Peroxisome Proliferator-Activated Receptor Signaling-Mediated 13-S-Hydroxyoctadecenoic Acid Is Involved in Lipid Metabolic Disorder and Oxidative Stress in the Liver of Freshwater Drum, Aplodinotus grunniens
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Experimental Animals and Experimental Design
2.3. Sample Collection
2.4. Biochemical Index Measurement
2.5. RNA Extraction and De Novo High-throughput Sequencing
2.6. Metabolome Sample Processing and LC-MS Detection
2.7. Interaction Analysis
2.8. RT-PCR Analysis
2.9. Statistical Analysis
3. Results
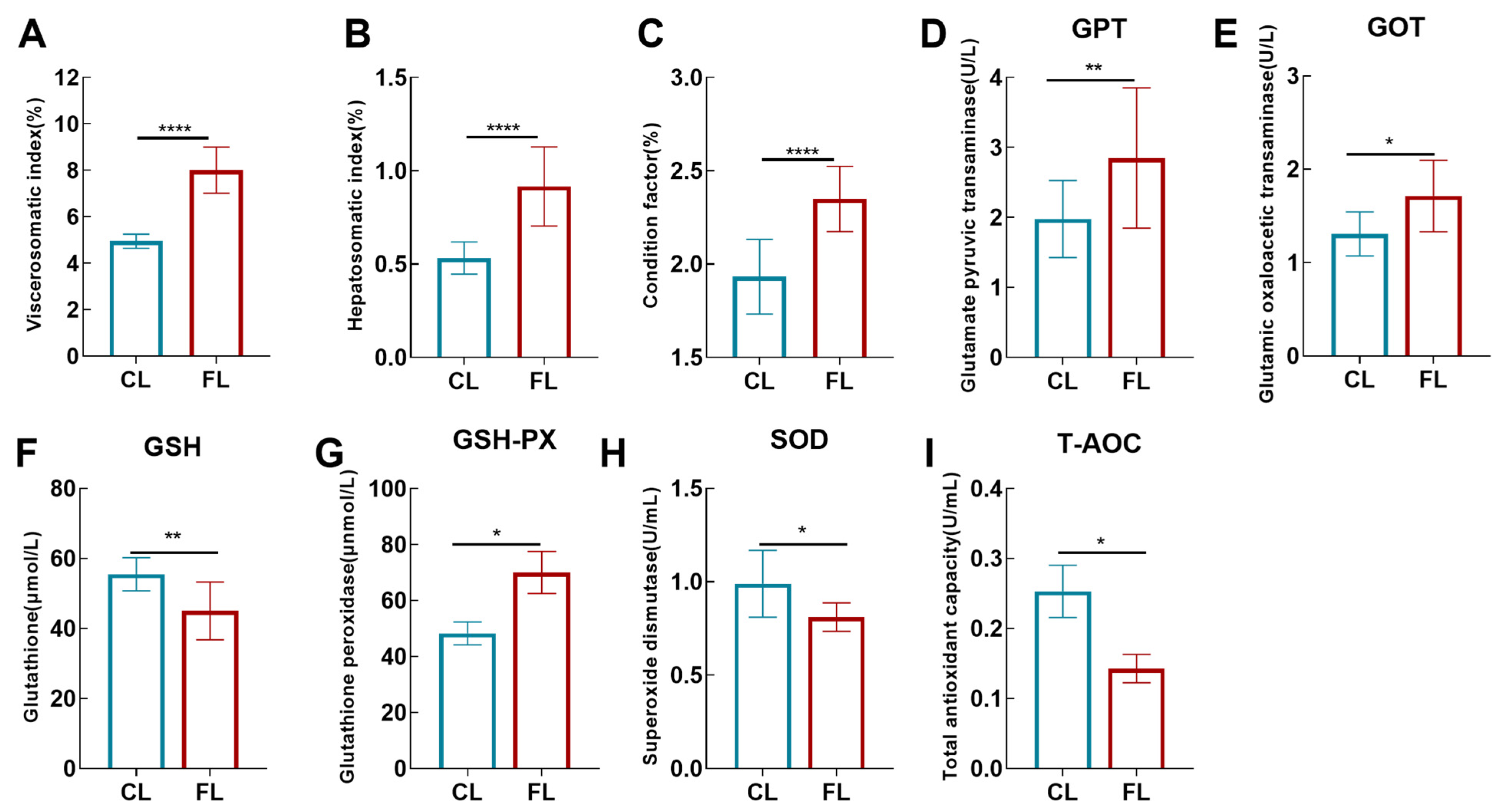
3.1. High-Fat Diet Induced Lipid Deposition and Destroyed Physiological Homeostasis in the Liver
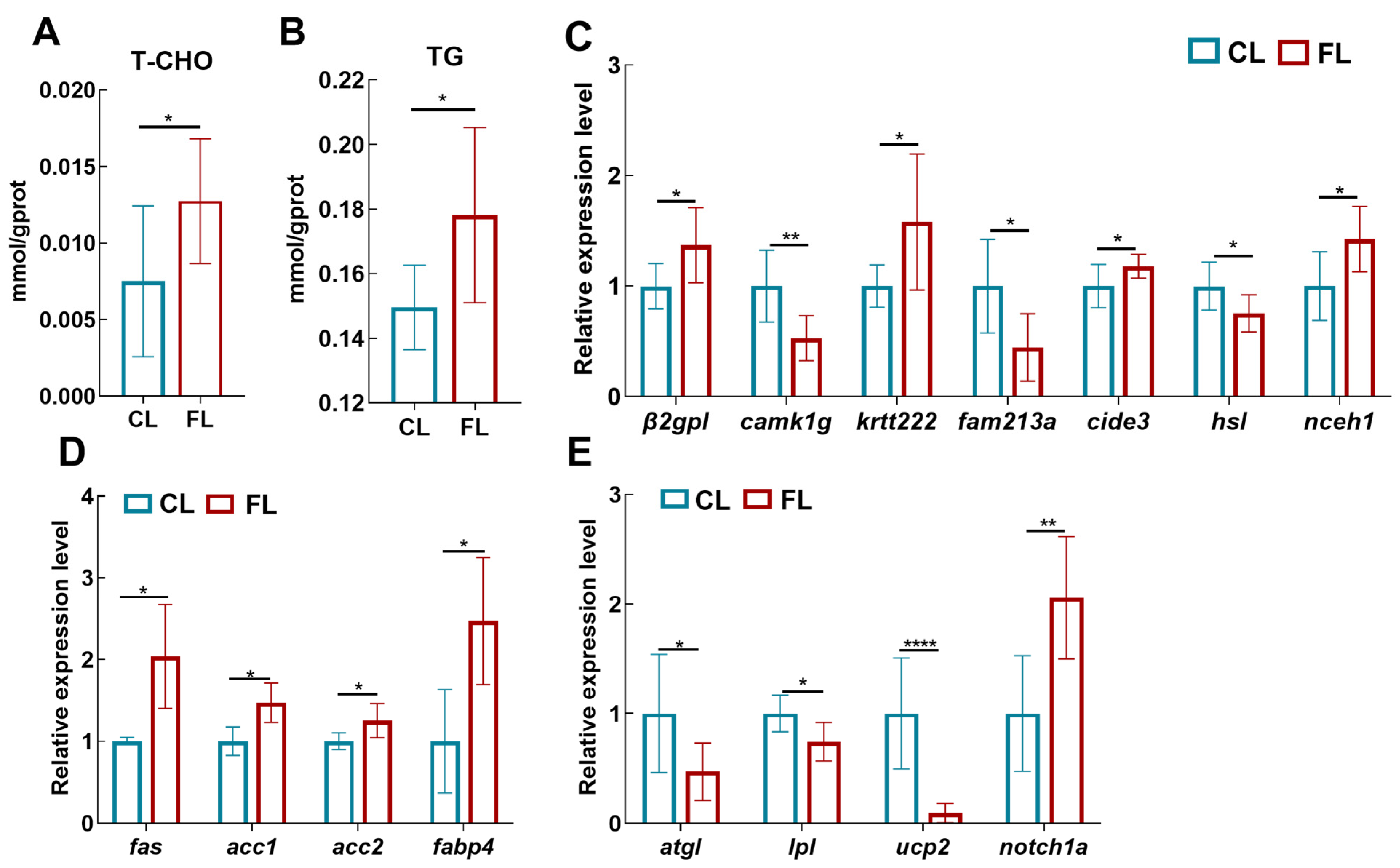
3.2. High-Fat Diet Led to Lipid Metabolism Disorder in the Liver of Freshwater Drum
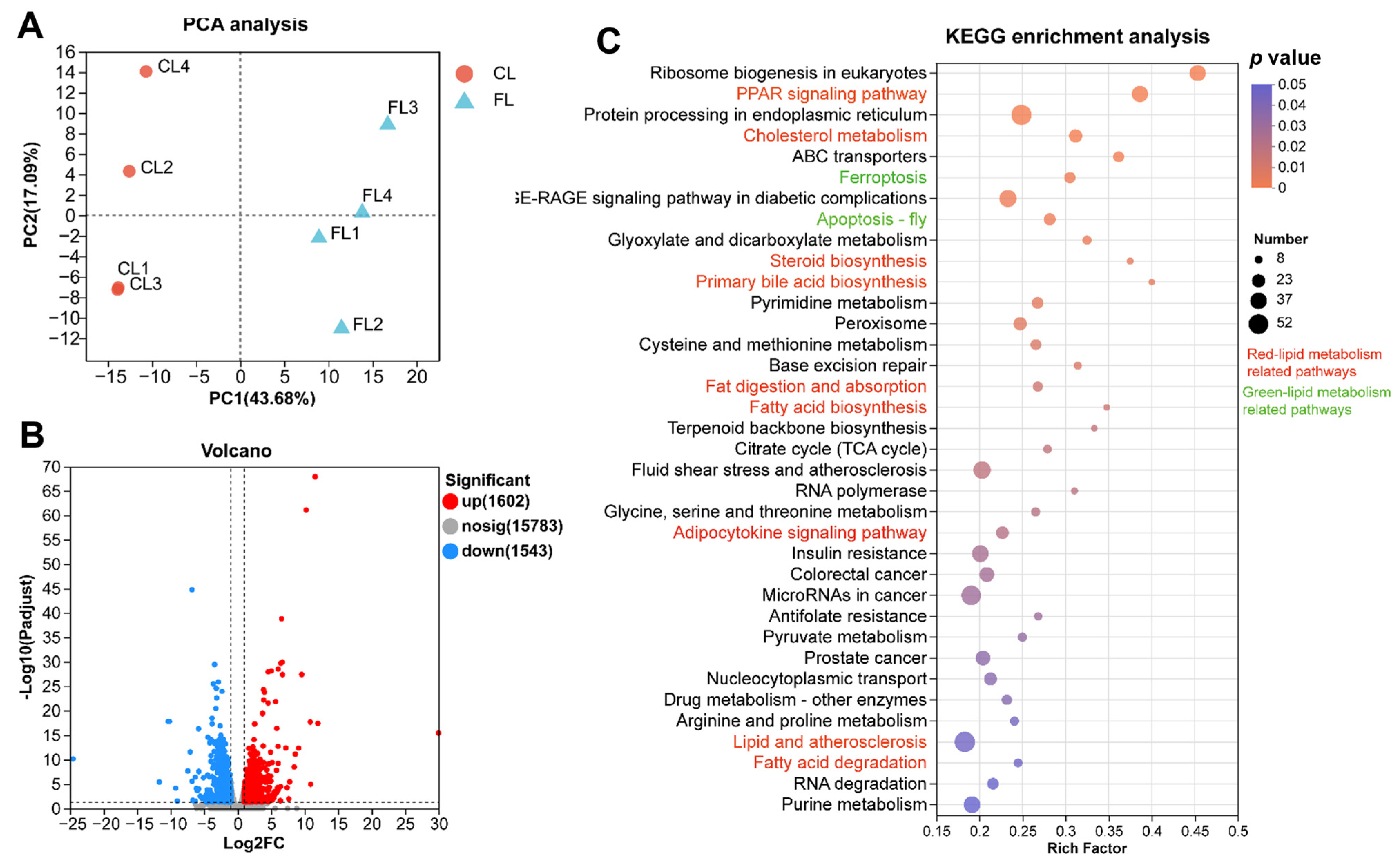
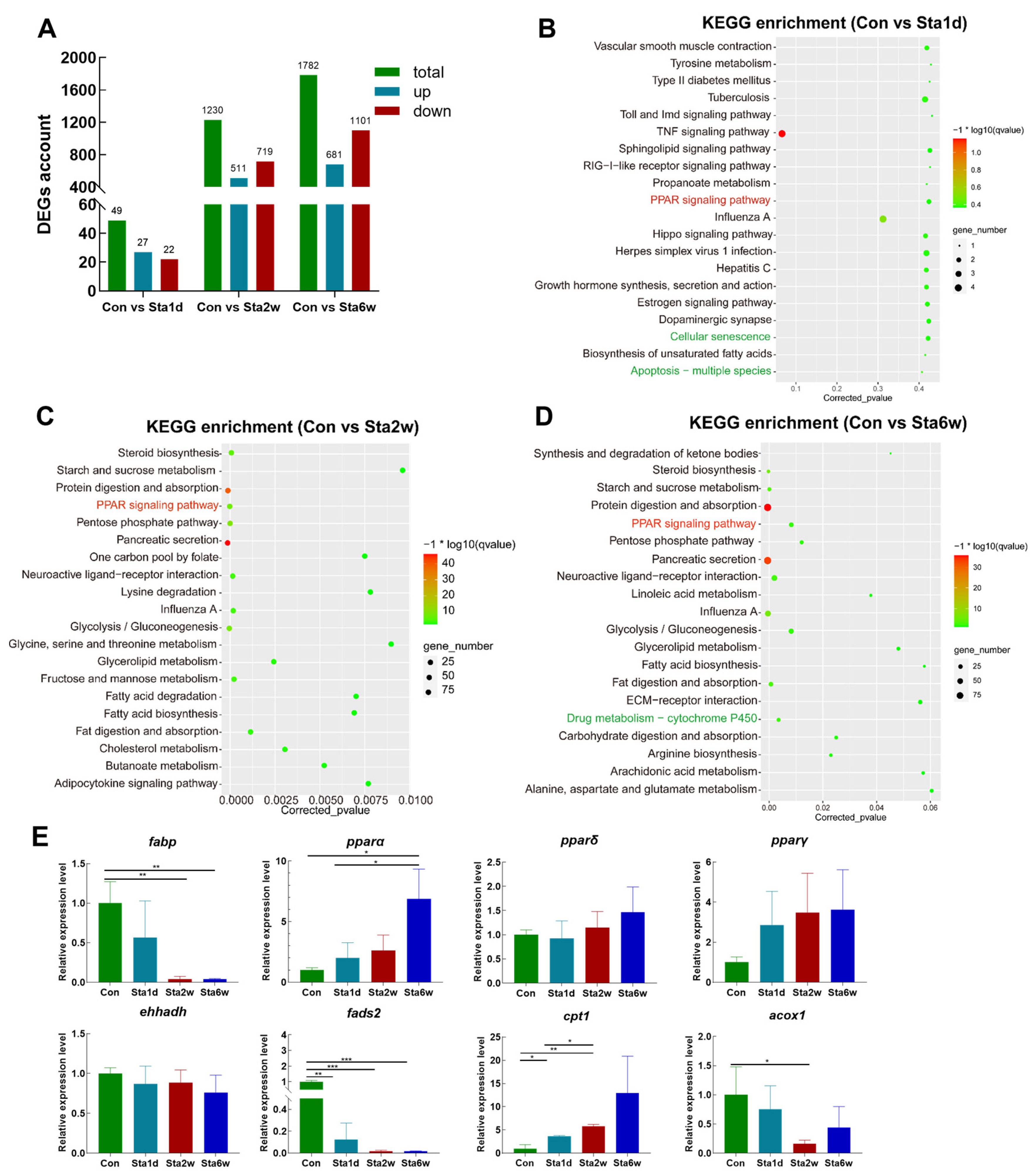
3.3. Transcriptomic Analysis Revealed Lipid Metabolism and Antioxidant Pathways Are Significantly Enriched in Livers with Lipid Deposition
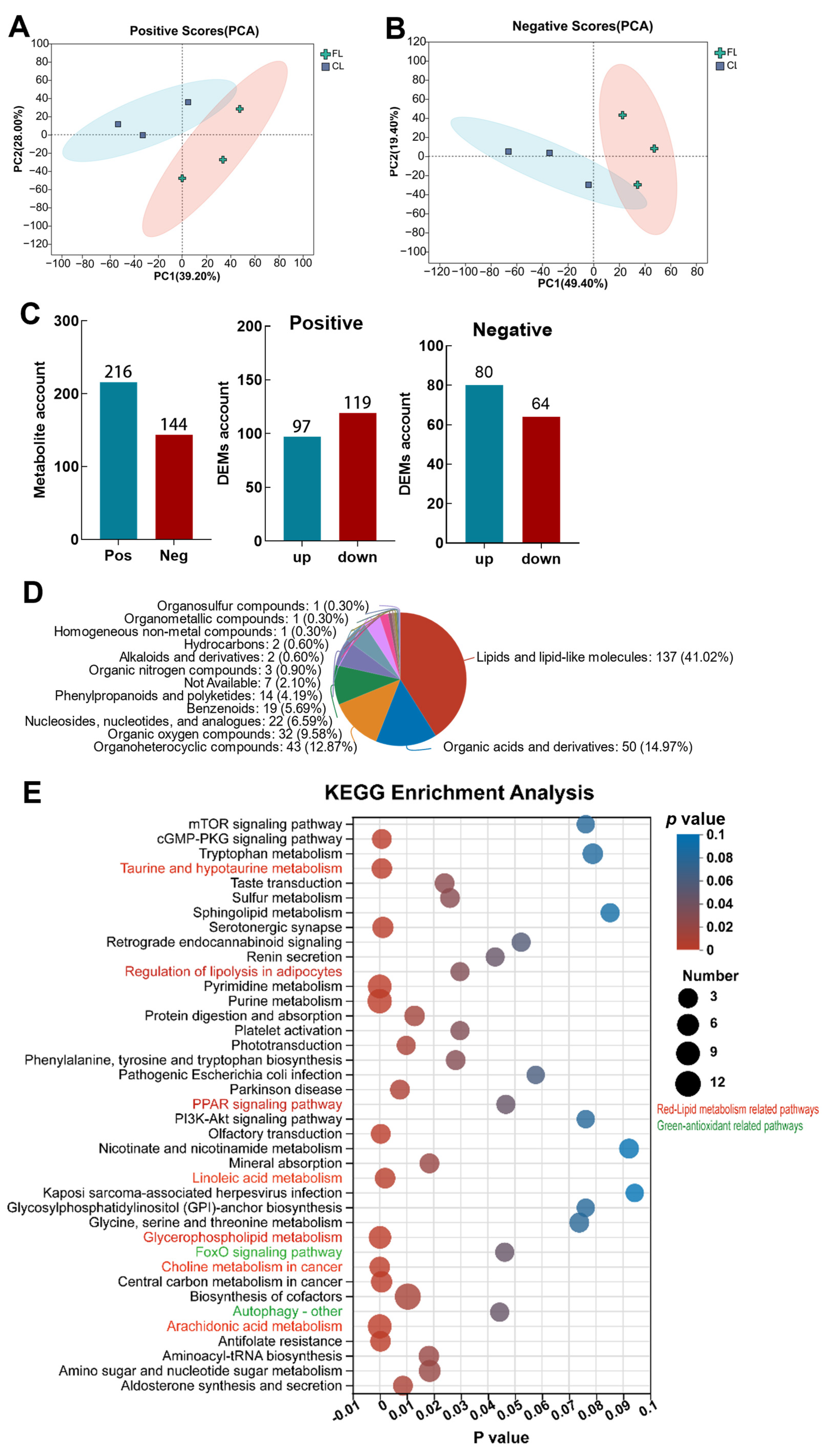
3.4. Metabolomic Analysis Revealed Lipid Metabolism and Antioxidant Pathways Are Significantly Enriched in Livers with Lipid Deposition
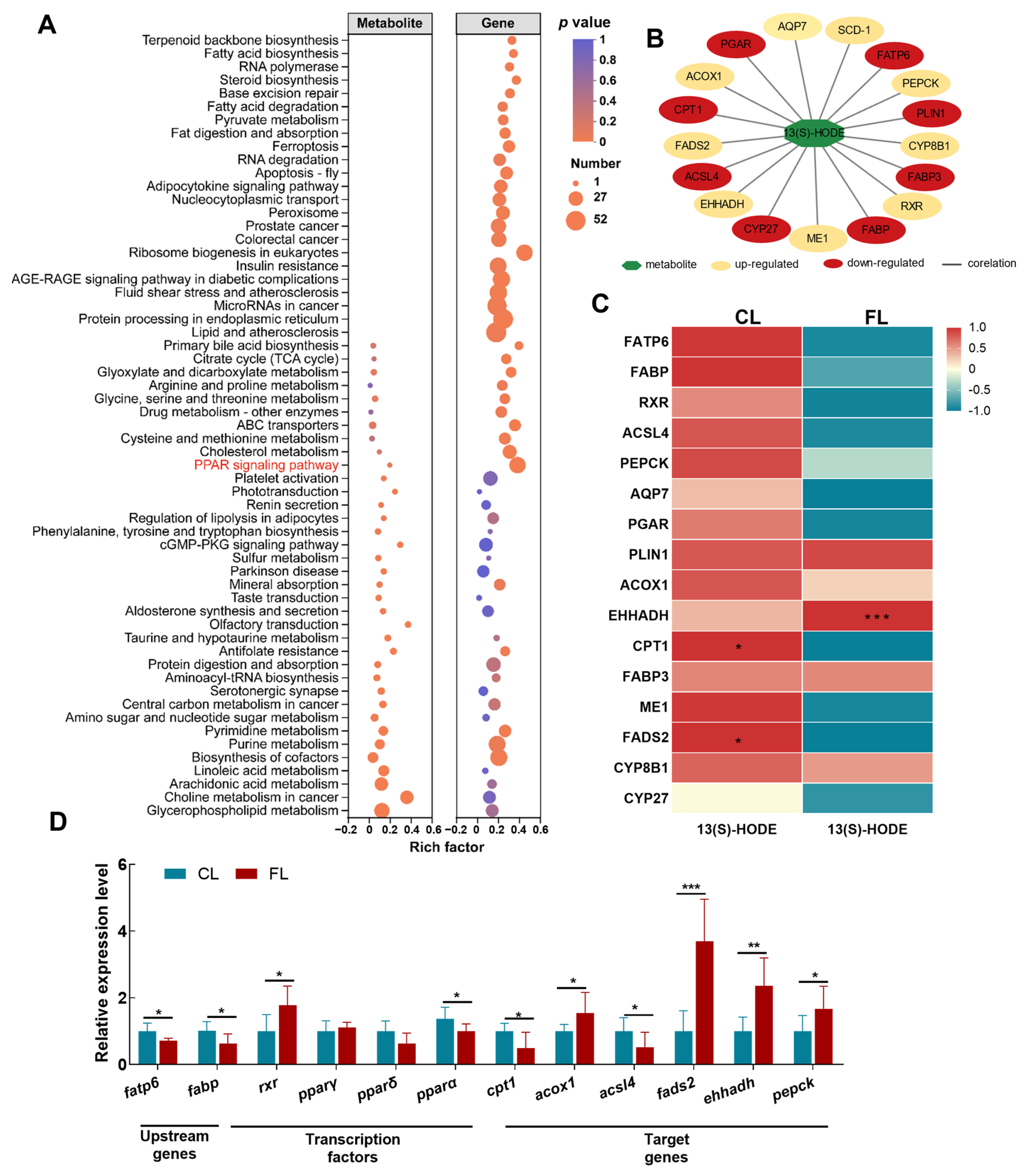
3.5. DEM and DEG Interaction Analysis Revealed that PPAR Signaling Is Involved in the Regulation of Lipid Deposition
3.6. Transcriptomic Analysis Revealed that PPAR Signaling Is Involved in Regulation of Lipid Consumption
3.7. Hypothesized Regulatory Mechanisms of Freshwater Drum
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yaakob, Z.; Ali, E.; Zainal, A.; Mohamad, M.; Takriff, M.S. An Overview: Biomolecules from Microalgae for Animal Feed and Aquaculture. J. Biol. Res. Thessalon. 2014, 21, 6. [Google Scholar] [CrossRef]
- Kamalam, B.S.; Medale, F.; Panserat, S. Utilisation of Dietary Carbohydrates in Farmed Fishes: New Insights on Influencing Factors, Biological Limitations and Future Strategies. Aquaculture 2017, 467, 3–27. [Google Scholar] [CrossRef]
- Wilson, R.P. Utilization of Dietary Carbohydrate by Fish. Aquaculture 1994, 124, 67–80. [Google Scholar] [CrossRef]
- Xie, R.; Amenyogbe, E.; Chen, G.; Huang, J. Effects of Feed Fat Level on Growth Performance, Body Composition and Serum Biochemical Indices of Hybrid Grouper (Epinephelus fuscoguttatus × Epinephelus polyphekadion). Aquaculture 2021, 530, 735813. [Google Scholar] [CrossRef]
- Craig, S.R.; Helfrich, L.A.; Kuhn, D.; Schwarz, M.H. Understanding Fish Nutrition, Feeds, and Feeding; Communications and Marketing, College of Agriculture and Life Sciences, Virginia Tech: Blacksburg, VA, USA, 2017; pp. 1–6. [Google Scholar]
- Watanabe, T. Lipid Nutrition in Fish. Comp. Biochem. Physiol. Part B Comp. Biochem. 1982, 73, 3–15. [Google Scholar] [CrossRef]
- Du, Z.-Y.; Liu, Y.-J.; Tian, L.-X.; Wang, J.-T.; Wang, Y.; Liang, G.-Y. Effect of Dietary Lipid Level on Growth, Feed Utilization and Body Composition by Juvenile Grass Carp (Ctenopharyngodon idella). Aquac. Nutr. 2005, 11, 139–146. [Google Scholar] [CrossRef]
- Jiang, S.; Wu, X.; Li, W.; Wu, M.; Luo, Y.; Lu, S.; Lin, H. Effects of Dietary Protein and Lipid Levels on Growth, Feed Utilization, Body and Plasma Biochemical Compositions of Hybrid Grouper (Epinephelus lanceolatus ♂ × Epinephelus fuscoguttatus ♀) Juveniles. Aquaculture 2015, 446, 148–155. [Google Scholar] [CrossRef]
