Root in the Treatment of Diabetic Nephropathy Based on Network Pharmacology. Evid.-Based Complement. Altern. Med. 2020, 2020, 6754761. [Google Scholar] [CrossRef]
- Lobanov, M.; Bogatyreva, N.S.; Galzitskaia, O.V. Radius of gyration is indicator of compactness of protein structure. Mol. Biol. 2008, 42, 701–706. [Google Scholar] [CrossRef]
- García-Sánchez, A.; Miranda-Díaz, A.G.; Cardona-Muñoz, E.G. The Role of Oxidative Stress in Physiopathology and Pharmacological Treatment with Pro- and Antioxidant Properties in Chronic Diseases. Oxid. Med. Cell. Longev. 2020, 2020, 2082145. [Google Scholar] [CrossRef]
- Liang, B.; Zhu, Y.C.; Lu, J.; Gu, N. Effects of Traditional Chinese Medication-Based Bioactive Compounds on Cellular and Molecular Mechanisms of Oxidative Stress. Oxid. Med. Cell. Longev. 2021, 2021, 3617498. [Google Scholar] [CrossRef]
- Maestri, D.M.; Nepote, V.; Lamarque, A.L.; Zygadlo, J.A. Natural products as antioxidants. Adv. Res. 2006, 37, 105–135. [Google Scholar]
- Ismail, B.B.; Pu, Y.F.; Guo, M.M.; Ma, X.B.; Liu, D.H. LC-MS/QTOF identification of phytochemicals and the effects of solvents on phenolic constituents and antioxidant activity of baobab (Adansonia digitata) fruit pulp. Food Chem. 2019, 277, 279–288. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, G.; Wang, T.; Cao, W.; Zhang, L.X.; Chen, X.Y. Nrf2-Keap1 pathway-mediated effects of resveratrol on oxidative stress and apoptosis in hydrogen peroxide-treated rheumatoid arthritis fibroblast-like synoviocytes. Ann. N. Y. Acad. Sci. 2019, 1457, 166–178. [Google Scholar] [CrossRef] [PubMed]
- Qiang, Z.; Pan, J.K.; Liu, H.; Jiao, Z.G. Characterization of the Synergistic Antioxidant Activity of Epigallocatechin Gallate (EGCG) and Kaempferol. Molecules 2023, 28, 5265. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Yin, D.X.; Zhang, T.; Hou, X.; Qiao, Q.; Song, P. Major Fatty Acid Compositions and Antioxidant Activity of Cultivated Paeonia ostii under Different Nitrogen Fertilizer Application. Chem. Biodivers. 2020, 17, e2000617. [Google Scholar] [CrossRef]
- Zhang, J.F.; Yang, Y.X.; Han, H.L.; Zhang, L.L.; Wang, T. Bisdemethoxycurcumin Protects Small Intestine from Lipopolysaccharide-Induced Mitochondrial Dysfunction via Activating Mitochondrial Antioxidant Systems and Mitochondrial Biogenesis in Broiler Chickens. Oxid. Med. Cell. Longev. 2021, 2021, 9927864. [Google Scholar] [CrossRef]
- Shabani, M.; Jamali, Z.; Bayrami, D.; Salimi, A. Vanillic acid alleviates methamphetamine-induced mitochondrial toxicity in cardiac mitochondria via antioxidant activity and inhibition of MPT Pore opening: An in-vitro study. BMC Pharmacol. Toxicol. 2023, 24, 33. [Google Scholar] [CrossRef]
