In Silico Analysis of Non-Conventional Oxidative Stress-Related Enzymes and Their Potential Relationship with Carcinogenesis
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Mining and Processing
2.2. Protein–Protein Interaction (PPI) Network and Hub Gene Identification
2.3. Differential Gene Expression Analysis
2.4. Differential Protein Expression Analysis
2.5. Genomic Alterations
3. Results
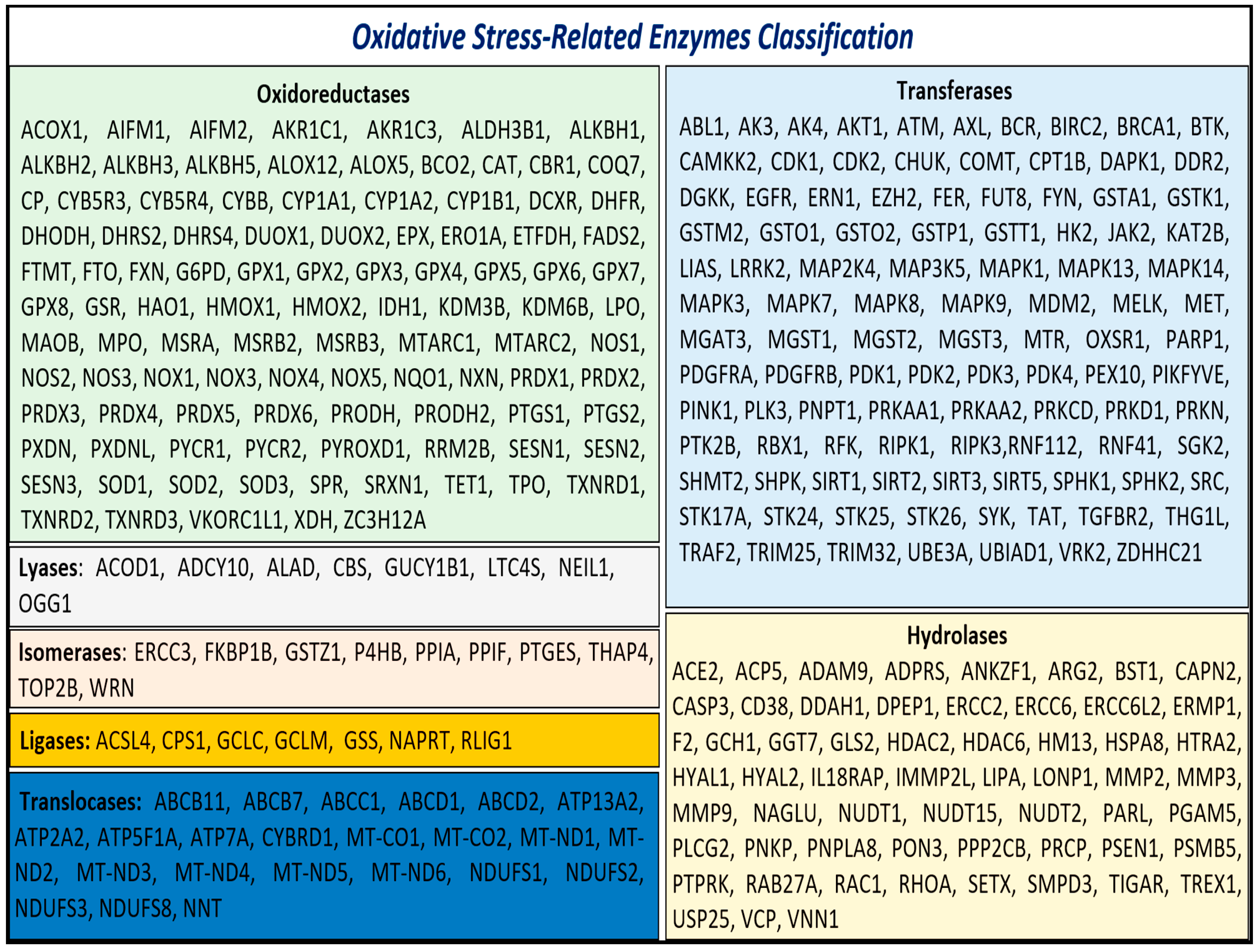
3.1. Identification and Enrichment of Genes Associated with Oxidative Stress
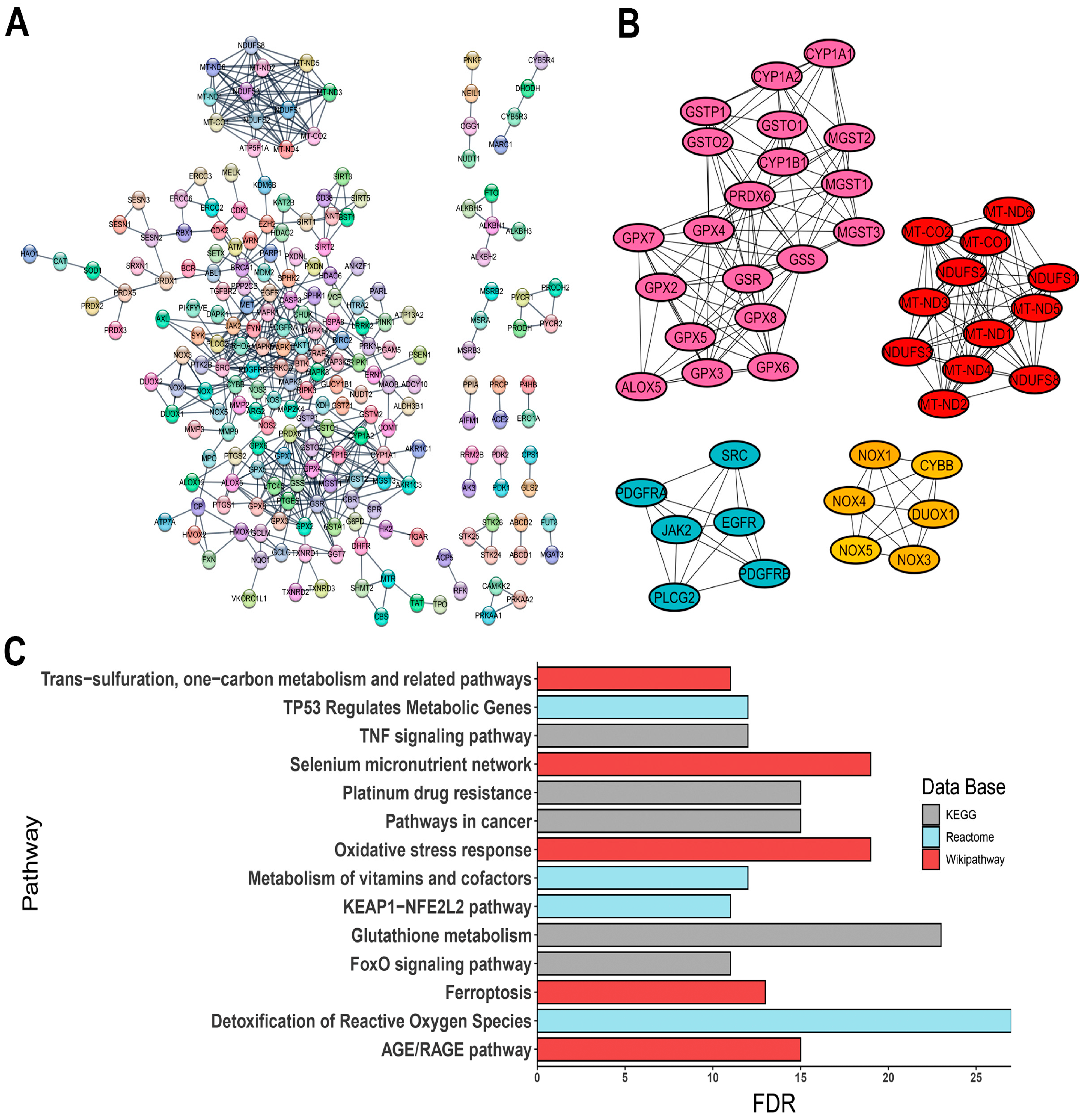
3.2. Protein–Protein Interaction (PPI) Network and Hub Gene Identification
- -
- ALOX5, encoding 5-lipoxygenase (5-LO);
- -
- CYBB, a component of the microbicidal oxidase system in phagocytes;
- -
- DUOX1, encoding a dual-function oxidase;
- -
- Several cytochrome P450 genes, such as CYP1A1, CYP1A2, and CYP1B1;
