Abstract
Polyamine (PA) catabolism mediated by amine oxidases is an important process involved in fine-tuning PA homeostasis and related mechanisms during salt stress. The significance of these amine oxidases in short-term responses to salt stress is, however, not well understood. In the present study, the effects of L-aminoguanidine (AG) on tomato roots treated with short-term salt stress induced by NaCl were studied. AG is usually used as a copper amine oxidase (CuAO or DAO) inhibitor. In our study, other alterations of PA catabolism, such as reduced polyamine oxidase (PAO), were also observed in AG-treated plants. Salt stress led to an increase in the reactive oxygen and nitrogen species in tomato root apices, evidenced by in situ fluorescent staining and an increase in free PA levels. Such alterations were alleviated by AG treatment, showing the possible antioxidant effect of AG in tomato roots exposed to salt stress. PA catabolic enzyme activities decreased, while the imbalance of hydrogen peroxide (H2O2), nitric oxide (NO), and hydrogen sulfide (H2S) concentrations displayed a dependence on stress intensity. These changes suggest that AG-mediated inhibition could dramatically rearrange PA catabolism and related reactive species backgrounds, especially the NO-related mechanisms. More studies are, however, needed to decipher the precise mode of action of AG in plants exposed to stress treatments.
1. Introduction
Salt stress is one of the most important abiotic stress factors threatening agriculture and food security globally [1]. Under increasingly challenging climate conditions, more efforts are needed to understand the mechanisms of salt stress and salt tolerance of crop plants [2]. Plants have evolved many different strategies to cope with salinity stress, for example, by synthesizing compounds that alleviate salt stress injury [3].
Polyamines (PAs) are hub molecules in drought and salt stress in plants [4,5]. These essential N-containing molecules occur in different forms in plants, can be free, bound to high-molecular-weight compounds or conjugated to low-molecular-weight ones [6,7]. Many studies have revealed that the accumulation of PAs is a general effect of salt stress on plants, and the extent depends on the plant species, the duration of the stress, or the plant organs affected [8,9,10,11]. PAs are able to reprogram oxidative and nitrosative statuses and the proteome in citrus plants during salt stress [12].
Their involvement in development and abiotic stress response is crucial, as they can interact with and influence other signal molecules, such as nitric oxide (NO) or hydrogen sulfide (H2S), both of which are important players in plant abiotic stress signaling [13,14,15].
The biosynthesis of PAs occurs via two pathways: from L-arginine via the enzyme arginine decarboxylase (ADC) or from L-ornithine via the enzyme ornithine decarboxylase (ODC). The function of these enzymes is developmental stage- and organ-dependent in plants during abiotic stress conditions [10].
Besides biosynthesis, PA catabolism is one of the most important processes for fine-tuning the PA levels in salt stress [15,16]. The enzymes involved in catabolic reactions are diamine oxidases (DAOs) and polyamine oxidases (PAOs) [17,18]. Recently, the prominent role of DAOs in the control of plant development and abiotic stress tolerance has been emphasized [19,20]. The precise role of these enzymes in salt stress responses and their interplay with other signaling pathways are, however, not well understood. Fast activation of the genes involved in PA catabolism was shown to happen within one hour after salt treatment [21]. Reduced cytoplasmic PAO activities led to enhanced tolerance to salt and drought stress in Arabidopsis by suppressing reactive oxygen species (ROS) production and inducing the expression defense genes [22]. Deregulation of apoplastic PAO could affect tobacco development and influence responses to salt stress [23].
ROS are crucial in plant abiotic stress signaling [24]. Their production, transport, and scavenging by enzymatic and non-enzymatic antioxidants were shown to be tightly connected to various signaling pathways. ROS were shown to influence PA homeostasis, whereas PAs are known to modulate redox homeostasis during salt stress [25]. Spermidine oxidation could induce hydrogen peroxide (H2O2) production in abiotic stress conditions [26]. Copper amine oxidases (CuAOs) were shown to control NO levels, as the Arabidopsis cuao mutant (AtCuAO) plants displayed impaired NO production [27,28]. Copper amine oxidase 8 (CuAO8) could regulate the arginine-dependent NO production in Arabidopsis thaliana [29]. Nitric oxide (NO) is an important gaseous signal molecule and one of the most crucial reactive nitrogen species (RNS) in plants. NO production is connected to PA homeostasis, as its production from L-arginine is enhanced by PAs [30,31,32]. NO levels could be under feed-back regulation, and NO can also influence abiotic stress responses through the modulation of ROS signals [33] However, there is a gap in our knowledge about the significance of PA catabolism in inducing ROS/RNS production during short-term salt stress in tomatoes.
In order to study the significance of the enzymes of PA catabolism in salt tolerance, different inhibitors which can suppress enzyme activities have been used [34,35]. L-aminoguanidine (AG) is one of those inhibitors which can block the activities of DAOs [34,36]. Efficiency of enzyme inhibition depends on the time of application, the concentration of the inhibitor, and the accompanying stress treatments [34]. Despite being one of the most commonly used inhibitors, controversial effects of AG have been observed. We have observed cultivar-dependent effects of AG treatment on tomato germination [37].
PA metabolism is strongly dependent on the nitrogen (N) balance in plants [38,39]. N metabolism was shown to be regulated by interacting PA and NO signals [40]. Nitrite is not only a N source but is also involved in NO production through a reductive biosynthetic pathway [41]. This process contributes to S-nitrosothiol (SNO) production and subsequent S-nitrosylation of proteins, which is an efficient post-translational modification in plants [42,43,44]. Salinity could induce changes in the S-nitrosylation pattern of mitochondrial proteins in Pisum sativum [45]. Additionally, S-nitrosylation or denitrosylation could act as a regulatory mechanism of salt-stress sensing in sunflower seedlings [46].
Our knowledge about the involvement of PAs and their catabolism in short-term salt stress is limited [47], and their contribution to triggering other signaling pathways has not been studied. The objective of this research was to study and give an overview of the short-term effect of salt stress and AG treatment on PA catabolism and related pathways to decipher the significance of PA catabolism and related pathways in the regulation of the fast stress responses of tomato roots.
2. Materials and Methods
2.1. Plant Material and Growth Conditions
The tomato plants used in this study were Solanum lycopersicum Mill. L. cv. Rio Fuego. Seeds were germinated at 26 °C for 3 d in the dark, and the seedlings were subsequently transferred to perlite for 2 weeks. The plants were then placed in a hydroponic culture, as described [8]. The plants were grown for 6 weeks in a controlled environment under 200 µmol m−2 s−1 photon flux density (F36W/GRO lamps, OSRAM SYLVANIA, Danvers, MA, USA), with a 12/12-h light/dark period, day/night temperatures of 24/22 °C, and relative humidity of 55–60%.
2.2. NaCl- and AG-Inhibitor Treatments
Salinity treatment was applied to 6-week-old tomato plants by supplementing the hydroponic solution with 100 and 250 mM NaCl for 1 h in the absence or presence of 1 mM L-aminoguanidine (AG). The concentration of AG was defined to inhibit DAO activity as described [37]. Whole root samples were used for biochemical analysis, whereas root apical tips were used for fluorescent staining. The experiments were repeated three times. In order to avoid the diurnal changes of PA homeostasis, sampling was performed at the same time in all experiments.
2.3. Polyamine Catabolic Enzyme Activities
DAO (EC 1.4.3.6) and PAO (EC 1.4.3.4) activities were estimated spectrophotometrically, as described by [26] with some modification. Approximately 200 mg of root tissue was homogenized in liquid N2, and 0.6 mL extraction buffer was added to each sample. The extraction buffer contained 0.2 M TRIS (hydroxymethyl)aminomethane (pH 8.0); 10% glycerol; 0.25% Triton X-100; 0.5 mM phenylmethanesulfonyl fluoride (PMSF); 0.01 mM leupeptin in 100 mM potassium phosphate buffer (pH 6.6). The homogenates were left on ice for 20 min and centrifuged for 10 min at 7000× g at 4 °C (Eppendorf centrifuge 5424R, Eppendorf GMBH, Hamburg, Germany). A 150 μL aliquot of the supernatant was combined with 600 μL 100 mM potassium phosphate buffer (pH 6.6), and then the reaction was started by adding 22.5 μL of 2-aminobenzaldehyde (from 10 mg/mL stock solution) and 1 M putrescine (Put) for DAO and 1 M spermidine (Spd) for PAO activity measurements. The reaction mixture was incubated for 1.5 h at 37 °C, and the reaction was stopped by adding 50 μL of 20% (w/v) trichloroacetic acid (TCA). The absorbance was determined at 430 nm (KONTRON, Milan, Italy). The enzyme activity was expressed as specific activity (U g−1 FW), where one unit (U) represents the amount of enzyme catalyzing the formation of 1 μmol of Δ1-pyrroline min−1.