- Zhang, M.L.; He, X.H. Role of fat in the nutrition and feeds of fishes. Reserv. Fish. 2003, 23, 62–63. (In Chinese) [Google Scholar]
- Chatzifotis, S.; Panagiotidou, M.; Papaioannou, N.; Pavlidis, M.; Nengas, I.; Mylonas, C.C. Effect of Dietary Lipid Levels on Growth, Feed Utilization, Body Composition and Serum Metabolites of Meagre (Argyrosomus regius) Juveniles. Aquaculture 2010, 307, 65–70. [Google Scholar] [CrossRef]
- Morais, S.; Bell, J.G.; Robertson, D.A.; Roy, W.J.; Morris, P.C. Protein/Lipid Ratios in Extruded Diets for Atlantic Cod (Gadus Morhua L.): Effects on Growth, Feed Utilisation, Muscle Composition and Liver Histology. Aquaculture 2001, 203, 101–119. [Google Scholar] [CrossRef]
- Wang, J.-T.; Liu, Y.-J.; Tian, L.-X.; Mai, K.-S.; Du, Z.-Y.; Wang, Y.; Yang, H.-J. Effect of Dietary Lipid Level on Growth Performance, Lipid Deposition, Hepatic Lipogenesis in Juvenile Cobia (Rachycentron canadum). Aquaculture 2005, 249, 439–447. [Google Scholar] [CrossRef]
- Xie, S.; Lin, Y.; Wu, T.; Tian, L.; Liang, J.; Tan, B. Dietary Lipid Levels Affected Growth Performance, Lipid Accumulation, Inflammatory Response and Apoptosis of Japanese Seabass (Lateolabraxjaponicus). Aquac. Nutr. 2021, 27, 807–816. [Google Scholar] [CrossRef]
- Park, K.-H.; Ye, Z.-W.; Zhang, J.; Kim, S.-H. Palmitic Acid-Enriched Diet Induces Hepatic Steatosis and Injury in Adult Zebrafish. Zebrafish 2019, 16, 497–504. [Google Scholar] [CrossRef] [PubMed]
- Tran, V.C.; Cho, S.-Y.; Kwon, J.; Kim, D. Alginate Oligosaccharide (AOS) Improves Immuno-Metabolic Systems by Inhibiting STOML2 Overexpression in High-Fat-Diet-Induced Obese Zebrafish. Food Funct. 2019, 10, 4636–4648. [Google Scholar] [CrossRef]
- Du, Z. Causes of fatty liver in farmed fish: A review and new perspectives. J. Fish. China 2014, 38, 1628–1638. (In Chinese) [Google Scholar]
- Yuan, X.; Liang, X.-F.; Liu, L.; Fang, J.; Li, J.; Li, A.; Cai, W.; Xue, M.; Wang, J.; Wang, Q. Fat Deposition Pattern and Mechanism in Response to Dietary Lipid Levels in Grass Carp, Ctenopharyngodon Idellus. Fish Physiol. Biochem. 2016, 42, 1557–1569. [Google Scholar] [CrossRef]
- Alves-Bezerra, M.; Cohen, D.E. Triglyceride Metabolism in the Liver. Compr. Physiol. 2017, 8, 1–22. [Google Scholar] [CrossRef]
- Trefts, E.; Gannon, M.; Wasserman, D.H. The Liver. Curr. Biol. 2017, 27, R1147–R1151. [Google Scholar] [CrossRef]
- Ferré, P.; Foufelle, F. Hepatic Steatosis: A Role for de Novo Lipogenesis and the Transcription Factor SREBP-1c. Diabetes Obes. Metab. 2010, 12, 83–92. [Google Scholar] [CrossRef]
- Lu, K.-L.; Xu, W.-N.; Li, X.-F.; Liu, W.-B.; Wang, L.-N.; Zhang, C.-N. Hepatic Triacylglycerol Secretion, Lipid Transport and Tissue Lipid Uptake in Blunt Snout Bream (Megalobrama amblycephala) Fed High-Fat Diet. Aquaculture 2013, 408–409, 160–168. [Google Scholar] [CrossRef]
- Ma, Q.; Li, L.-Y.; Le, J.-Y.; Lu, D.-L.; Qiao, F.; Zhang, M.-L.; Du, Z.-Y.; Li, D.-L. Dietary Microencapsulated Oil Improves Immune Function and Intestinal Health in Nile Tilapia Fed with High-Fat Diet. Aquaculture 2018, 496, 19–29. [Google Scholar] [CrossRef]