- -
- Protein tyrosine kinase genes, including EGFR, JAK2, PDGFRA, PDGFRB, and SRC;
- -
- Various isoforms of glutathione peroxidase: GPX2, GPX3, GPX4, GPX5, GPX6, GPX7, and GPX8;
- -
- Genes involved in glutathione metabolism, such as GSR, GSS, GSTO1, GSTO2, GSTP1, MGST1, MGST2, and MGST3;
- -
- PLCG2, a phospholipase tyrosine kinase;
- -
- PRDX6, a classic antioxidant enzyme;
- -
- ROS-generating enzymes NOX1, NOX3, NOX4, and NOX5;
- -
- Mitochondrial respiratory complex genes, including MT-CO1, MT-CO2, MT-ND1, MT-ND2, MT-ND3, MT-ND4, MT-ND5, MT-ND6, NDUFS1, NDUFS2, NDUFS3, and NDUFS8.
3.3. Enrichment Pathway Analysis
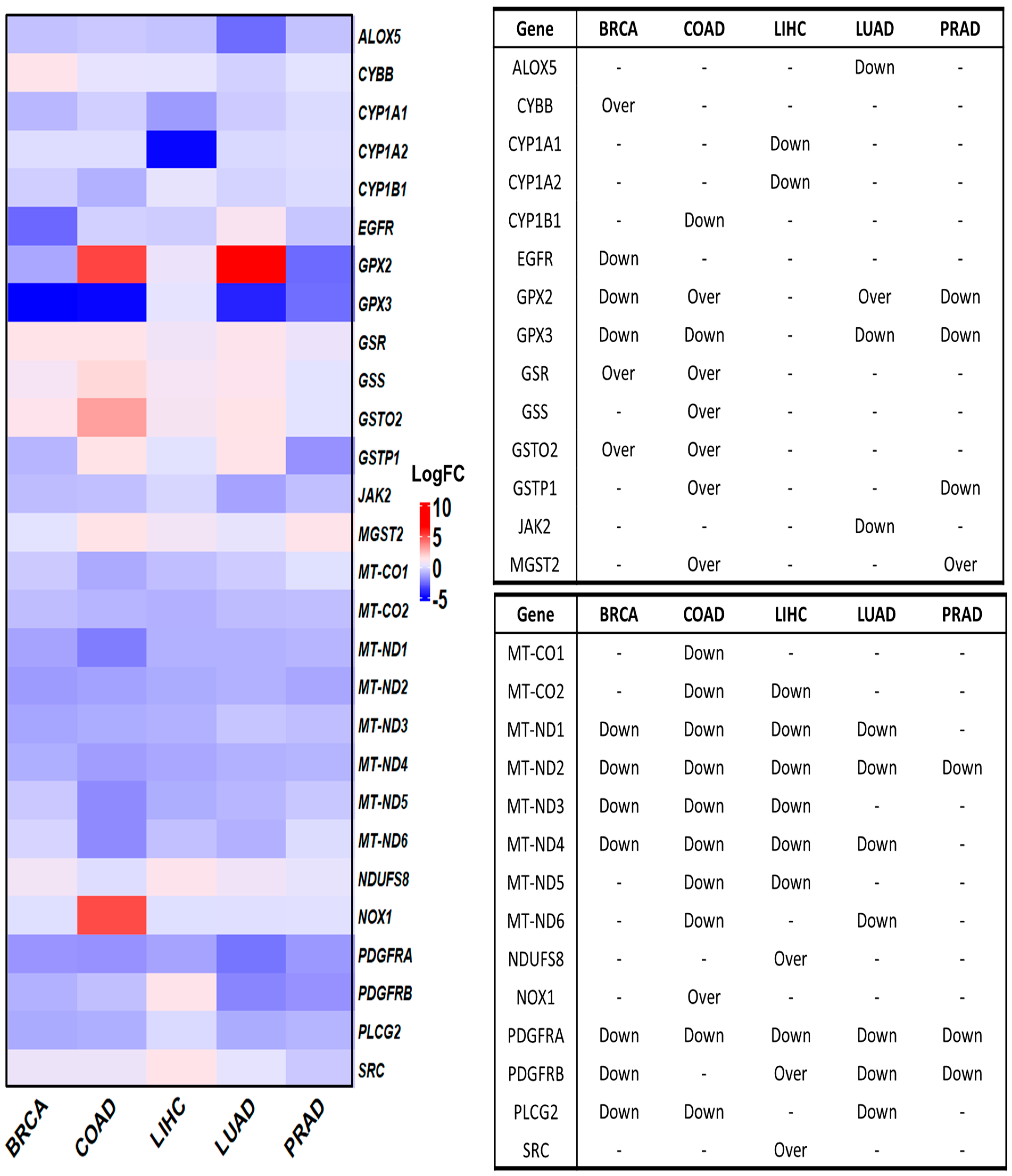
3.4. Deregulated OSRE in Cancer Samples
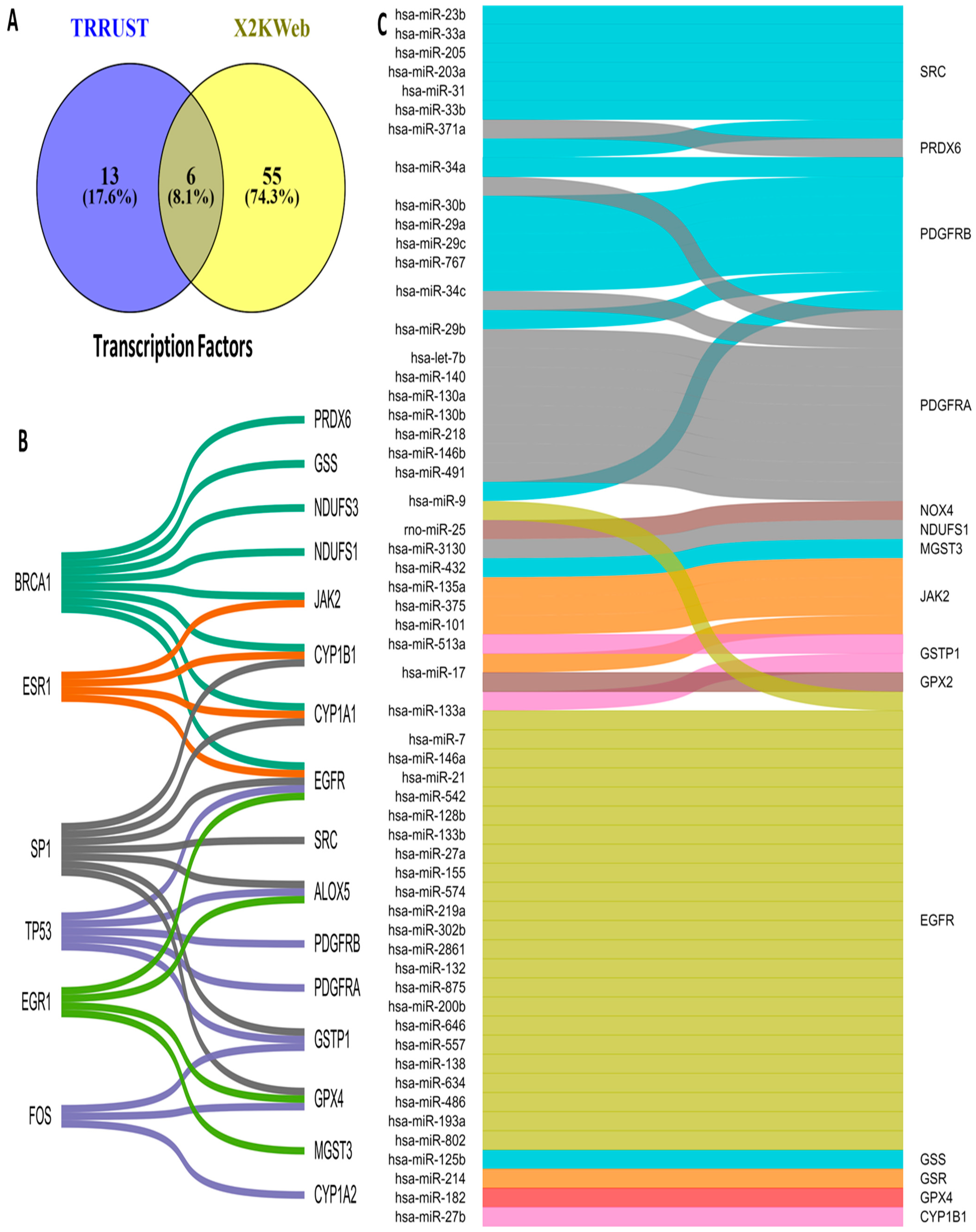
3.5. Upstream Regulators of OSRE Genes
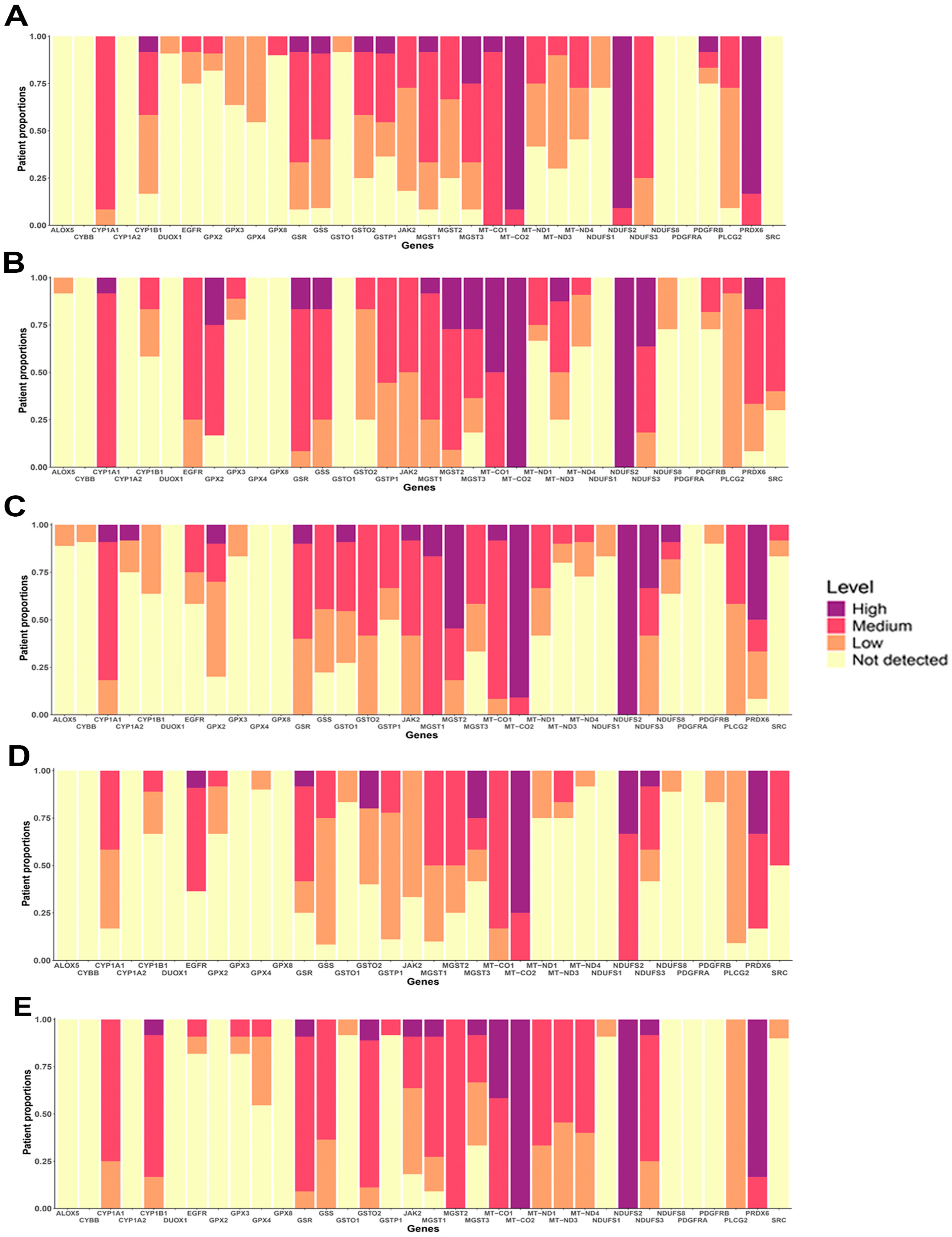