2.4. Determination of Free PA Content Using HPLC
Free PA contents were determined as described by [37]. In brief, 200 mg of root was homogenized in 5% perchloric acid. After centrifugation, 2.5 mL of the supernatant was neutralized with 1 mL of 2 M NaOH, and PAs were then derivatized with 10 µL of benzoyl chloride and separated using HPLC. The applied standards were Put, Spd, and spermine (Spm) in the form of hydrochlorides. The results are the means of three independent biological samples expressed in nmol g−1 fresh weight−1.
2.5. RNA Purification and Gene Expression Analyses with Quantitative Real-Time PCR
Total RNA was extracted from 100 mg of tomato root using GeneJET Plant RNA Purification Kit (Thermo Scientific™, K0801 from Thermo Fisher Scientific, Waltham, MA, USA) as recommended by the manufacturer. The isolated RNA was DNase-treated with a TURBO DNA-free™ Kit (Invitrogen by Thermo Fisher Scientific), and first-strand cDNA synthesis of 1 µg of total RNA was carried out with a High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems by Thermo Fisher Scientific), using random hexamers. Real-time PCR was carried out with an ABI 7900 Fast Real-Time System (Applied Biosystems by Thermo Fisher Scientific) with the following protocol: 40 cycles at 95 °C for 15 s, 60 °C for 1 min, using Maxima SYBR Green qPCR Master Mix (Thermo Fisher Scientific). The relative expression levels were normalized to both the SlEF1 and SlUBI3 reference genes. The normalized relative transcript levels were calculated according to [48,49] using the 2−ΔΔCt method, where the relative gene expression level of the untreated control was 1. The specific primers for each examined gene are described in Table S1 and related references are cited in the Supplementary Materials.
2.6. Hydrogen Peroxide Determination
The H2O2 levels of the tomato tissues were measured in six-week-old plants using an Amplex Red Hydrogen Peroxide/Peroxidase Assay Kit (Thermo Fisher Scientific, A22188) as recommended by the manufacturer. Approximately 100 mg of fresh plant material was harvested, ground in liquid N2, and diluted in 20 mM potassium phosphate buffer (pH 6.5). The homogenates were centrifuged, and the supernatant was used to measure the H2O2 content. The accumulation of resorufin was determined spectrophotometrically at 560 nm (Thermo Scientific, Multiscan Go Microplate Spectrophotometer). The amount of H2O2 was calculated using a standard curve.
2.7. Superoxide Dismutase Enzyme Activity Measurement
Enzyme extracts were prepared as described by [50]. SOD (EC 1.15.1.1) activity measurement was based on the ability of the enzyme to inhibit the photochemical reduction in p-nitro-blue tetrazolium chloride (Sigma-Aldrich, St. Louis, MO, USA) in the presence of riboflavin in the light. One enzyme unit (U) of SOD represents the amount of enzyme causing a 50% inhibition of p-nitro-blue tetrazolium chloride reduction, and its activity was calculated as U g–1 fresh weight.
2.8. Histochemical In Situ Detection of Reactive N, O, and S Species
NO production was visualized as described by [51]. NO was visualized in tomato root tips using 4-amino-5-methylamino-2′,7′-difluorofluorescein diacetate (DAF-FM DA) dye. The samples were incubated for 30 min at room temperature in the dark in 10 μM DAF-FM DA dissolved in 10 mM TRIS-HCl buffer (pH 7.4). Superoxide anion (O2•− levels were investigated using 10 µM fluorescent DHE (dihydroethidium) [52]. H2S determination was visualized by WSP-1 (Washington State Probe-1). Root tips were stained for 40 min in WSP-1 solution, washed three times and examined by a microscope as described by [53]. After staining, the samples were rinsed twice with 10 mM TRIS-HCl buffer (pH 7.4). Fluorescence intensity (pixel intensity) was detected with a Zeiss Axiowert 200M-type fluorescence microscope (Carl Zeiss Inc., Jena, Germany), as described earlier [53]. Filter set 10 (exc.: 450–490, em.:515–565 nm) was used for DAF-FM and WSP-1, and filter set 9 (exc.: 450–490, em.: 515–∞ nm) for DHE [54].
2.9. Determination of SNOs and Nitrite Content
Approximately 100 mg of plant material was harvested immediately after the treatments. After freezing in liquid N2, the samples were homogenized twice for 10 s using a Silamat S6 tissue homogenizer (Ivoclar Vivadent, Schaan, Liechtenstein) and 1.7–2.0 mm glass beads (Roth). The homogenized root material was extracted in 500 µL 1× phosphate-buffered saline (PBS) and incubated on ice for 10 min, followed by centrifugation for 10 min at 12,435 rpm. The protein content of the plant extract was determined using the Protein Assay Dye Reagent Concentrate (Bio-Rad, Hercules, CA, USA). The quantification of nitrite and SNOs in tomato root tissues was performed using Sievers’ Nitric Oxide Analyzer NOA 280i (GE Water & Process Technologies, Ratingen, Germany) as described by [41]. Endogenous nitrite was reduced to gaseous NO by the injection of root extracts into a reaction vessel containing triiodide solution (28.5 mM I2, 66.9 mM KI in 77.7% acetic acid) at 30 °C. For SNO detection, endogenous nitrite was scavenged by adding 5% sulfanilamide (w/v, in 1 M HCl) at a dilution of 1:9 to the sample before injection into the triiodide solution.
2.10. GC-MS Analysis of TCA Cycle Metabolites
After sample collection, 0.1 g portions of the samples were ground in liquid N2 and transferred into 2.0 mL safety Eppendorf tubes, containing 30 µL ribitol (1 mg/mL) as an internal standard (ISTD), in 0.5 mL of 60% (v/v) methanol. The tubes were vortex-mixed for 30 s, then placed in an ultrasound bath for 5 min, vortex-mixed again for 15 s, and centrifuged for 5 min at 10,000 rpm, and the supernatant was collected. The extraction was repeated with 0.5 mL of 60% (v/v) methanol and again with 0.5 mL of 90% (v/v) methanol. The supernatant was collected and mixed well. An aliquot (100 µL) was dried in vacuum. For derivatization, methoxyamine hydrochloride dissolved in pyridine (20 mg/mL) was added, incubated at 37 °C for 90 min., then N-trimethylsilyl-N-methyl trifluoroacetamide was added and incubated for 30 min at the same temperature.
The samples were transferred to vials and injected split-mode into the Shimadzu GCMS-TQ equipped with GC column (Phase: HP-5MS length 30 m; ID 0.25 mm; Film thickness: 0.25 µm). A total of 1 µL was injected into the column at 230 °C, and the transfer line and ion source were 250 °C. The carrier gas was He, and a constant flow rate (1 mL/min) was used. The thermal program was 70 °C for 1 min, which increased to 320 °C at a rate of 7 °C/min, and the high temperature was maintained for 5 min. The Kovats retention index was used to identify the standards, and GCMSSolution 4.16 for Shimadzu was used for data processing. Both the GC analyses and data processing were carried out with GCMSSolution 4.16 for Shimadzu using the Wiley and Nist databases.
2.11. Statistical Analysis
The experiments were carried out in three independent biological repetitions. The data are given as the mean ± standard deviation (SD), calculated from the combination of biological repetitions. Two-way analysis of variance (ANOVA) was carried out using GraphPad Prism version 8.0.1.244 for Windows (GraphPad Software, La Jolla, CA, USA), with significance level 0.05 (p ≤ 0.05) (interaction analysis can be seen in Table S3). Different letters on the bars denote significant differences (p = 0.05) based on Tukey’s post hoc test for multiple comparisons.