- Boeing, T.; Souza, P.D.; Speca, S.; Somensi, L.B.; Mariano, L.N.B.; Cury, B.J.; Anjos, M.F.D.; Quintão, N.L.M.; Dubuqoy, L.; Desreumax, P.; et al. Luteolin prevents irinotecan-induced intestinal mucositis in mice through antioxidant and anti-inflammatory properties. Br. J. Pharmacol. 2020, 177, 2393–2408. [Google Scholar] [CrossRef]
- Ma, X.L.; Tian, Y.; Xue, K.Y.; Huai, Y.; Patil, S.Y.; Deng, X.N.; Hao, Q.; Li, D.M.; Miao, Z.P.; Zhang, W.J.; et al. Kaempferide enhances antioxidant capacity to promote osteogenesis through FoxO1/β-catenin signaling pathway. Eur. J. Pharmacol. 2021, 911, 174555. [Google Scholar] [CrossRef]
- Imran, M.; Rauf, A.; Shah, Z.A.; Saeed, F.; Imran, A.; Arshad, M.U.; Ahmad, B.; Bawazeer, S.; Atif, M.; Peters, D.G.; et al. Chemo-preventive and therapeutic effect of the dietary flavonoid kaempferol: A comprehensive review. Phytother. Res. 2019, 33, 263–275. [Google Scholar] [CrossRef]
- Akihiro, T.; Atsuko, I.; Hideyuki, I. Structural evidence for the DPPH radical-scavenging mechanism of 2-Oa-D-glucopyranosyl-L ascorbic acid. Bioorg. Med. Chem. 2017, 25, 5303–5310. [Google Scholar]
- Zhen, J.; Villani, T.S.; Guo, Y.; Qi, Y.; Chin, K.; Pan, M.H.; Wu, Q. Phytochemistry, antioxidant capacity, total phenolic content and anti-inflammatory activity of Hibiscus sabdariffa leaves. Food Chem. 2016, 190, 673–680. [Google Scholar] [CrossRef]
- Yang, S.; Lian, G. ROS and diseases: Role in metabolism and energy supply. Mol. Cell. Biochem. 2020, 467, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Vahdati, S.; Lashkari, A.; Navasatli, A.S.; Ardestani, K.S. and M. Safavi. Butylated hydroxyl-toluene, 2,4-Di-tert-butylphenol, and phytol of Chlorella sp. protect the PC12 cell line against H2O2-induced neurotoxicity. Biomed. Pharmacother. 2020, 145, 112415. [Google Scholar]
- Wen, L.; Zheng, G.; You, L.; Abbasi, A.M.; Li, T.; Fu, X.; Liu, R.H. Phytochemical profiles and cellular antioxidant activity of Malus doumeri (bois) chevalier on 2,2′-azobis (2-amidinopropane) dihydrochloride (ABAP)-induced oxidative stress. J. Funct. Foods 2016, 25, 242–256. [Google Scholar] [CrossRef]
- Jiang, J.; Zhuang, J.Y.; Fan, Y.Y.; Shen, B. Mapping of QTLs for leaf malondialdehyde content associated with stress tolerance in rice. Rice Sci. 2009, 16, 72–74. [Google Scholar] [CrossRef]
- Pham-Huy, L.A.; He, H.; Pham-Huy, C. Free radicals, antioxidants in disease and health. Int. J. Biomed. Sci. 2008, 4, 89–96. [Google Scholar] [CrossRef]







| No. | RT (min) | Molecular Name | Molecular Formula | [M + H]+ | [M − H]− | Fragment Ions (m/z) |
|---|---|---|---|---|---|---|
| 1 | 5.21 | 4-methoxybenzyl-O-β-D-glucopyranoside | C14H20O7 | 301.1282 | 205.0153, 160.9889 | |
| 2 | 8.26 | bisdemethoxycurcumin | C19H16O4 | 309.1134 | 217.0764 | |