3.6. Protein Expression of OSRE
4. Discussion
5. Conclusions
6. Limitations
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Baiskhanova, D.; Schäfer, H. The Role of Nrf2 in the Regulation of Mitochondrial Function and Ferroptosis in Pancreatic Cancer. Antioxidants 2024, 13, 696. [Google Scholar] [CrossRef] [PubMed]
- Forman, H.J.; Zhang, H. Targeting oxidative stress in disease: Promise and limitations of antioxidant therapy. Nat. Rev. Drug Discov. 2021, 20, 689–709. [Google Scholar] [CrossRef] [PubMed]
- Panieri, E.; Pinho, S.A.; Afonso, G.J.M.; Oliveira, P.J.; Cunha-Oliveira, T.; Saso, L. NRF2 and Mitochondrial Function in Cancer and Cancer Stem Cells. Cells 2022, 11, 2401. [Google Scholar] [CrossRef] [PubMed]
- Kozakiewicz, P.; Grzybowska-Szatkowska, L.; Ciesielka, M.; Rzymowska, J. The Role of Mitochondria in Carcinogenesis. Int. J. Mol. Sci. 2021, 22, 5100. [Google Scholar] [CrossRef] [PubMed]
- Senga, S.S.; Grose, R.P. Hallmarks of cancer-the new testament. Open Biol. 2021, 11, 200358. [Google Scholar] [CrossRef]
- Abdel Hadi, N.; Reyes-Castellanos, G.; Carrier, A. Targeting Redox Metabolism in Pancreatic Cancer. Int. J. Mol. Sci. 2021, 22, 1534. [Google Scholar] [CrossRef]
- Iqbal, M.J.; Kabeer, A.; Abbas, Z.; Siddiqui, H.A.; Calina, D.; Sharifi-Rad, J.; Cho, W.C. Interplay of oxidative stress, cellular communication and signaling pathways in cancer. Cell Commun. Signal. 2024, 22, 7. [Google Scholar] [CrossRef]
- Ferlay, J.; Ervik, M.; Lam, F.; Laversanne, M.; Colombet, M.; Mery, L.; Piñeros, M.; Znaor, A.; Soerjomataram, I.; Bray, F. Global Cancer Observatory: Cancer Today. Available online: https://gco.iarc.who.int/today (accessed on 26 June 2024).