3. Results
3.1. AG Has a Nonspecific Inhibitory Effect on NaCl-Induced Concentration-Dependent Alterations in DAO and PAO Activities of Tomato Roots
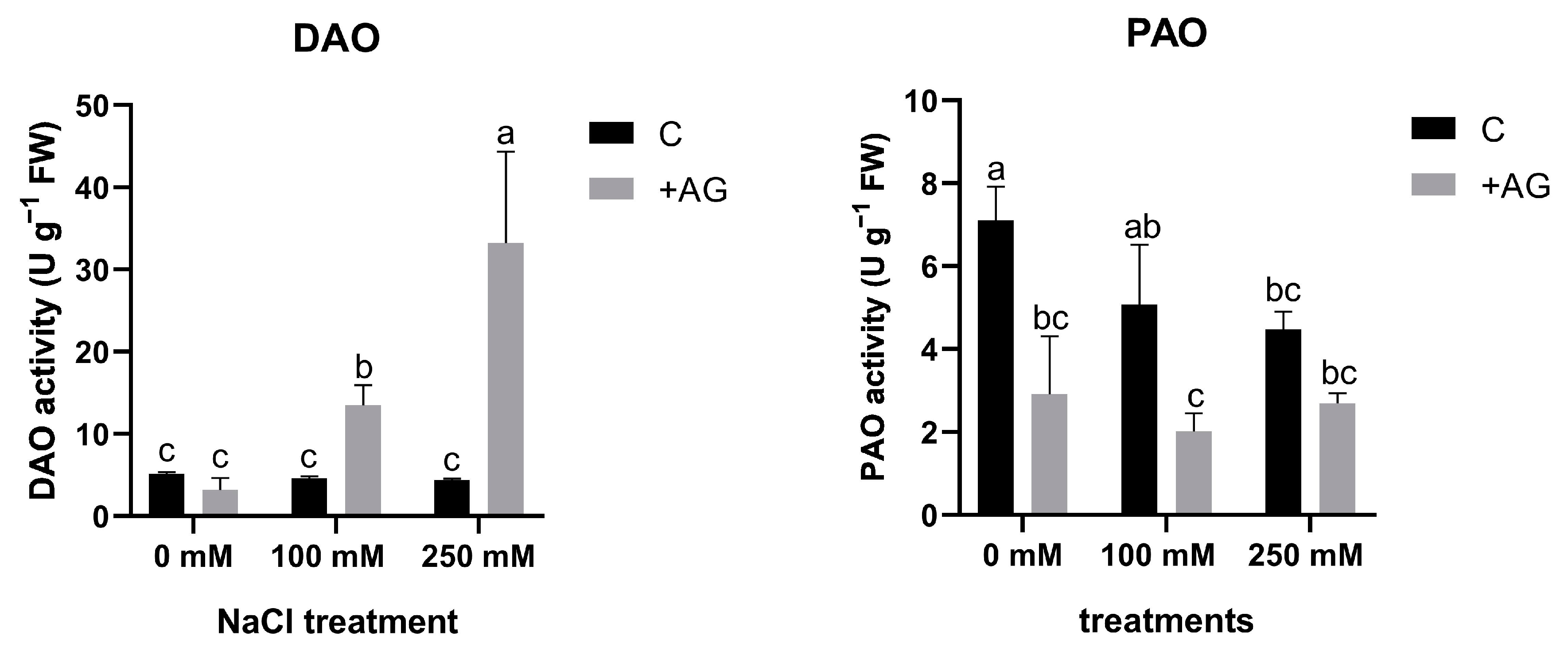
In order to investigate the capacity of AG as a DAO inhibitor, 1 mM AG was added to the nutrient solution, and tomato roots were treated for 1 h. NaCl treatment alone did not affect DAO activity, but a significant reduction in PAO activity was detected by salt stress, which was concentration-dependent (Figure 1). In the presence of AG, DAO activities in tomato roots were enhanced by increasing the concentration of salt (Figure 1).
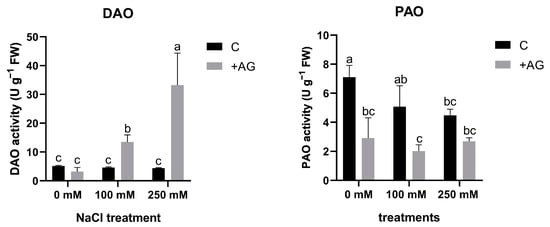
Figure 1.
DAO and PAO activities in AG-treated tomato roots with and without salt stress (100 and 250 mM NaCl). Data of enzyme activities are the mean ± SD of three biological replicates. Different letters denote significant differences, two-way ANOVA, following Tukey’s post hoc test p = 0.05.
3.2. Free PA Levels Are Reduced by AG Treatment Independent of NaCl Concentration
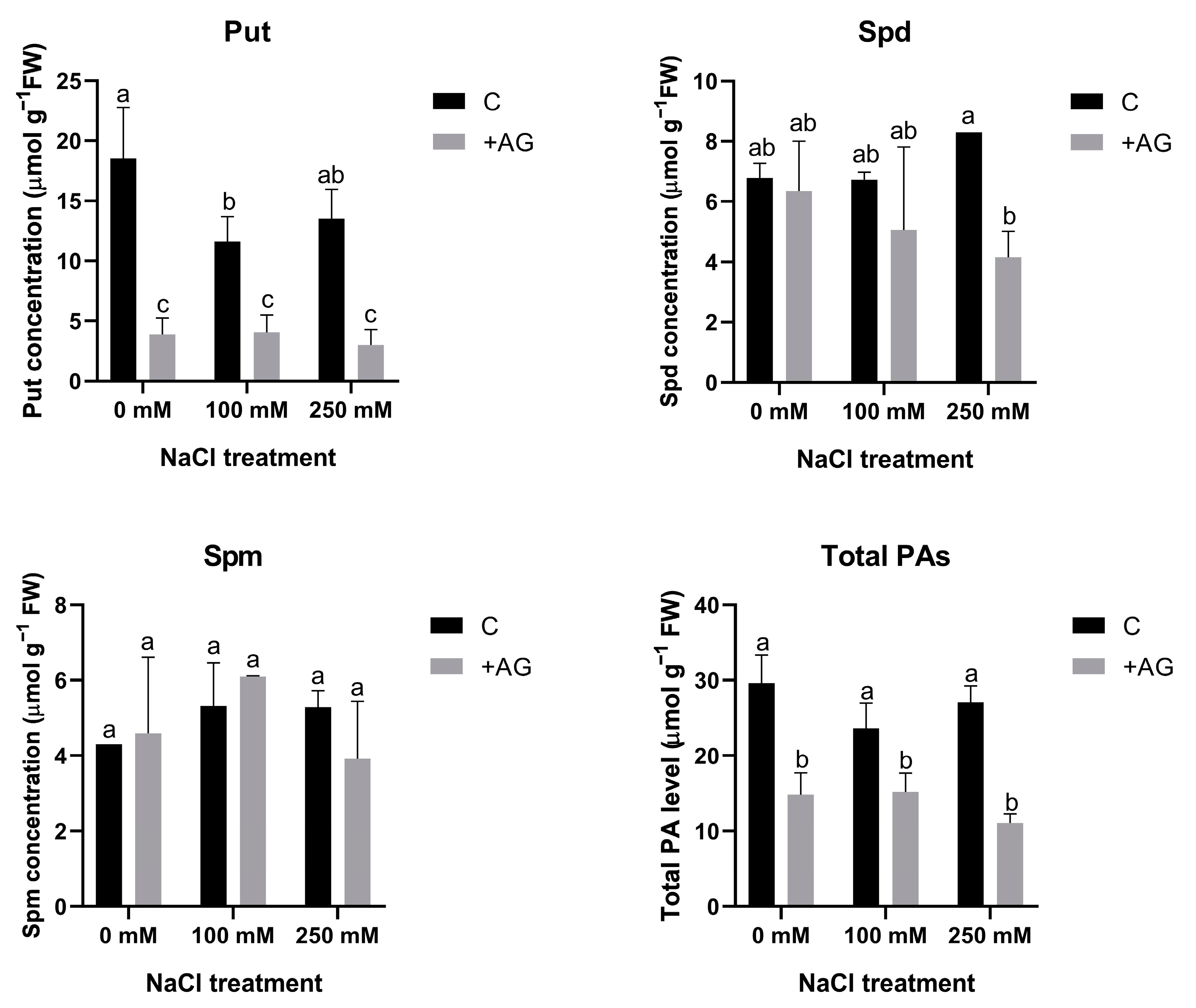
In order to determine the effect of AG on PA metabolism, free PA concentrations were determined in salt- and AG-treated tomato roots. Total PA contents were lower in AG-treated roots, which were not influenced by salt stress. Put levels were considerably reduced by AG with or without salt treatment. Spd concentration was reduced by AG only in the presence of 250 mM NaCl, whereas Spm contents were not influenced by either AG or salt (Figure 2).