- Tang, T.; Hu, Y.; Peng, M.; Chu, W.; Hu, Y.; Zhong, L. Effects of High-Fat Diet on Growth Performance, Lipid Accumulation and Lipid Metabolism-Related MicroRNA/Gene Expression in the Liver of Grass Carp (Ctenopharyngodon idella). Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2019, 234, 34–40. [Google Scholar] [CrossRef]
- Jia, Y.; Jing, Q.; Niu, H.; Huang, B. Ameliorative Effect of Vitamin E on Hepatic Oxidative Stress and Hypoimmunity Induced by High-Fat Diet in Turbot (Scophthalmus maximus). Fish Amp; Shellfish Immunol. 2017, 67, 634–642. [Google Scholar] [CrossRef]
- Jin, M.; Pan, T.; Cheng, X.; Zhu, T.T.; Sun, P.; Zhou, F.; Ding, X.; Zhou, Q. Effects of Supplemental Dietary L-Carnitine and Bile Acids on Growth Performance, Antioxidant and Immune Ability, Histopathological Changes and Inflammatory Response in Juvenile Black Seabream (Acanthopagrus schlegelii) Fed High-Fat Diet. Aquaculture 2019, 504, 199–209. [Google Scholar] [CrossRef]
- Jia, R.; Cao, L.-P.; Du, J.-L.; He, Q.; Gu, Z.-Y.; Jeney, G.; Xu, P.; Yin, G.-J. Effects of High-Fat Diet on Antioxidative Status, Apoptosis and Inflammation in Liver of Tilapia (Oreochromis Niloticus) via Nrf2, TLRs and JNK Pathways. Fish Shellfish Immunol. 2020, 104, 391–401. [Google Scholar] [CrossRef]
- Shao, Y.; Xie, Z.; Liang, S.; Chen, C.; Tocher, D.R.; Lin, L.; Huang, Y.; Li, Y.; Xie, D.; Hong, Y.; et al. Dietary Calcium Pyruvate Could Improve Growth Performance and Reduce Excessive Lipid Deposition in Juvenile Golden Pompano (Trachinotus ovatus) Fed a High Fat Diet. Fish Physiol. Biochem. 2022, 48, 555–570. [Google Scholar] [CrossRef]
- Qiang, J.; Tao, F.; Bao, W.; He, J.; Li, X.; Chen, J.; Xu, P. Responses of Functional miRNA-mRNA Regulatory Modules to a High-Fat Diet in the Liver of Hybrid Yellow Catfish (Pelteobagrus fulvidraco × P. vachelli). Genomics 2021, 113, 1207–1220. [Google Scholar] [CrossRef]
- Mourente, G.; Tocher, D.R.; Diaz, E.; Grau, A.; Pastor, E. Relationships between Antioxidants, Antioxidant Enzyme Activities and Lipid Peroxidation Products during Early Development in Dentex Dentex Eggs and Larvae. Aquaculture 1999, 179, 309–324. [Google Scholar] [CrossRef]
- Christofides, A.; Konstantinidou, E.; Jani, C.; Boussiotis, V.A. The Role of Peroxisome Proliferator-Activated Receptors (PPAR) in Immune Responses. Metabolism 2021, 114, 154338. [Google Scholar] [CrossRef]
- Poulsen, L.L.C.; Siersbæk, M.; Mandrup, S. PPARs: Fatty Acid Sensors Controlling Metabolism. Semin. Cell Dev. Biol. 2012, 23, 631–639. [Google Scholar] [CrossRef]
- Song, C.; Liu, B.; Xu, P.; Ge, X.; Zhang, H. Emodin Ameliorates Metabolic and Antioxidant Capacity Inhibited by Dietary Oxidized Fish Oil through PPARs and Nrf2-Keap1 Signaling in Wuchang Bream (Megalobrama amblycephala). Fish Shellfish Immunol. 2019, 94, 842–851. [Google Scholar] [CrossRef]
- Wang, Y.; Nakajima, T.; Gonzalez, F.J.; Tanaka, N. PPARs as Metabolic Regulators in the Liver: Lessons from Liver-Specific PPAR-Null Mice. Int. J. Mol. Sci. 2020, 21, 2061. [Google Scholar] [CrossRef]
- Bougarne, N.; Weyers, B.; Desmet, S.J.; Deckers, J.; Ray, D.W.; Staels, B.; De Bosscher, K. Molecular Actions of PPARα in Lipid Metabolism and Inflammation. Endocr. Rev. 2018, 39, 760–802. [Google Scholar] [CrossRef]
- Toyama, T.; Nakamura, H.; Harano, Y.; Yamauchi, N.; Morita, A.; Kirishima, T.; Minami, M.; Itoh, Y.; Okanoue, T. PPARα Ligands Activate Antioxidant Enzymes and Suppress Hepatic Fibrosis in Rats. Biochem. Biophys. Res. Commun. 2004, 324, 697–704. [Google Scholar] [CrossRef]