- Duan, G.; Huang, C.; Zhao, J.; Zhang, Y.; Zhao, W.; Dai, H. Investigating subtypes of lung adenocarcinoma by oxidative stress and immunotherapy related genes. Sci. Rep. 2023, 13, 20930. [Google Scholar] [CrossRef]
- Bel’skaya, L.V.; Dyachenko, E.I. Oxidative Stress in Breast Cancer: A Biochemical Map of Reactive Oxygen Species Production. Curr. Issues Mol. Biol. 2024, 46, 4646–4687. [Google Scholar] [CrossRef]
- Diakite, M.T.; Diakite, B.; Kone, A.; Balam, S.; Fofana, D.; Diallo, D.; Kassogue, Y.; Traore, C.B.; Kamate, B.; Ba, D.; et al. Relationships between gut microbiota, red meat consumption and colorectal cancer. J. Carcinog. Mutagen. 2022, 13, 696. [Google Scholar]
- Aloufi, A.S.; Habotta, O.A.; Abdelfattah, M.S.; Habib, M.N.; Omran, M.M.; Ali, S.A.; Abdel Moneim, A.E.; Korany, S.M.; Alrajhi, A.M. Resistomycin Suppresses Prostate Cancer Cell Growth by Instigating Oxidative Stress, Mitochondrial Apoptosis, and Cell Cycle Arrest. Molecules 2023, 28, 7871. [Google Scholar] [CrossRef] [PubMed]
- de Morais, J.M.B.; Cruz, E.M.S.; Concato, V.M.; de Souza, M.C.; Santos, Y.M.; Quadreli, D.H.; Inoue, F.S.R.; Ferreira, F.B.; Fernandes, G.S.A.; Bidoia, D.L.; et al. Unraveling the impact of melatonin treatment: Oxidative stress, metabolic responses, and morphological changes in HuH7.5 hepatocellular carcinoma cells. Pathol. Res. Pract. 2024, 253, 155056. [Google Scholar] [CrossRef]
- Barrera, G.; Cucci, M.A.; Grattarola, M.; Dianzani, C.; Muzio, G.; Pizzimenti, S. Control of Oxidative Stress in Cancer Chemoresistance: Spotlight on Nrf2 Role. Antioxidants 2021, 10, 510. [Google Scholar] [CrossRef]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Xu, H.; Feng, Y.; Kahlert, U.D.; Du, R.; Torres-de la Roche, L.A.; Xu, K.; Shi, W.; Meng, F. Oxidative stress genes define two subtypes of triple-negative breast cancer with prognostic and therapeutic implications. Front. Genet. 2023, 14, 1230911. [Google Scholar] [CrossRef]
- Liu, Y.; Li, Y.; Chen, L.; Zha, W.; Zhang, J.; Wang, K.; Hao, C.; Gan, J. Construction of an Oxidative Stress Risk Model to Analyze the Correlation Between Liver Cancer and Tumor Immunity. Curr. Cancer Drug Targets 2024. [CrossRef]
- Tan, Z.; Deng, Y.; Cai, Z.; He, H.; Tang, Z.; Feng, Y.; Ye, J.; Liu, R.; Cai, S.; Huang, H.; et al. ACOX2 Serves as a Favorable Indicator Related to Lipid Metabolism and Oxidative Stress for Biochemical Recurrence in Prostate Cancer. J. Cancer 2024, 15, 3010–3023. [Google Scholar] [CrossRef]
- Yang, L.; Fang, C.; Zhang, R.; Zhou, S. Prognostic value of oxidative stress-related genes in colorectal cancer and its correlation with tumor immunity. BMC Genom. 2024, 25, 8. [Google Scholar] [CrossRef] [PubMed]
- Caputo, W.L.; de Souza, M.C.; Basso, C.R.; Pedrosa, V.A.; Seiva, F.R.F. Comprehensive Profiling and Therapeutic Insights into Differentially Expressed Genes in Hepatocellular Carcinoma. Cancers 2023, 15, 5653. [Google Scholar] [CrossRef]
- Gene Ontology, C.; Aleksander, S.A.; Balhoff, J.; Carbon, S.; Cherry, J.M.; Drabkin, H.J.; Ebert, D.; Feuermann, M.; Gaudet, P.; Harris, N.L.; et al. The Gene Ontology knowledgebase in 2023. Genetics 2023, 224, iyad031. [Google Scholar] [CrossRef]
- Tweedie, S.; Braschi, B.; Gray, K.; Jones, T.E.M.; Seal, R.L.; Yates, B.; Bruford, E.A. Genenames.org: The HGNC and VGNC resources in 2021. Nucleic Acids Res. 2021, 49, D939–D946. [Google Scholar] [CrossRef] [PubMed]
- Durinck, S.; Spellman, P.T.; Birney, E.; Huber, W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat. Protoc. 2009, 4, 1184–1191. [Google Scholar] [CrossRef] [PubMed]
- Carlson, M. org.Hs.eg.db: Genome Wide Annotation for Human, Version 3.8.2. 2019. Available online: https://bioconductor.org/packages/release/data/annotation/html/org.Hs.eg.db.html (accessed on 20 May 2024).