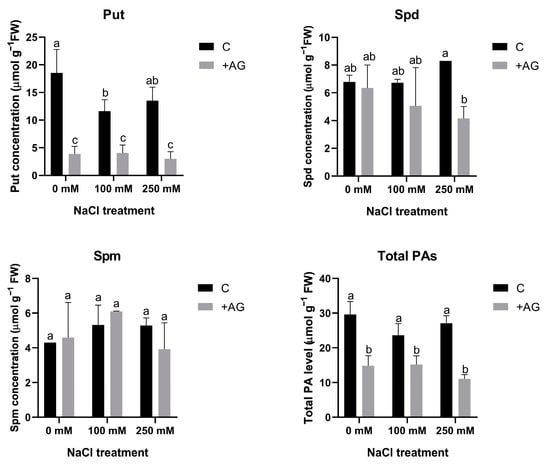
Figure 2.
Free polyamine contents of control and AG-treated tomato roots with and without NaCl treatment (100 and 250 mM). Data of PA contents are the mean ± SD of three biological replicates. Different letters denote significant differences, two-way ANOVA, following Tukey’s post hoc test p = 0.05.
3.3. AG Induces Expression of PA Biosynthetic and Catabolic Genes
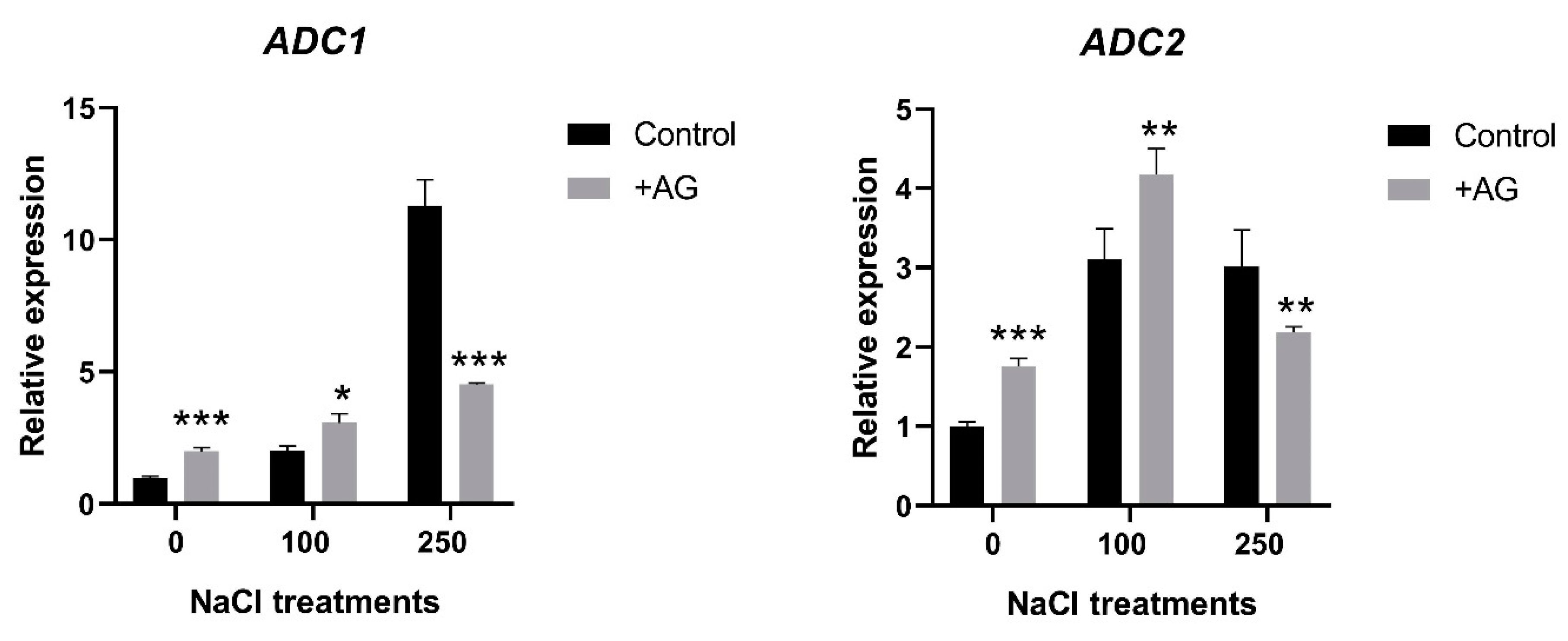
In order to study the effect of AG on the activity of genes implicated in the biosynthesis of PAs, transcript levels of ADC1 and ADC2 genes encoding arginine decarboxylase, and ODC1 coding for ornithine decarboxylase were determined. Both ADC1 and ADC2 were induced by salt, to different degrees. ADC1 transcript levels were 2 and 12 times higher in 100 mM and 250 mM NaCl-treated roots, respectively. AG treatment slightly enhanced ADC1 expression in control roots and in the presence of 100 mM NaCl (two and three times, respectively), whereas it reduced ADC1 expression by 60% compared to its 250 mM NaCl-treated control when stronger salt stress was applied. ADC2 expression was increased three-fold in salt-treated roots. AG slightly enhanced ADC2 transcription in control and 100 mM NaCl-treated roots (1.75- and 4.1-fold, respectively), whereas expression of this gene was slightly reduced only by 28% with AG at stronger salt stress (Figure 3). ODC1 expression was 1.5-fold increased by AG treatment in roots treated with no or 100 mM NaCl, but it was reduced by 250 mM NaCl with or without AG (Figure S1).
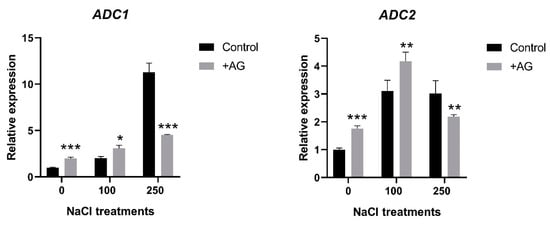
Figure 3.
Expression of ADC genes of salt- and AG-treated tomato roots. Relative expression levels are shown where 1 represents the untreated control. Data of each bar are the mean ± SD of three biological replicates. Asterisks denote significant differences from untreated control, Student t test, * p < 0.05, ** p < 0.01, *** p < 0.001.