| 3 | 9.37 | ponkanetin | C20H20O7 | 373.1295 | 131.0482, 147.0440, 163.0751, 175.0756 | |
| 4 | 10.01 | 3-caren-5-one | C10H14O | 151.1117 | 109.0640, 107.0494 | |
| 5 | 12.70 | feruloylputrescine | C14H20N2O3 | 265.1554 | 265.0985, 247.0877, 235,0876, 219.0926, 218.0849, 206.0843, 167.0672 | |
| 6 | 16.14 | dehydrocholic acid | C24H34O5 | 403.2493 | 117.0697, 373.0926 | |
| 7 | 19.51 | 3′,4′,5,7-tetramethylquercetin | C19H18O7 | 359.1143 | 359.2045, 197.0978, 177.0516, 167.0790, 137.0500, 135.0440, 121.1015, 107.0480 | |
| 8 | 20.88 | 5-methoxypodophyllotoxin | C23H24O9 | 445.1478 | 117.0695 | |
| 9 | 20.94 | ethyl p-methoxycinnamate | C12H14O3 | 207.1015 | 161.0595, 133.0641, 103.0537 | |
| 10 | 27.42 | quercetin 3-(6-O-acetyl-beta-glucoside) | C23H22O13 | 507.1158 | 105.0332, 117.0698 | |
| 11 | 1.22 | p-hydroxybenzoic acid | C7H6O3 | 137.0244 | 137.0226, 108.0207 | |
| 12 | 1.76 | Methyl 3,4-dihydroxybenzoate | C8H8O4 | 167.0349 | 107.0506 | |
| 13 | 2.4 | vanillic acid | C8H8O4 | 167.0349 | 123.0420 | |
| 14 | 3.77 | phenylmethanol | C7H8O | 107.0502 | 107.0479, 106.0404 | |
| 15 | 4.39 | p-hydroxycinnamic acid | C9H8O3 | 163.0400 | 119.0497, 117.0345 | |
| 16 | 4.51 | 1-O-4-Carboxylphenyl-(6-O-4-hydroxybenzoyl)-β- D-glucopyranoside | C20H20O10 | 419.0983 | 281.0646, 137.0235 | |
| 17 | 4.72 | ferulic acid | C10H10O4 | 193.0506 | 134.0359, 133.0281, 132.0196, 117.0331, 106.0411 | |
| 18 | 4.96 | benzoic acid | C7H6O2 | 121.0295 | 121.0288, 120.0209, 108.0207 | |
| 19 | 5.25 | hedycoropyran B | C20H24O7 | 375.1449 | 177.0548, 163.0393, 135.0442 | |
| 20 | 5.29 | kaempsulfonic acid A | C20H24O8S | 423.1119 | 423.1984, 423.1706, 287.0566, 267.1585, 243.0304, 229.1095, 135.0441 | |
| 21 | 5.29 | kaempsulfonic acid B | C20H24O8S | 423.1119 | 423.1984, 423.1706, 287.0566, 267.1585, 243.0304, 229.1095, 135.0441 | |
| 22 | 5.64 | (3R,5S)-3,5-dihydroxy-1,7-bis(3,4-dihydroxyphenyl) heptane | C19H24O6 | 347.1500 | 347.1477, 165.0547, 163.0757, 137.0600 | |
| 23 | 6.11 | (1R,3R,5R)-1,5-epoxy-3-hydroxy-1-(3,4-dihydroxyphenyl)-7-(3,4-dihydroxyphenyl) heptane | C19H22O6 | 345.1343 | 209.0814, 165.0549, 161.0600, 135.0440 | |
| 24 | 6.21 | p-methoxybenzoic acid | C8H8O3 | 151.0400 | 150.2345, 122.0327, 108.0205 | |
| 25 | 6.67 | (1R,2S,4R)-p-menth-5-ene-1,2,8-triol | C10H18O3 | 185.1183 | 139.1110 | |
| 26 | 7.72 | phaeoheptanoxide | C19H22O5 | 329.1394 | 161.0601, 159.0443, 134.0368, 135.0438 | |
| 27 | 8.33 | p-methoxycinnamic acid | C10H10O3 | 177.0557 | 161.0592, 119.0453, 117.0336 | |
| 28 | 8.86 | ethyl cinnamate | C11H12O2 | 175.0764 | 159.0443, 131.0478 | |
| 29 | 9.76 | (3R,4R,6S)-3,6-dihydroxy-1-menthene | C10H18O2 | 169.1234 | 125.0235, 107.0129 | |