- Lautenbacher, L.; Samaras, P.; Muller, J.; Grafberger, A.; Shraideh, M.; Rank, J.; Fuchs, S.T.; Schmidt, T.K.; The, M.; Dallago, C.; et al. ProteomicsDB: Toward a FAIR open-source resource for life-science research. Nucleic Acids Res. 2022, 50, D1541–D1552. [Google Scholar] [CrossRef] [PubMed]
- Tran, A.N.; Dussaq, A.M.; Kennell, T., Jr.; Willey, C.D.; Hjelmeland, A.B. HPAanalyze: An R package that facilitates the retrieval and analysis of the Human Protein Atlas data. BMC Bioinform. 2019, 20, 463. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Mahmood, M.; Reznik, E.; Gammage, P.A. Mitochondrial DNA is a major source of driver mutations in cancer. Trends Cancer 2022, 8, 1046–1059. [Google Scholar] [CrossRef]
- Piantadosi, C.A. Mitochondrial DNA, oxidants, and innate immunity. Free Radic. Biol. Med. 2020, 152, 455–461. [Google Scholar] [CrossRef]
- Carew, J.S.; Huang, P. Mitochondrial defects in cancer. Mol. Cancer 2002, 1, 9. [Google Scholar] [CrossRef]
- Liu, Y.; Zhao, W.; Hu, W.; Xu, J.; Zhang, H.; Huang, T.; Wu, C.; Yang, J.; Mao, W.; Yao, X.; et al. Exploring the relationship between anal fistula and colorectal cancer based on Mendelian randomization and bioinformatics. J. Cell Mol. Med. 2024, 28, e18537. [Google Scholar] [CrossRef]
- Zou, X.; Tang, X.Y.; Qu, Z.Y.; Sun, Z.W.; Ji, C.F.; Li, Y.J.; Guo, S.D. Targeting the PDGF/PDGFR signaling pathway for cancer therapy: A review. Int. J. Biol. Macromol. 2022, 202, 539–557. [Google Scholar] [CrossRef]
- Heldin, C.H. Targeting the PDGF signaling pathway in tumor treatment. Cell Commun. Signal. 2013, 11, 97. [Google Scholar] [CrossRef]
- El-Far, Y.M.; Khodir, A.E.; Emarah, Z.A.; Ebrahim, M.A.; Al-Gayyar, M.M.H. Chemopreventive and hepatoprotective effects of genistein via inhibition of oxidative stress and the versican/PDGF/PKC signaling pathway in experimentally induced hepatocellular carcinoma in rats by thioacetamide. Redox Rep. Commun. Free Radic. Res. 2022, 27, 9–20. [Google Scholar] [CrossRef] [PubMed]
- Pandey, P.; Khan, F.; Upadhyay, T.K.; Seungjoon, M.; Park, M.N.; Kim, B. New insights about the PDGF/PDGFR signaling pathway as a promising target to develop cancer therapeutic strategies. Biomed. Pharmacother. 2023, 161, 114491. [Google Scholar] [CrossRef] [PubMed]
- Baumgartner, V.; Schaer, D.; Eberli, D.; Salemi, S. Targeting Metabolic Vulnerabilities to Overcome Prostate Cancer Resistance: Dual Therapy with Apalutamide and Complex I Inhibition. Cancers 2023, 15, 5612. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.C.; Su, J.; Zhou, J.J.; Yuan, Q.; Han, J.S. Roles of MT-ND1 in Cancer. Curr. Med. Sci. 2023, 43, 869–878. [Google Scholar] [CrossRef]
- Cavalcante, G.C.; Ribeiro-Dos-Santos, A.; de Araujo, G.S. Mitochondria in tumour progression: A network of mtDNA variants in different types of cancer. BMC Genom. Data 2022, 23, 16. [Google Scholar] [CrossRef]
- Jayasekera, L.P.; Ranasinghe, R.; Senathilake, K.S.; Kotelawala, J.T.; de Silva, K.; Abeygunasekara, P.H.; Goonesinghe, R.; Tennekoon, K.H. Mitochondrial genome in sporadic breast cancer: A case control study and a proteomic analysis in a Sinhalese cohort from Sri Lanka. PLoS ONE 2023, 18, e0281620. [Google Scholar] [CrossRef]
- Li, Y.; Beckman, K.B.; Caberto, C.; Kazma, R.; Lum-Jones, A.; Haiman, C.A.; Le Marchand, L.; Stram, D.O.; Saxena, R.; Cheng, I. Association of Genes, Pathways, and Haplogroups of the Mitochondrial Genome with the Risk of Colorectal Cancer: The Multiethnic Cohort. PLoS ONE 2015, 10, e0136796. [Google Scholar] [CrossRef]
- Zhang, N.; Liao, H.; Lin, Z.; Tang, Q. Insights into the Role of Glutathione Peroxidase 3 in Non-Neoplastic Diseases. Biomolecules 2024, 14, 689. [Google Scholar] [CrossRef]
- Li, L.; Lu, M.; Peng, Y.; Huang, J.; Tang, X.; Chen, J.; Li, J.; Hong, X.; He, M.; Fu, H.; et al. Oxidatively stressed extracellular microenvironment drives fibroblast activation and kidney fibrosis. Redox Biol. 2023, 67, 102868. [Google Scholar] [CrossRef]
- Arner, E.S.J.; Schmidt, E.E. Unresolved questions regarding cellular cysteine sources and their possible relationships to ferroptosis. Adv. Cancer Res. 2024, 162, 1–44. [Google Scholar] [CrossRef]
- Santric, V.; Dragicevic, D.; Matic, M.; Djokic, M.; Pljesa-Ercegovac, M.; Radic, T.; Suvakov, S.; Nikitovic, M.; Stankovic, V.; Milojevic, B.; et al. Polymorphisms in Genes Encoding Glutathione Transferase Pi and Glutathione Transferase Omega Influence Prostate Cancer Risk and Prognosis. Front. Mol. Biosci. 2021, 8, 620690. [Google Scholar] [CrossRef] [PubMed]