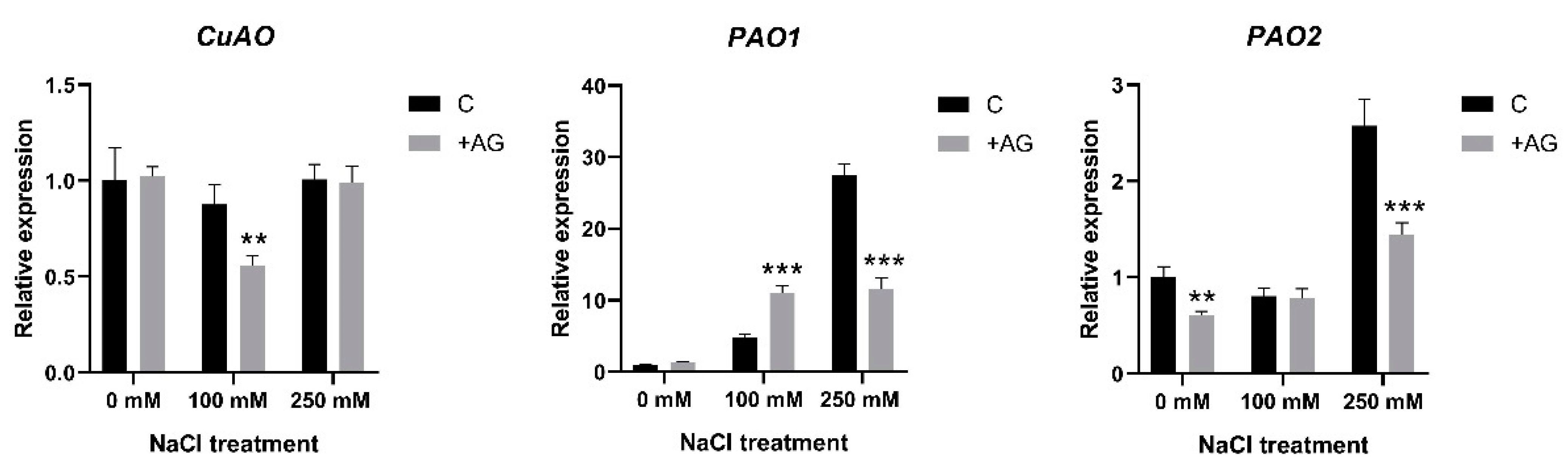
To test whether the altered enzyme activities were derived from the changes in gene expression, we determined the transcript levels of the PA catabolism genes. CuAO expression was slightly reduced, only by 30% after AG treatment and only in 100 mM NaCl-treated roots (Figure 4). Considerable variation was observed when the expression of PAO genes [21] was tested. Transcript levels of PAO1 were enhanced by salt in a concentration-dependent manner, displaying 30-fold induction in 250 mM NaCl-treated roots. AG enhanced PAO1 expression at moderate salt stress but reduced it when high salt stress was applied. Expression of PAO2 was not affected either by 100 mM NaCl or by AG at this salt concentration, while 250 mM NaCl induced PAO2 activity 2.5-fold, which was reduced in the presence of AG. Transcript levels of PAO4 were reduced to half by salt treatments but were not affected by AG (Figure 4 and Figure S2). Collectively, the PAO genes showed higher expression in the 250 mM NaCl-treated plants, which was reduced by AG, except for PAO4 and PAO5.
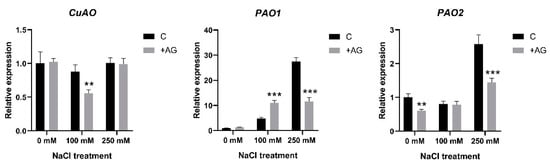
Figure 4.
Expression of CuAO and PAO genes of control and AG-treated tomato roots with and without NaCl treatment. Relative expression levels are shown as in Figure 3. Data of each bar are the mean ± SD of three biological replicates. Asterisks denote significant differences from untreated control, Student t test, ** p < 0.01, *** p < 0.001.
3.4. ROS Levels Are Reduced by AG
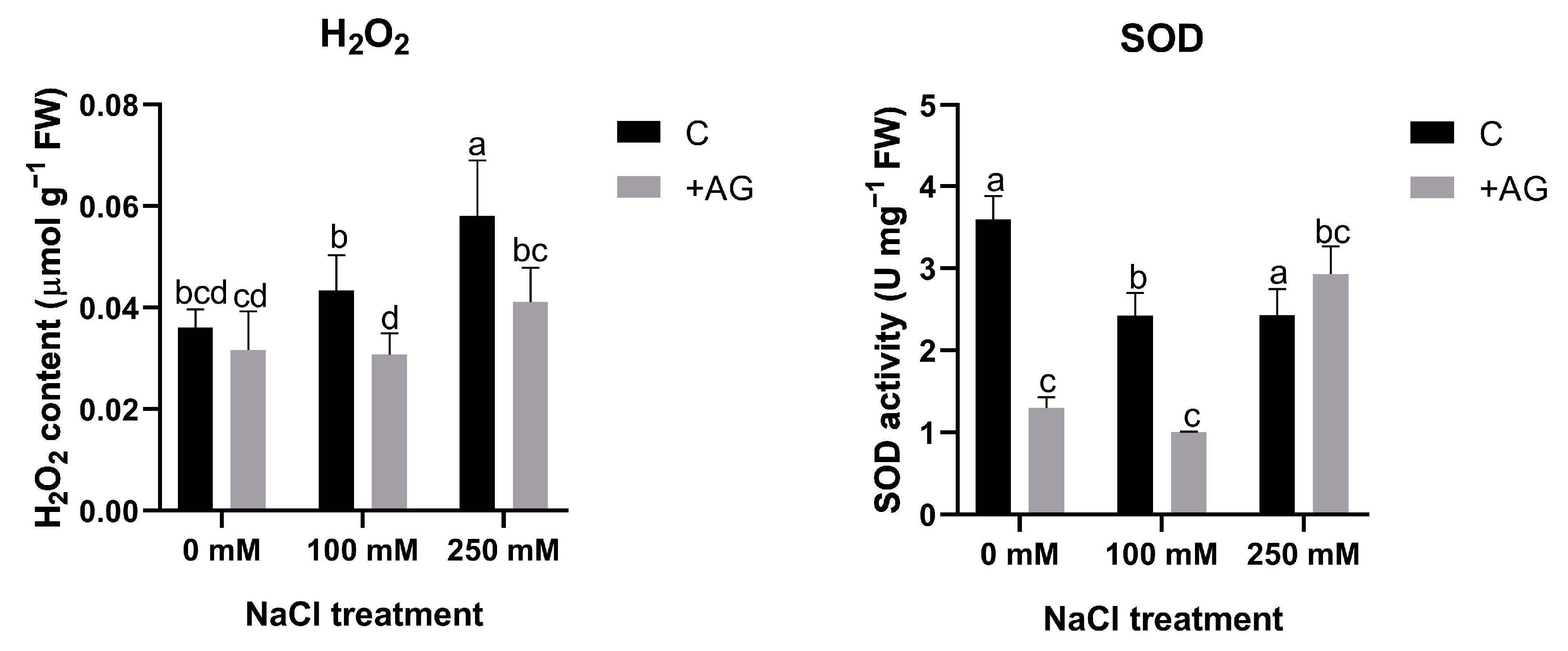
To test whether H2O2 could be altered by reduced PA catabolism, H2O2 levels were determined in AG- and salt-treated roots. Salt stress enhanced H2O2 content in a concentration-dependent manner, which was blocked by AG treatment. To further investigate peroxide-derived H2O2 generation, we subsequently tested the influence of salt and AG on SOD activity. Salt stress slightly reduced SOD, which was inhibited by AG in the absence of salt or when roots were treated by 100 mM NaCl. At higher salt stress, AG had no influence on SOD (Figure 5). These results suggested that SOD activity can be affected by AG in tomato roots, but its influence on H2O2 content is negligible.
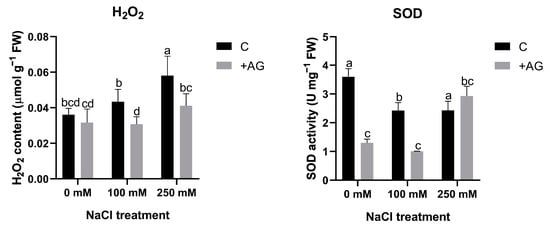
Figure 5.
H2O2 contents and SOD activities of control and AG-treated tomato roots with and without NaCl treatment. Data of each bar are the mean ± SD of three biological replicates. Different letters denote significant differences (two-way ANOVA, Tukey’s post hoc test p = 0.05).