- Gao, Z.; Li, Y.-H. Antioxidant Stress and Anti-Inflammation of PPARαon Warm Hepatic Ischemia-Reperfusion Injury. PPAR Res. 2012, 2012, 738785. [Google Scholar] [CrossRef]
- Chen, J.; Montagner, A.; Tan, N.; Wahli, W. Insights into the Role of PPARβ/δ in NAFLD. Int. J. Mol. Sci. 2018, 19, 1893. [Google Scholar] [CrossRef] [PubMed]
- Barlaka, E.; Görbe, A.; Gáspár, R.; Pálóczi, J.; Ferdinandy, P.; Lazou, A. Activation of PPARβ/δ Protects Cardiac Myocytes from Oxidative Stress-Induced Apoptosis by Suppressing Generation of Reactive Oxygen/Nitrogen Species and Expression of Matrix Metalloproteinases. Pharmacol. Res. 2015, 95–96, 102–110. [Google Scholar] [CrossRef]
- Walczak, R.; Tontonoz, P. PPARadigms and PPARadoxes: Expanding Roles for PPARγ in the Control of Lipid Metabolism. J. Lipid Res. 2002, 43, 177–186. [Google Scholar] [CrossRef] [PubMed]
- Corona, J.C.; Duchen, M.R. PPARγ as a Therapeutic Target to Rescue Mitochondrial Function in Neurological Disease. Free Radic. Biol. Med. 2016, 100, 153–163. [Google Scholar] [CrossRef] [PubMed]
- Zuo, X.; Wu, Y.; Morris, J.S.; Stimmel, J.B.; Leesnitzer, L.M.; Fischer, S.M.; Lippman, S.M.; Shureiqi, I. Oxidative Metabolism of Linoleic Acid Modulates PPAR-Beta/Delta Suppression of PPAR-Gamma Activity. Oncogene 2005, 25, 1225–1241. [Google Scholar] [CrossRef]
- Coleman, J.D.; Prabhu, K.S.; Thompson, J.T.; Reddy, P.S.; Peters, J.M.; Peterson, B.R.; Reddy, C.C.; Vanden Heuvel, J.P. The Oxidative Stress Mediator 4-Hydroxynonenal Is an Intracellular Agonist of the Nuclear Receptor Peroxisome Proliferator-Activated Receptor-β/δ (PPARβ/δ). Free Radic. Biol. Med. 2007, 42, 1155–1164. [Google Scholar] [CrossRef] [PubMed]
- Nixon, J.B.; Kamitani, H.; Baek, S.J.; Eling, T.E. Evaluation of Eicosanoids and NSAIDs as PPARγ Ligands in Colorectal Carcinoma Cells. Prostaglandins Leukot. Essent. Fat. Acids 2003, 68, 323–330. [Google Scholar] [CrossRef]
- Hernández-Gómez, R.E.; Contreras-Sánchez, W.M.; Hernández-Franyutti, A.; Perera-García, M.A.; Torres-Martínez, A. Testicular Structure and Development of the Male Germinal Epithelium in the Freshwater Drumv. Aplodinotus grunniens (Perciformes: Sciaenidae) from the Usumacinta River, Southern Mexico. Acta Zool. 2021, 103, 414–432. [Google Scholar] [CrossRef]
- Zhang, L.; Wen, H.; Zheng, B. Artificial spawning and embryonic development of freshwater drum, Aplodinotus grunnien. J. Fish. Sci. China 2021, 28, 569–578. (In Chinese) [Google Scholar]
- Chen, J.; Li, H.; Xu, P.; Tang, Y.; Su, S.; Liu, G.; Wu, N.; Xue, M.; Yu, F.; Feng, W.; et al. Hypothermia-Mediated Apoptosis and Inflammation Contribute to Antioxidant and Immune Adaption in Freshwater Drum, Aplodinotus grunniens. Antioxidants 2022, 11, 1657. [Google Scholar] [CrossRef]
- Chen, J.; Song, C.; Wen, H.; Liu, G.; Wu, N.; Li, H.; Xue, M.; Xu, P. miR-1/AMPK-Mediated Glucose and Lipid Metabolism under Chronic Hypothermia in the Liver of Freshwater Drum, Aplodinotus grunniens. Metabolites 2022, 12, 697. [Google Scholar] [CrossRef] [PubMed]
- Wu, N.; Wen, H.; Xu, P.; Chen, J.; Xue, M.; Li, J.; Wang, M.; Song, C.; Li, H. PPAR Signaling Maintains Metabolic Homeostasis under Hypothermia in Freshwater Drum (Aplodinotus grunniens). Metabolites 2023, 13, 102. [Google Scholar] [CrossRef]