- Dvash, E.; Har-Tal, M.; Barak, S.; Meir, O.; Rubinstein, M. Leukotriene C4 is the major trigger of stress-induced oxidative DNA damage. Nat. Commun. 2015, 6, 10112. [Google Scholar] [CrossRef] [PubMed]
- Yuan, H.; Xia, P.; Sun, X.; Ma, J.; Xu, X.; Fu, C.; Zhou, H.; Guan, Y.; Li, Z.; Zhao, S.; et al. Photothermal Nanozymatic Nanoparticles Induce Ferroptosis and Apoptosis through Tumor Microenvironment Manipulation for Cancer Therapy. Small 2022, 18, e2202161. [Google Scholar] [CrossRef]
- Wang, G.; Zhong, W.C.; Bi, Y.H.; Tao, S.Y.; Zhu, H.; Zhu, H.X.; Xu, A.M. The Prognosis Of Peroxiredoxin Family In Breast Cancer. Cancer Manag. Res. 2019, 11, 9685–9699. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Grueso, M.J.; Lagal, D.J.; Garcia-Jimenez, A.F.; Tarradas, R.M.; Carmona-Hidalgo, B.; Peinado, J.; Requejo-Aguilar, R.; Barcena, J.A.; Padilla, C.A. Knockout of PRDX6 induces mitochondrial dysfunction and cell cycle arrest at G2/M in HepG2 hepatocarcinoma cells. Redox Biol. 2020, 37, 101737. [Google Scholar] [CrossRef]
- Kim, S.S.; Shin, H.; Ahn, K.G.; Park, Y.M.; Kwon, M.C.; Lim, J.M.; Oh, E.K.; Kim, Y.; Han, S.M.; Noh, D.Y. Quantifiable peptide library bridges the gap for proteomics based biomarker discovery and validation on breast cancer. Sci. Rep. 2023, 13, 8991. [Google Scholar] [CrossRef]
- Behera, M.; Jiang, R.; Huang, Z.; Bunn, B.; Wynes, M.W.; Switchenko, J.; Scagliotti, G.V.; Belani, C.P.; Ramalingam, S.S. Natural History and Real-World Treatment Outcomes for Patients With NSCLC Having EGFR Exon 20 Insertion Mutation: An International Association for the Study of Lung Cancer-American Society of Clinical Oncology CancerLinQ Study. JTO Clin. Res. Rep. 2024, 5, 100592. [Google Scholar] [CrossRef]
- Ewald, C.Y.; Pulous, F.E.; Lok, S.W.Y.; Pun, F.W.; Aliper, A.; Ren, F.; Zhavoronkov, A. TNIK’s emerging role in cancer, metabolism, and age-related diseases. Trends Pharmacol. Sci. 2024, 45, 478–489. [Google Scholar] [CrossRef]
- Guo, J.; Liang, J.; Wang, Y.; Guo, T.; Liao, Y.; Zhong, B.; Guo, S.; Cao, Q.; Li, J.; Flores-Morales, A.; et al. TNIK drives castration-resistant prostate cancer via phosphorylating EGFR. iScience 2024, 27, 108713. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, C.; Dai, J.; Xu, Q.; Chen, Y.; Hu, Z.; Wang, Y.; Wang, C. PLCG2, A Regulator of Lung Adenocarcinoma Proliferation and Migration Associated with Immune Infiltration. Curr. Cancer Drug Targets 2024. [CrossRef]
- Ji, Y.; Wang, Y.; Zou, J.; Liu, G.; Xia, M.; Ren, J.; Wang, D. Methyltransferase DNMT3B promotes colorectal cancer cell proliferation by inhibiting PLCG2. Acta Biochim. Biophys. Sin. 2024. [CrossRef] [PubMed]
- Jackson, J.T.; Mulazzani, E.; Nutt, S.L.; Masters, S.L. The role of PLCgamma2 in immunological disorders, cancer, and neurodegeneration. J. Biol. Chem. 2021, 297, 100905. [Google Scholar] [CrossRef] [PubMed]
- Hong, J.; Behar, J.; Wands, J.; Resnick, M.; Wang, L.J.; Delellis, R.A.; Lambeth, D.; Cao, W. Bile acid reflux contributes to development of esophageal adenocarcinoma via activation of phosphatidylinositol-specific phospholipase Cgamma2 and NADPH oxidase NOX5-S. Cancer Res. 2010, 70, 1247–1255. [Google Scholar] [CrossRef]
- Hussain, M.; Ikram, W.; Ikram, U. Role of c-Src and reactive oxygen species in cardiovascular diseases. Mol. Genet. Genom. MGG 2023, 298, 315–328. [Google Scholar] [CrossRef]
- Mathur, S.; Srivastava, P.; Srivastava, A.; Rai, N.K.; Abbas, S.; Kumar, A.; Tiwari, M.; Sharma, L.K. Regulation of metastatic potential by drug repurposing and mitochondrial targeting in colorectal cancer cells. BMC Cancer 2024, 24, 323. [Google Scholar] [CrossRef] [PubMed]
- Peng, W.T.; Sun, W.Y.; Li, X.R.; Sun, J.C.; Du, J.J.; Wei, W. Emerging Roles of G Protein-Coupled Receptors in Hepatocellular Carcinoma. Int. J. Mol. Sci. 2018, 19, 1366. [Google Scholar] [CrossRef]
- Lu, F.; Zhou, J.; Chen, Q.; Zhu, J.; Zheng, X.; Fang, N.; Qiao, L. PSMA5 contributes to progression of lung adenocarcinoma in association with the JAK/STAT pathway. Carcinogenesis 2022, 43, 624–634. [Google Scholar] [CrossRef]
- Xie, S.; Li, X.; Yan, J.; Yu, H.; Chen, S.; Chen, K. Knockdown of liver cancer cell-secreted exosomal PSMA5 controls macrophage polarization to restrain cancer progression by blocking JAK2/STAT3 signaling. Immun. Inflamm. Dis. 2024, 12, e1146. [Google Scholar] [CrossRef]
- Seo, S.U.; Kim, T.H.; Kim, D.E.; Min, K.J.; Kwon, T.K. NOX4-mediated ROS production induces apoptotic cell death via down-regulation of c-FLIP and Mcl-1 expression in combined treatment with thioridazine and curcumin. Redox Biol. 2017, 13, 608–622. [Google Scholar] [CrossRef]