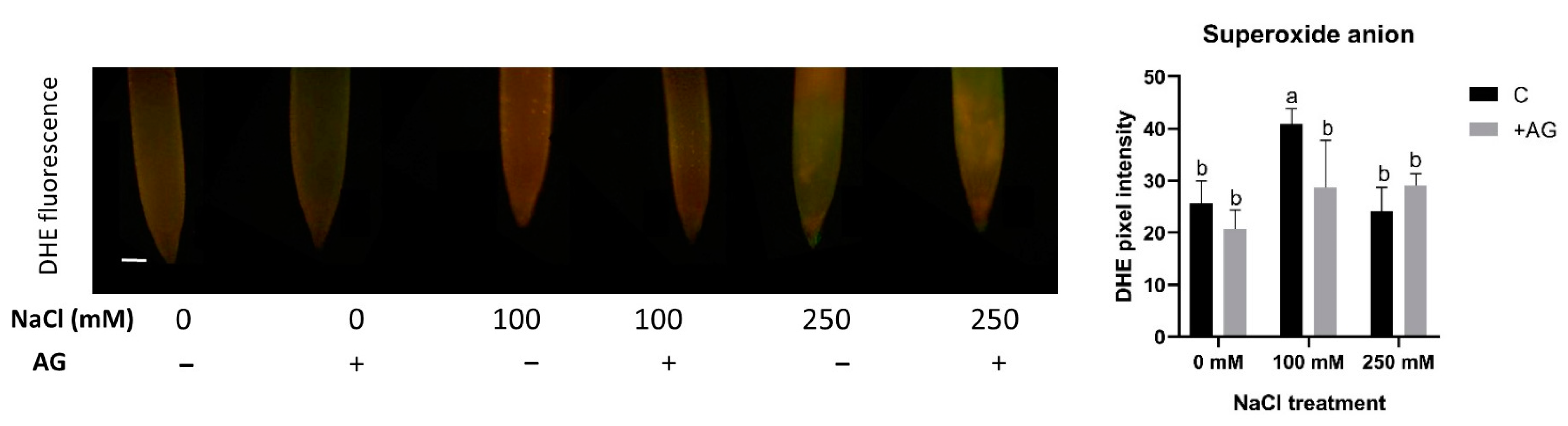
To test the influence of AG on superoxide production, the accumulation of superoxide anions was tested using in situ DHE assay. Superoxide levels were enhanced by 100 mM but not by 250 mM NaCl. AG had either no or a slightly negative effect on superoxide levels in the root tips (Figure 6).
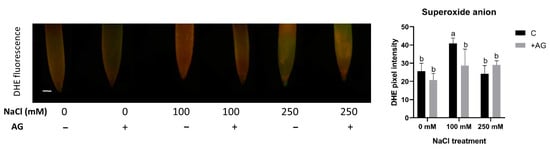
Figure 6.
Detection of superoxide anions in control and AG-treated tomato root apices with and without NaCl treatment (100 and 250 mM). Superoxide anion content was measured using a fluorescence probe DHE (dihydroethidium) (scale bar: 100 µm) and expressed as pixel intensity. DHE fluorescence was measured at 500 µm away from the root tips and averaged. Significant reduction occurred in the 100 mM treatment after AG treatment. Bars indicate ±SE of measurements performed with at least 10 seedlings per each treatment, different letters denote significant differences (two-way ANOVA, Tukey’s post hoc test p = 0.05).
3.5. SNO Increase and Nitrite Reduction Could Be Responsible for Absence of Detectable NO
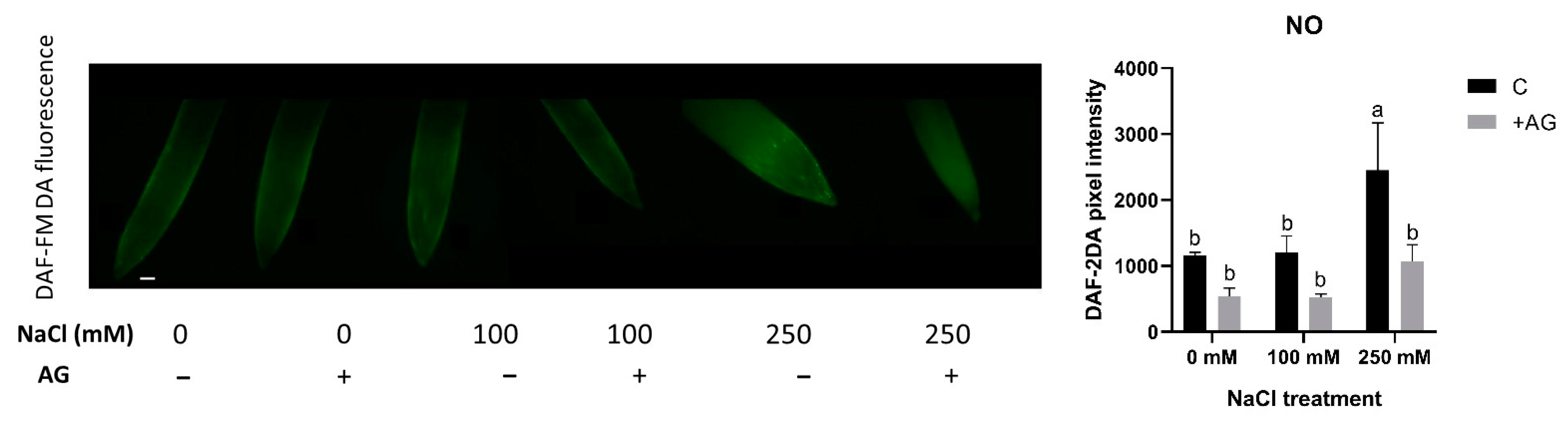
PAs are known to induce NO production in plants. We therefore determined NO levels in salt- and AG-treated tomato root tips using in situ DAF-FM DA fluorescent reaction. While 100 mM NaCl had no effect, 250 mM NaCl treatment led to significantly higher NO accumulation. AG treatment reduced the NO levels only in 250 mM NaCl-treated samples (Figure 7).
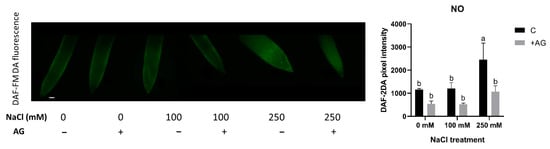
Figure 7.
Detection of NO in control and AG-treated tomato root apices with and without NaCl treatment. NO content was detected using a fluorescence probe DAF-FM-DA (scale bar: 100 µm). Quantitative data are expressed as pixel intensity. DAF-FM fluorescence was measured at 500 µm away from the root tips and averaged. Note that AG reduced the NO contents of root tips in all cases. Bars indicate ±SE of measurements performed with at least 10 seedlings per each treatment, different letters denote significant differences (two-way ANOVA, Tukey’s post hoc test p = 0.05).
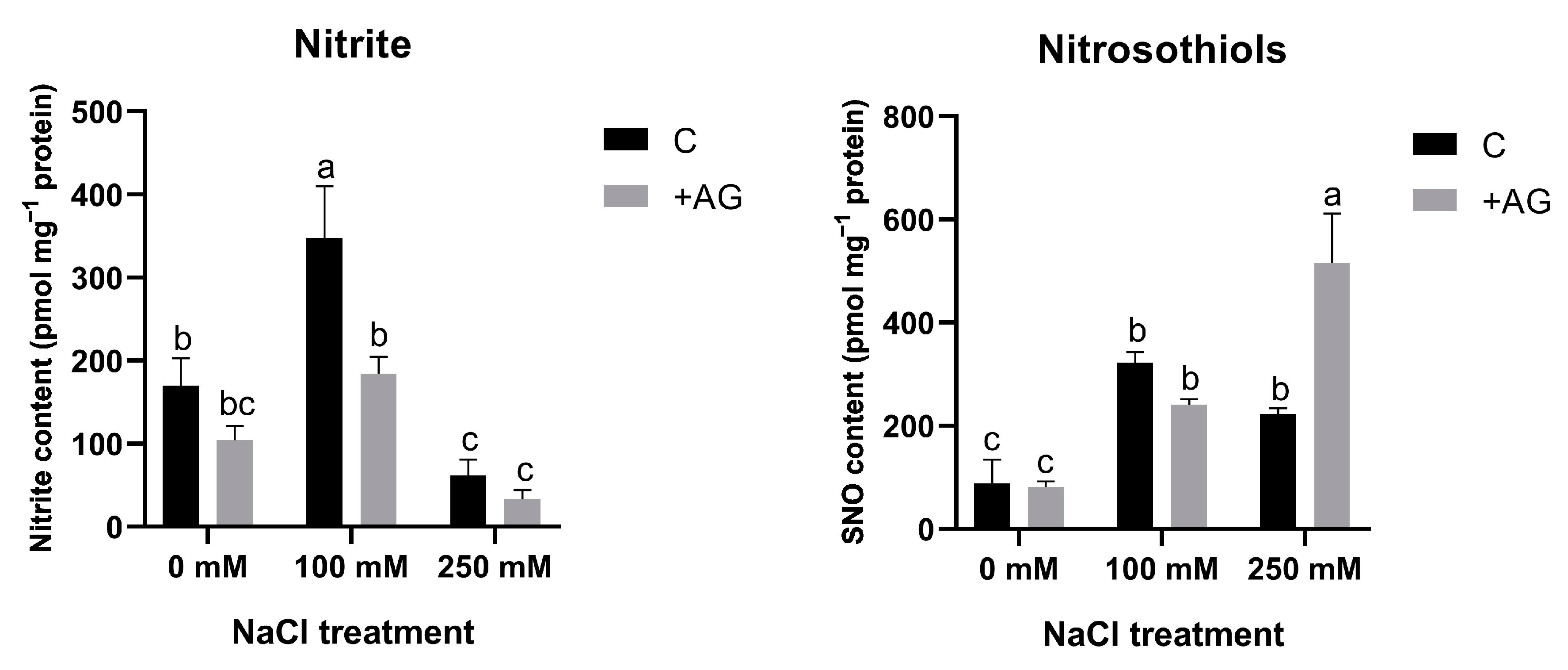
NO can be generated from nitrite and nitrosothiols (SNO). In order to compare NO levels with such precursors, nitrite and SNO levels were determined in salt- and AG-treated tomato roots. Nitrite content was higher in roots treated by 100 mM NaCl but was reduced by high salt. Nitrite concentrations were reduced by AG in all samples. SNO levels were enhanced by the NaCl treatments. SNO contents were similar in AG-treated roots in the absence of salt or in the presence of 100 mM NaCl, whereas AG enhanced SNO accumulation in plants stressed by a high concentration of salt (Figure 8).