- Song, C.; Wen, H.; Liu, G.; Ma, X.; Lv, G.; Wu, N.; Chen, J.; Xue, M.; Li, H.; Xu, P. Gut Microbes Reveal Pseudomonas Medicates Ingestion Preference via Protein Utilization and Cellular Homeostasis under Feed Domestication in Freshwater Drum, Aplodinotus grunniens. Front. Microbiol. 2022, 13, 861705. [Google Scholar] [CrossRef]
- Zhang, Y.-K.; Yang, B.-K.; Zhang, C.-N.; Xu, S.-X.; Sun, P. Effects of Polystyrene Microplastics Acute Exposure in the Liver of Swordtail Fish (Xiphophorus helleri) Revealed by LC-MS Metabolomics. Sci. Total Environ. 2022, 850, 157772. [Google Scholar] [CrossRef] [PubMed]
- Lu, K.-L.; Xu, W.-N.; Wang, L.-N.; Zhang, D.-D.; Zhang, C.-N.; Liu, W.-B. Hepatic β-Oxidation and Regulation of Carnitine Palmitoyltransferase (CPT) I in Blunt Snout Bream Megalobrama amblycephala Fed a High Fat Diet. PLoS ONE 2014, 9, e93135. [Google Scholar] [CrossRef]
- Tao, Y.; Qiang, J.; He, J.; Zhu, H.; Bao, J.; Xu, P. Untargeted LC–MS Metabolomics Approach Reveals Metabolic Changes in Genetically Improved Farmed Tilapia (Oreochromis niloticus) with Fatty Liver Induced by a High-fat Diet. Aquac. Res. 2020, 52, 724–735. [Google Scholar] [CrossRef]
- Jobling, M.; Koskela, J.; Savolainen, R. Influence of Dietary Fat Level and Increased Adiposity on Growth and Fat Deposition in Rainbow Trout, Oncorhynchus Mykiss (Walbaum). Aquac. Res. 1998, 29, 601–607. [Google Scholar] [CrossRef]
- Peng, M.; Xu, W.; Mai, K.; Zhou, H.; Zhang, Y.; Liufu, Z.; Zhang, K.; Ai, Q. Growth Performance, Lipid Deposition and Hepatic Lipid Metabolism Related Gene Expression in Juvenile Turbot (Scophthalmus maximus L.) Fed Diets with Various Fish Oil Substitution Levels by Soybean Oil. Aquaculture 2014, 433, 442–449. [Google Scholar] [CrossRef]
- Zhou, W.; Rahimnejad, S.; Lu, K.; Wang, L.; Liu, W. Effects of Berberine on Growth, Liver Histology, and Expression of Lipid-Related Genes in Blunt Snout Bream (Megalobrama amblycephala) Fed High-Fat Diets. Fish Physiol. Biochem. 2018, 45, 83–91. [Google Scholar] [CrossRef] [PubMed]
- Tao, Y.-F.; Qiang, J.; Bao, J.-W.; Chen, D.-J.; Yin, G.-J.; Xu, P.; Zhu, H.-J. Changes in Physiological Parameters, Lipid Metabolism, and Expression of MicroRNAs in Genetically Improved Farmed Tilapia (Oreochromis niloticus) with Fatty Liver Induced by a High-Fat Diet. Front. Physiol. 2018, 9, 1521. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.-L.; Guo, J.-L.; Tang, R.-J.; Ma, H.-J.; Chen, Y.-J.; Lin, S.-M. High Dietary Lipid Level Alters the Growth, Hepatic Metabolism Enzyme, and Anti-Oxidative Capacity in Juvenile Largemouth Bass Micropterus salmoides. Fish Physiol. Biochem. 2019, 46, 125–134. [Google Scholar] [CrossRef]
- Fang, H.; Xie, J.; Zhao, W.; Liu, Z.; Liu, Y.; Tian, L.; Niu, J. Study Supplementation of Astaxanthin in High-fat Diet on Growth Performance, Antioxidant Ability, Anti-inflammation, Non-specific Immunity and Intestinal Structure of Juvenile Trachinotus Ovatus. Aquac. Nutr. 2021, 27, 2575–2586. [Google Scholar] [CrossRef]
- Arfin, S.; Jha, N.K.; Jha, S.K.; Kesari, K.K.; Ruokolainen, J.; Roychoudhury, S.; Rathi, B.; Kumar, D. Oxidative Stress in Cancer Cell Metabolism. Antioxidants 2021, 10, 642. [Google Scholar] [CrossRef]
- Ma, Y.; Lee, G.; Heo, S.-Y.; Roh, Y.-S. Oxidative Stress Is a Key Modulator in the Development of Nonalcoholic Fatty Liver Disease. Antioxidants 2021, 11, 91. [Google Scholar] [CrossRef]