- Arlt, A.; Bauer, I.; Schafmayer, C.; Tepel, J.; Muerkoster, S.S.; Brosch, M.; Roder, C.; Kalthoff, H.; Hampe, J.; Moyer, M.P.; et al. Increased proteasome subunit protein expression and proteasome activity in colon cancer relate to an enhanced activation of nuclear factor E2-related factor 2 (Nrf2). Oncogene 2009, 28, 3983–3996. [Google Scholar] [CrossRef]
- Lau, A.; Villeneuve, N.F.; Sun, Z.; Wong, P.K.; Zhang, D.D. Dual roles of Nrf2 in cancer. Pharmacol. Res. 2008, 58, 262–270. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Y.; Yin, B.; Ye, Y.; Dekhel, O.; Xiong, X.; Jian, Z.; Gu, L. The bidirectional role of the JAK2/STAT3 signaling pathway and related mechanisms in cerebral ischemia-reperfusion injury. Exp. Neurol. 2021, 341, 113690. [Google Scholar] [CrossRef] [PubMed]
- Chuffa, L.G.A.; de Souza, M.C.; Cruz, E.M.S.; Ferreira, F.B.; de Morais, J.M.B.; Seiva, F.R.F. Hepatocellular carcinoma and miRNAs: An in silico approach revealing potential therapeutic targets for polyphenols. Phytomedicine Plus 2022, 2, 100259. [Google Scholar] [CrossRef]
- Chuang, Y.T.; Yen, C.Y.; Tang, J.Y.; Wu, K.C.; Chang, F.R.; Tsai, Y.H.; Chien, T.M.; Chang, H.W. Marine anticancer drugs in modulating miRNAs and antioxidant signaling. Chem. Biol. Interact. 2024, 399, 111142. [Google Scholar] [CrossRef]
- Klimczak-Tomaniak, D.; Haponiuk-Skwarlinska, J.; Kuch, M.; Paczek, L. Crosstalk between microRNA and Oxidative Stress in Heart Failure: A Systematic Review. Int. J. Mol. Sci. 2022, 23, 5013. [Google Scholar] [CrossRef]
- Mishan, M.A.; Khazeei Tabari, M.A.; Mahrooz, A.; Bagheri, A. Role of microRNAs in the anticancer effects of the flavonoid luteolin: A systematic review. Eur. J. Cancer Prev. Off. J. Eur. Cancer Prev. Organ. 2021, 30, 413–421. [Google Scholar] [CrossRef]

| GO | GO ID | GO Description | FDR |
|---|---|---|---|
| BP | GO:0034599 | Response to oxidative stress | 2.14−184 |
| BP | GO:0062197 | Cellular response to chemical stress | 1.33−98 |
| BP | GO:1901700 | Metabolic process | 7.11−82 |
| BP | GO:0000302 | Response to reactive oxygen species | 8.51−80 |
| BP | GO:0010035 | Response to oxygen-containing compound | 1.12−73 |
| MF | GO:0016491 | Catalytic activity | 2.67−153 |
| MF | GO:0016209 | Oxidoreductase activity | 2.54−88 |
| MF | GO:0003824 | Antioxidant activity | 3.52−50 |
| MF | GO:0004601 | Small molecule binding | 1.96−47 |
| MF | GO:0019899 | Peroxidase activity | 9.80−41 |
| CC | GO:0005739 | Mitochondrion | 1.65−46 |
| CC | GO:0005737 | Cytoplasm | 1.76−32 |
| CC | GO:0043227 | Membrane-bound organelle | 1.20−25 |
| CC | GO:0005740 | Mitochondrial envelope | 7.96−18 |
| CC | GO:0031967 | Oxidoreductase complex | 1.07−15 |
| Term | p Value | Odds Ratio | Combined Score |
|---|---|---|---|
| Hepatoma HepG2 | 2.98 × 10−8 | 13.38 | 231.97 |
| Prostate LNCaP | 3.73 × 10−7 | 10.07 | 149.14 |
| Lung A-549 | 1.45 × 10−4 | 11.24 | 99.36 |
| Colon RKO | 0.001 | 7.11 | 48.54 |
| Breast MDA-MB | 0.004 | 6.36 | 33.84 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Seiva, F.R.F.; Agneis, M.L.G.; de Almeida, M.R.; Caputo, W.L.; de Souza, M.C.; das Neves, K.A.; Oliveira, É.N.; Justulin, L.A., Jr.; Chuffa, L.G.d.A. In Silico Analysis of Non-Conventional Oxidative Stress-Related Enzymes and Their Potential Relationship with Carcinogenesis. Antioxidants 2024, 13, 1279. https://doi.org/10.3390/antiox13111279
Seiva FRF, Agneis MLG, de Almeida MR, Caputo WL, de Souza MC, das Neves KA, Oliveira ÉN, Justulin LA Jr., Chuffa LGdA. In Silico Analysis of Non-Conventional Oxidative Stress-Related Enzymes and Their Potential Relationship with Carcinogenesis. Antioxidants. 2024; 13(11):1279. https://doi.org/10.3390/antiox13111279
Chicago/Turabian StyleSeiva, Fábio Rodrigues Ferreira, Maria Luisa Gonçalves Agneis, Matheus Ribas de Almeida, Wesley Ladeira Caputo, Milena Cremer de Souza, Karoliny Alves das Neves, Érika Novais Oliveira, Luis Antônio Justulin, Jr., and Luiz Gustavo de Almeida Chuffa. 2024. "In Silico Analysis of Non-Conventional Oxidative Stress-Related Enzymes and Their Potential Relationship with Carcinogenesis" Antioxidants 13, no. 11: 1279. https://doi.org/10.3390/antiox13111279
APA StyleSeiva, F. R. F., Agneis, M. L. G., de Almeida, M. R., Caputo, W. L., de Souza, M. C., das Neves, K. A., Oliveira, É. N., Justulin, L. A., Jr., & Chuffa, L. G. d. A. (2024). In Silico Analysis of Non-Conventional Oxidative Stress-Related Enzymes and Their Potential Relationship with Carcinogenesis. Antioxidants, 13(11), 1279. https://doi.org/10.3390/antiox13111279