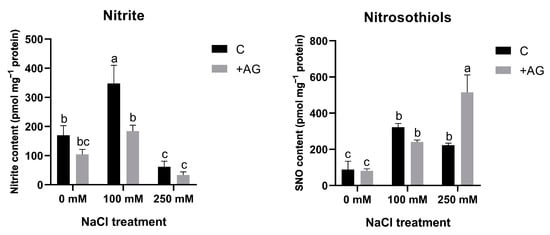
Figure 8.
Nitrite and S-nitrosothiol contents of control and AG-treated tomato roots with and without NaCl treatment. Data of each bar are the mean ± SD of three biological replicates. Different letters denote significant differences (two-way ANOVA, Tukey’s post hoc test p = 0.05).
3.6. H2S Levels Displayed Different Changes during Salt Stress
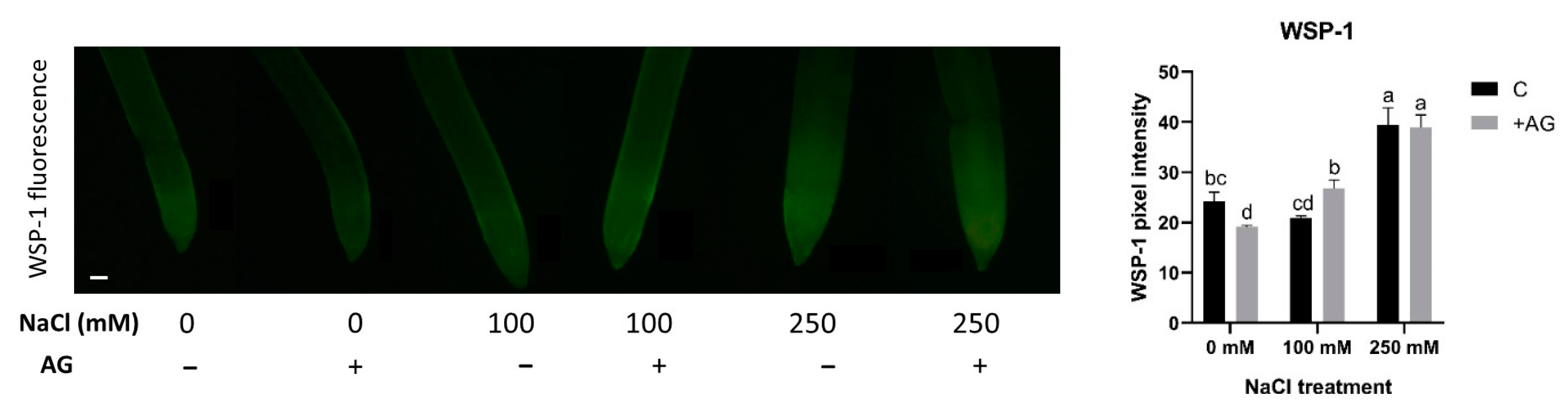
Recent studies have suggested that H2S could interact with NO and H2O2 metabolic pathways [55]. Therefore, H2S concentrations were measured in salt- and AG-treated root tips, using a specific H2S staining method, WSP-1. H2S content was not affected by 100 mM NaCl but was increased nearly two-fold by 250 mM NaCl. AG treatment had no or only a slight influence on H2S concentrations (Figure 9).
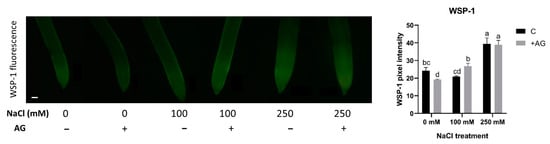
Figure 9.
Detection of hydrogen sulfide in control and AG-treated tomato root apices with and without NaCl treatment (100 and 250 mM). Hydrogen sulfide content was measured using a fluorescence probe WSP-1 (Washington State Probe-1) (scale bar: 100 µm) and expressed as pixel intensity. WSP-1 fluorescence was measured at 500 µm away from the root tips and averaged. Bars indicate ±SE of measurements performed with at least 10 seedlings per each treatment, different letters denote significant differences (two-way ANOVA, following Tukey’s post hoc test p = 0.05).
3.7. GABA and the TCA Cycle as a Possible Explanation of Decreased PA Levels
Put degradation could result in enhanced GABA production in plants [56]. As reduced PA levels could occur during AG application, we determined the concentrations of GABA and several TCA cycle metabolites in salt- and AG-treated roots (Figure S3). GABA contents slightly increased after 1 h of salt stress, but the difference was not significant. AG reduced the GABA levels in control roots but had no effect on salt-treated roots.
To test possible changes in TCA cycle metabolites, concentrations of citrate, succinate, fumarate, and malate were measured in salt- and AG-treated tomato roots. Moderate salt stress (100 mM NaCl) had only a slight effect on these metabolites, with the exception of fumaric acid, whose concentration was elevated two-fold. Higher salt stress reduced concentrations of succinic, fumaric, and citric acids. AG treatment alone led to reduced amounts of all TCA metabolites tested. In combination with salt stress, AG treatment had no or only a minor influence on the concentrations of these metabolites (Figure S3 and Table S2).
4. Discussion
The function of PA catabolism and the enzymes implicated in biosynthesis and catabolism are often studied in relation to specific inhibitors. AG is one of the exogenously applied inhibitor compounds, which are known to block the activity of copper amine oxidase or diamine oxidase. This inhibitor has been demonstrated to decrease DAO enzyme activity, but the effects were found to be different depending on the tested plant species, organ, or the applied dose [36,37]. Short-term salt stress altered PA catabolic responses, similarly to results obtained with tobacco or cucumber, demonstrating a feed-forward ROS amplification loop between ROS-producing NADPH oxidase and the apoplastic PAO [57]. In this study, we tested the specificity of AG when used as a DAO inhibitor in salt-stressed tomato plants. Interestingly, in our experimental conditions, AG was unspecific for DAO and suppressed PAO activity as well (Figure 1). Similar results were reported by Kabala et al. [57]: a 1-h salt treatment did not significantly affect DAO enzyme activity in cucumber, and lower H2O2 contents were maintained. Although inhibition of DAO enzyme activity was expected to increase PA concentrations, especially Put levels, in our study, reduction in PAO activity by AG treatment correlated with reduced PA levels under both saline and non-saline conditions (Figure 2 and Table S3). To decipher the possible mechanisms of this reduction, the expression of genes which encode PA biosynthesis enzymes such as ADCs and ODC1 and genes which encode DAO and PAO enzymes was studied. We found that salt stress induced ADC expression, especially that of ADC2. Interestingly, ODC1 expression was reduced by 250 mM NaCl and further decreased after AG treatment (Figure S1). Additionally, 100 mM NaCl increased ADC and ODC expression, which was further enhanced by AG treatment, suggesting that PA levels were increased in these plants (Figure 3). ADC and ODC could compensate for each other’s activity, as revealed by [58]. Meanwhile, 250 mM NaCl was shown to reduce PA content. The expression of genes implicated in PA catabolism in response to salt stress [21] was also tested. PAO1 of tomato is similar to the Arabidopsis AtPAO1, which belongs to Clade 1, and is localized in the cytoplasm. PAO2, PAO4, and PAO5 are in Clade 4, showing peroxisome localization. PAO2 was reported to contribute to ABA-mediated plant developmental processes in Arabidopsis [59]. Transcript levels of PAO4 were reduced to half in salt-treated plants, whereas PAO5 expression was reduced by 100 mM NaCl but increased in higher salt-stress conditions (Figure S2). PAO5 expression was previously shown to be unchanged by salt stress, showing a role in xylem differentiation [60]. To explain the differences between these genes, further studies are needed.