- Latunde-Dada, G.O. Ferroptosis: Role of Lipid Peroxidation, Iron and Ferritinophagy. Biochim. et Biophys. Acta (BBA)—Gen. Subj. 2017, 1861, 1893–1900. [Google Scholar] [CrossRef]
- Mello, T.; Zanieri, F.; Ceni, E.; Galli, A. Oxidative Stress in the Healthy and Wounded Hepatocyte: A Cellular Organelles Perspective. Oxidative Med. Cell. Longev. 2016, 2016, 8327410. [Google Scholar] [CrossRef] [PubMed]
- Ojha, A.; Yaduvanshi, S.K.; Srivastava, N. Effect of combined exposure of commonly used organophosphate pesticides on lipid peroxidation and antioxidant enzymes in rat tissues. Pestic. Biochem. Physiol. 2010, 99, 148–156. [Google Scholar] [CrossRef]
- Kosower, N.S.; Kosower, E.M. The Glutathione Status of Cells. In International Review of Cytology; Elsevier: Amsterdam, The Netherlands, 1978; pp. 109–160. [Google Scholar]
- Yin, P.; Xie, S.; Zhuang, Z.; He, X.; Tang, X.; Tian, L.; Liu, Y.; Niu, J. Dietary Supplementation of Bile Acid Attenuate Adverse Effects of High-Fat Diet on Growth Performance, Antioxidant Ability, Lipid Accumulation and Intestinal Health in Juvenile Largemouth Bass (Micropterus salmoides). Aquaculture 2021, 531, 735864. [Google Scholar] [CrossRef]
- Adjoumani, J.-J.Y.; Wang, K.; Zhou, M.; Liu, W.; Zhang, D. Effect of Dietary Betaine on Growth Performance, Antioxidant Capacity and Lipid Metabolism in Blunt Snout Bream Fed a High-Fat Diet. Fish Physiol. Biochem. 2017, 43, 1733–1745. [Google Scholar] [CrossRef] [PubMed]
- Mazzolini, G.; Sowa, J.-P.; Atorrasagasti, C.; Kücükoglu, Ö.; Syn, W.-K.; Canbay, A. Significance of Simple Steatosis: An Update on the Clinical and Molecular Evidence. Cells 2020, 9, 2458. [Google Scholar] [CrossRef]
- Wang, A.; Meng, D.; Hao, Q.; Xia, R.; Zhang, Q.; Ran, C.; Yang, Y.; Li, D.; Liu, W.; Zhang, Z.; et al. Effect of Supplementation of Solid-State Fermentation Product of Bacillus Subtilis HGcc-1 to High-Fat Diet on Growth, Hepatic Lipid Metabolism, Epidermal Mucus, Gut and Liver Health and Gut Microbiota of Zebrafish. Aquaculture 2022, 560, 738542. [Google Scholar] [CrossRef]
- Jin, M.; Pan, T.; Tocher, D.R.; Betancor, M.B.; Monroig, Ó.; Shen, Y.; Zhu, T.; Sun, P.; Jiao, L.; Zhou, Q. Dietary Choline Supplementation Attenuated High-Fat Diet-Induced Inflammation through Regulation of Lipid Metabolism and Suppression of NFκB Activation in Juvenile Black Seabream (Acanthopagrus schlegelii). J. Nutr. Sci. 2019, 8, e38. [Google Scholar] [CrossRef]
- Li, Y.; Liang, S.; Shao, Y.; Li, Y.; Chen, C.; You, C.; Monroig, Ó.; Rahimnejad, S.; Tocher, D.R.; Wang, S. Impacts of Dietary Konjac Glucomannan Supplementation on Growth, Antioxidant Capacity, Hepatic Lipid Metabolism and Inflammatory Response in Golden Pompano (Trachinotus ovatus) Fed a High Fat Diet. Aquaculture 2021, 545, 737113. [Google Scholar] [CrossRef]
- Xue, M.; Wen, H.; Xu, P.; Chen, J.; Wang, Q.; Tang, Y.; Ma, X.; Lv, G.; Li, H.; Song, C. Validation and Functional Analysis of Reference and Tissue-Specific Genes in Adipose Tissue of Freshwater Drum, Aplodinotus grunniens, under Starvation and Hypothermia Stress. Cells 2023, 12, 1328. [Google Scholar] [CrossRef]