| 30 | 12.18 | 4-methoxy-benzyl (E)-3-(4-methoxyp-henyl) acrylate | C18H18O4 | 297.1132 | 253.1220, 235.1648, 121.0434, 107.0487 | |
| 31 | 19.55 | kaempferide | C16H12O6 | 299.0561 | 299.0527, 271.0593, 271.0831, 263.2009, 257.0372, 163.0058 | |
| 32 | 26.71 | kaempferol | C15H10O6 | 285.0404 | 285.0390, 257.0428, 217.0507, 147.0777 | |
| 33 | 26.75 | luteolin | C15H10O6 | 285.0404 | 285.0377, 271.1278, 257.0431, 241.0483, 201.0543, 159.0404, 157.0656 | |
| 34 | 30.53 | dibutyl phthalate | C16H22O4 | 277.1445 | 277.1423, 233.1539, 217.1216, | |
| 35 | 31.22 | monopalmitin | C19H38O4 | 329.2697 | 329.2645, 329.2264, 257.1844 | |
| 36 | 31.42 | sandaracopimaradien-6β,9α-diol-l-one | C20H30O3 | 317.2122 | 273.2196, 271.2070, 149.0980 | |
| 37 | 32.22 | kaemgalangol A | C20H30O3 | 317.2122 | 273.2196 | |
| 38 | 34.44 | linolenic acid | C18H30O2 | 277.2173 | 277.2157, 147.0781 | |
| 39 | 36.59 | 6β-acetoxysandaracopimaradiene-1α,9α-diol | C22H34O4 | 361.2384 | 269.1882, 215.1805 | |
| 40 | 39.69 | 6β-acetoxy-1α-14α-dihydroxyisopimara-8(9),15-diene | C22H34O4 | 361.2384 | 283.2615, 269.1875 | |
| 41 | 39.47 | stearic acid | C18H36O2 | 283.2642 | 283.2621, 265.2518 | |
| 42 | 43.22 | 6β-hydroxypimara-8(14),15-diene-1-one | C20H30O2 | 301.2173 | 301.2138, 255.2591 | |
| 43 | 44.00 | linoleic acid | C18H32O2 | 279.2329 | 279.2314, 261.2201 |
| Samples | DPPH (EC50) | ABTS (IC50) |
|---|---|---|
| KGEA (mg/mL) | 4.88 ± 0.74 | 1.07 ± 0.06 |
| Ascorbic acid (μg/mL) | 22.64 ± 0.85 | 14.83 ± 0.74 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, S.; Yang, J.; Cai, L.; Li, H.; Han, X.; Liu, B.; Wu, J. Antioxidant Effect of Ethyl Acetate Fraction from Kaempferia galanga L.: Integrated Phytochemical Profiling, Network Analysis, and Experimental Validation. Antioxidants 2025, 14, 551. https://doi.org/10.3390/antiox14050551
Wang S, Yang J, Cai L, Li H, Han X, Liu B, Wu J. Antioxidant Effect of Ethyl Acetate Fraction from Kaempferia galanga L.: Integrated Phytochemical Profiling, Network Analysis, and Experimental Validation. Antioxidants. 2025; 14(5):551. https://doi.org/10.3390/antiox14050551
Chicago/Turabian StyleWang, Siyu, Jianzhan Yang, Lei Cai, Haoxiang Li, Xiaodong Han, Bo Liu, and Jianwei Wu. 2025. "Antioxidant Effect of Ethyl Acetate Fraction from Kaempferia galanga L.: Integrated Phytochemical Profiling, Network Analysis, and Experimental Validation" Antioxidants 14, no. 5: 551. https://doi.org/10.3390/antiox14050551
APA StyleWang, S., Yang, J., Cai, L., Li, H., Han, X., Liu, B., & Wu, J. (2025). Antioxidant Effect of Ethyl Acetate Fraction from Kaempferia galanga L.: Integrated Phytochemical Profiling, Network Analysis, and Experimental Validation. Antioxidants, 14(5), 551. https://doi.org/10.3390/antiox14050551