- Alves Martins, D.; Rocha, F.; Martínez-Rodríguez, G.; Bell, G.; Morais, S.; Castanheira, F.; Bandarra, N.; Coutinho, J.; Yúfera, M.; Conceição, L.E.C. Teleost Fish Larvae Adapt to Dietary Arachidonic Acid Supply through Modulation of the Expression of Lipid Metabolism and Stress Response Genes. Br. J. Nutr. 2011, 108, 864–874. [Google Scholar] [CrossRef]
- Valenti, L.; Mendoza, R.M.; Rametta, R.; Maggioni, M.; Kitajewski, C.; Shawber, C.J.; Pajvani, U.B. Hepatic Notch Signaling Correlates with Insulin Resistance and Nonalcoholic Fatty Liver Disease. Diabetes 2013, 62, 4052–4062. [Google Scholar] [CrossRef]
- Jia, R.; Wang, L.; Hou, Y.; Feng, W.; Li, B.; Zhu, J. Effects of Stocking Density on the Growth Performance, Physiological Parameters, Redox Status and Lipid Metabolism of Micropterus salmoides in Integrated Rice–Fish Farming Systems. Antioxidants 2022, 11, 1215. [Google Scholar] [CrossRef]
- Yu, L.; Tian, J.; Zhang, C.; Xue, Z.; Lu, X.; Wen, H.; Jiang, M.; Wu, F. Acetylferulic Paeonol Ester: A New Feed Additive Reduces Lipid Accumulation in the Liver of Nile Tilapia (Oreochromis niloticus) by Modulating Lipid and Glucose Metabolism. Aquaculture 2022, 561, 738671. [Google Scholar] [CrossRef]
- Hong, C.; Tontonoz, P. Coordination of Inflammation and Metabolism by PPAR and LXR Nuclear Receptors. Curr. Opin. Genet. Dev. 2008, 18, 461–467. [Google Scholar] [CrossRef]
- Liu, G.; Song, C.; Wen, H.; Wu, N.; Chen, J.; Li, H.; Xu, P. Effect of Starvation stress on physical indexes, muscle fatty acid composition and liver metabolism gene expression of Aplodinotus grunniens. Chin. J. Anim. Nutr. 2022, 34, 544–554. (In Chinese) [Google Scholar]
- Kersten, S.; Desvergne, B.; Wahli, W. Roles of PPARs in Health and Disease. Nature 2000, 405, 421–424. [Google Scholar] [CrossRef] [PubMed]
- Lamichane, S.; Dahal Lamichane, B.; Kwon, S.-M. Pivotal Roles of Peroxisome Proliferator-Activated Receptors (PPARs) and Their Signal Cascade for Cellular and Whole-Body Energy Homeostasis. Int. J. Mol. Sci. 2018, 19, 949. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Sun, S.; He, J.; Wang, Y.; Ren, T.; He, H.; Gao, J. Enoyl-CoA Hydratase/3-Hydroxyacyl CoA Dehydrogenase Is Essential for the Production of DHA in Zebrafish. J. Lipid Res. 2023, 64, 100326. [Google Scholar] [CrossRef] [PubMed]
- Eaton, S. Control of Mitochondrial β-Oxidation Flux. Prog. Lipid Res. 2002, 41, 197–239. [Google Scholar] [CrossRef] [PubMed]
- Vaittinen, M.; Männistö, V.; Käkelä, P.; Ågren, J.; Tiainen, M.; Schwab, U.; Pihlajamäki, J. Interorgan Cross Talk between Fatty Acid Metabolism, Tissue Inflammation, andFADS2genotype in Humans with Obesity. Obesity 2017, 25, 545–552. [Google Scholar] [CrossRef]
- Vaittinen, M.; Walle, P.; Kuosmanen, E.; Männistö, V.; Käkelä, P.; Ågren, J.; Schwab, U.; Pihlajamäki, J. FADS2 Genotype Regulates Delta-6 Desaturase Activity and Inflammation in Human Adipose Tissue. J. Lipid Res. 2016, 57, 56–65. [Google Scholar] [CrossRef] [PubMed]

| Accession No. | Gene | Primer Sequence (5′—3′) |
|---|---|---|
| XP_008328442.1 | β-actin | F: AGGCTGTGCTGTCCCTGTAT R: GCTGTGGTGGTGAAGGAGTAG |
| XP_010742647.2 | β2gp1 | F: GGCAGTATCCTCACCCCATC R: CCTTCTGAGGTCCATCCAGC |
| KKF21127.1 | camk1g | F: TACATGCTCGGCTCCACTCT R: TCTCCTTCACGCTCAACTCG |
| KKF23363.1 | krt222 | F: GAGAGTGCAGAAGGTCACGG R: GGGGAGGCTGTCCTGTTTAG |
| XP_018535573.1 | cide3 | F: ACCCCACATCCAAACAGCAT R: TTTTTGGCAGCGTAACAGCG |
| XP_019122735.1 | hsl | F: TTGCTGAGATGAGGGTGGA R: ACAGGCTGGTCTATGTTCC |
| XP_010730495.2 | nceh1 | F: TATTAACGGTGGCGTTCGCT R: AAAGAAGCCAGGTGCATCGT |
| XP_010741055.2 | fam213a | F: CCCGTGAAAGAAAGATGG R: GTCCAATGACGAACACCC |
| KKF24881.1 | fas | F: TGGCATCGAGTACAACAAGC R: TTGGCACGAAGTAGCATCAC |
| XP_019127403.1 | acc1 | F: CTGGAGGAGACGGTGAAAAG R: TGCGTATCTGCTTGAGGATG |
| XP_018523996.1 | acc2 | F: AGAGGACCATCCGTTTTGTG R: TTCAGAGGAATGACCCCATC |
| XP_010755203.2 | fabp4 | F: CAGACGGTCGAAAGACCAAG R: TCATGGCAACAACATCATCC |
| XP_010739642.2 | atgl | F: ACGGGGAGAACATACTGGTG R: GTGGAAGCTGGTGGAGTTGT |
| XP_010729007.2 | lpl | F: CAGCCGTGCAGTATGTGACT R: AGGTTTTGGAGGTGCTGTTG |
| XP_019132010.1 | ucp2 | F: ATTCGTGGTCTGTGGAAAGG R: CTGCACCAAAGGCTGATAGG |
| XP_019134682.1 | notch1a | F: ACTCAAATGGCTCCCTCCTT R: TCAGTTTCCCCATCTCTGCT |
| XP_019127721.1 | fatp6 | F: TCAGATCCAGCGTGTGTACG G: CAACAAGGCAACGCAGTCTC |
| XP_027137407.1 | acsl4 | F: GGCACCCGAGATGTACTGAG R: ACTCCGCTCTGGTTTCACAG |
| KAE8296399.1 | cpt1 | F: TCAGAGGCAGGAGCCCTATT R: GTGCATGTTCACCACGTTCC |
| XP_010744948.3 | fabp | F: TGGTGAAAACCCTGAGCACC R: GCACTTGCACCAGTTTGTCT |
| XP_019127104.1 | rxr | F: CAAGCTGTTGCTGCGGTTAC R: TCATTTGATGCGGGGCTTCT |
| XP_010746626.2 | acox1 | F: TTACCAGCGCATCAGTGGAG R: CTGCGTTGGTTGTCCATGTG |
| AGG69480.1 | fads2 | F: GAAACAGCTTACGCACTCTGC R: AAGTTGCTCTCCATCCACAGG |
| XP_019126127.1 | ehhadh | F: CCTGGTCATTGAGGCTGTGT R: GTTACGGGTTTGAGAGGCCA |
| XP_019131039.1 | pepck | F: CCACGTCAACTGGTTCAGGA R: CAGCCAGCCGATAATGCTCT |
| XP_010747326.2 | pparα | F: GTGCCTCTCTGTGGGAATGT R: GCTTCGTGGATCTGCCTTAC |
| XP_019110784.1 | pparγ | F: GCCTTTGTCTGCCTCTCAAC R: GACCTCGCTACCCTTTCCTC |
| XP_010746753.2 | pparδ | F: ATCACCGTCGCTGTCAGAAC R: CCCTTACAACCCTCACAGG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xue, M.; Xu, P.; Wen, H.; Chen, J.; Wang, Q.; He, J.; He, C.; Kong, C.; Song, C.; Li, H. Peroxisome Proliferator-Activated Receptor Signaling-Mediated 13-S-Hydroxyoctadecenoic Acid Is Involved in Lipid Metabolic Disorder and Oxidative Stress in the Liver of Freshwater Drum, Aplodinotus grunniens. Antioxidants 2023, 12, 1615. https://doi.org/10.3390/antiox12081615
Xue M, Xu P, Wen H, Chen J, Wang Q, He J, He C, Kong C, Song C, Li H. Peroxisome Proliferator-Activated Receptor Signaling-Mediated 13-S-Hydroxyoctadecenoic Acid Is Involved in Lipid Metabolic Disorder and Oxidative Stress in the Liver of Freshwater Drum, Aplodinotus grunniens. Antioxidants. 2023; 12(8):1615. https://doi.org/10.3390/antiox12081615
Chicago/Turabian StyleXue, Miaomiao, Pao Xu, Haibo Wen, Jianxiang Chen, Qingyong Wang, Jiyan He, Changchang He, Changxin Kong, Changyou Song, and Hongxia Li. 2023. "Peroxisome Proliferator-Activated Receptor Signaling-Mediated 13-S-Hydroxyoctadecenoic Acid Is Involved in Lipid Metabolic Disorder and Oxidative Stress in the Liver of Freshwater Drum, Aplodinotus grunniens" Antioxidants 12, no. 8: 1615. https://doi.org/10.3390/antiox12081615
APA StyleXue, M., Xu, P., Wen, H., Chen, J., Wang, Q., He, J., He, C., Kong, C., Song, C., & Li, H. (2023). Peroxisome Proliferator-Activated Receptor Signaling-Mediated 13-S-Hydroxyoctadecenoic Acid Is Involved in Lipid Metabolic Disorder and Oxidative Stress in the Liver of Freshwater Drum, Aplodinotus grunniens. Antioxidants, 12(8), 1615. https://doi.org/10.3390/antiox12081615