Salinity strongly affects ROS signaling in plants [61] by involving the catabolic processes of PAs. PA catabolism could induce H2O2 production in the roots of tomato plants exposed to salt stress [35]. AG did not change H2O2 levels compared to the control conditions, providing evidence that AG alone did not induce any additional stress in these plants (Figure 5). AG could alleviate salt-induced H2O2 accumulation, suggesting that AG might have antioxidant features similar to those reported in soybean [34,36]. To decipher the contribution of other potential biosynthetic processes to H2O2 production, SOD enzyme activity was measured in salt- and AG-treated roots. SOD activity was reduced by AG in control and 100 mM NaCl-treated roots, but not in the 250 mM NaCl-treated ones. To examine the possible contribution of stress-dependent superoxide production to H2O2 accumulation, superoxide levels were also measured. AG could reduce superoxide levels in roots subjected to mild salt stress, but it had no significant effect on superoxide production in control or 250 mM NaCl-treated roots (Figure 6). These results are consistent with [57], where short-term salt stress induced slight changes in H2O2 and SOD in cucumber. It may arise that other antioxidant defense mechanisms could play a role in alleviating oxidative stress during short-term conditions.
AG was suggested as functioning as an irreversible NOS (iNOS) inhibitor widely used for reducing NO production in plants [62]. PAs could be produced from L-arginine, affecting oxidative NO production in plants [63]. In our experimental system, AG reduced NO levels independently of salt stress (Figure 7). A similar effect of AG has been reported in [34], which agrees with other studies. Treatment with 250 mM NaCl could provoke nitrosative stress by inducing reductive or oxidative pathways for NO production, potentially generating reactive nitrogen species for tomatoes, which could be alleviated by AG. To confirm the source of these increased NO levels, we investigated the reductive pathway of NO production. Nitrite could be a source of NO [41,64] in plants. PAs were shown to modulate NR activity in wheat leaves, affecting NO production [65]. Moderate salt stress could enhance nitrite levels, whereas high salinity reduced them, and they were further reduced by AG. SNO is like a stored form of NO. Salinity induced SNO accumulation, which was further increased in 250 mM NaCl-treated tomato roots (Figure 8). There is some evidence for the involvement of SNOs and S-nitrosylation in salinity stress, but these need to be further investigated [66].
Recent results support the role of H2S as a signal molecule in salt-stress responses in plants [14,67,68]. Its connection to other signal molecules, such as NO or H2O2, is almost unknown [69,70]. An early study suggested that NO and H2S could interact with each other and regulate each other’s level; however, some other evidence indicates an inhibiting effect of H2S on NO production [70]. Additionally, it is known that H2S treatment could induce an increase in the expression of genes involved in PA biosynthesis during salt stress [70]. Our results revealed that 250 mM NaCl was able to increase the H2S levels, but AG had only a minor influence on H2S accumulation in tomato roots (Figure 9). These results confirm that H2S production depends on the stress intensity in short-term salt treatment.
To provide further evidence about the decreased PA levels, we also checked the other degradation routes from Put to GABA, which could connect with the TCA cycle as an alternative GABA shunt, contributing to the N balance in plants [71]. Our experiments, however, did not demonstrate any GABA increase or other shift in TCA cycle metabolites by metabolomic analysis (Figure S3).
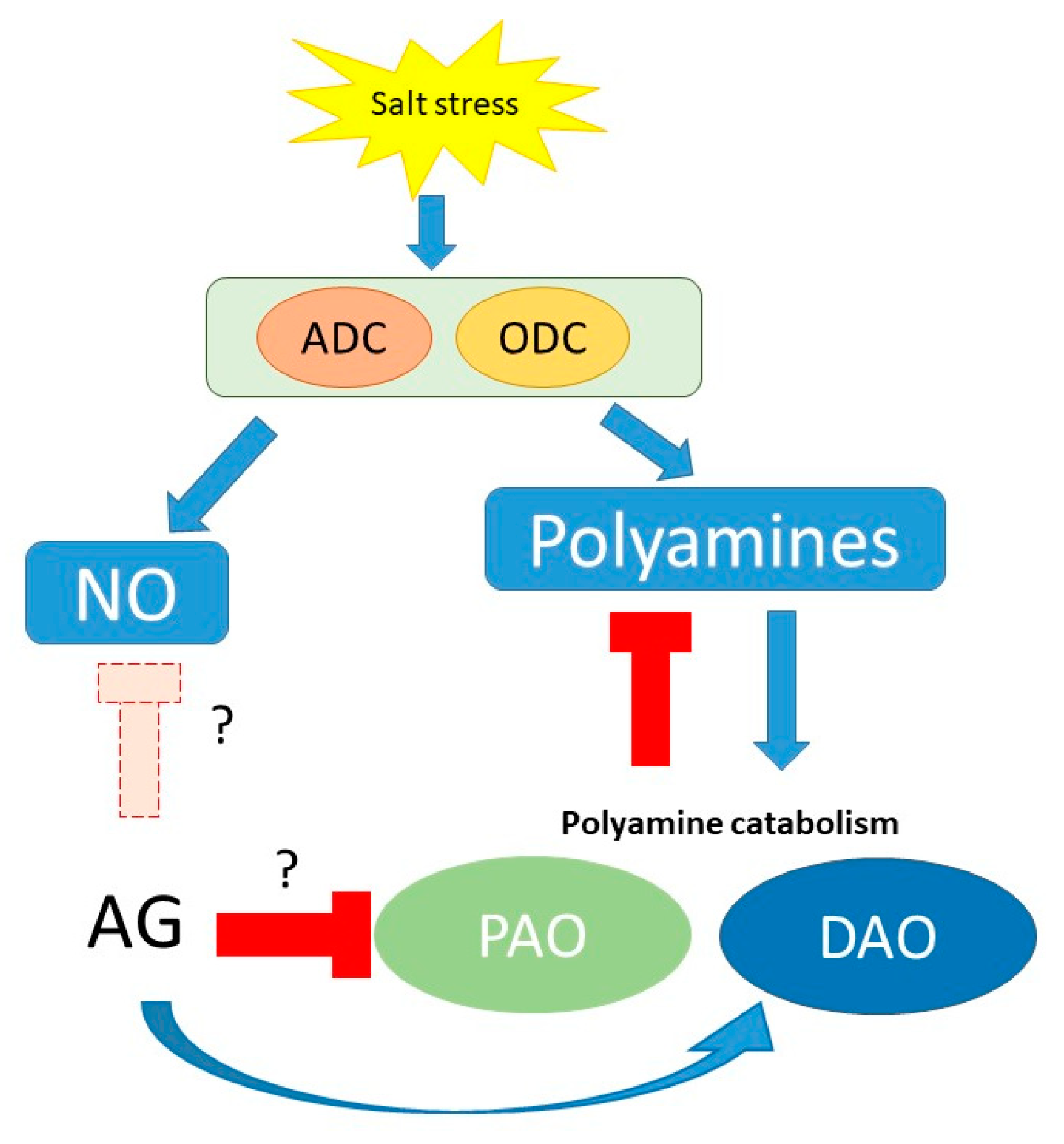
As a conclusion, our results confirm AG as a nonspecific inhibitor of PA catabolism in short-term conditions, with implications in ROS and NO signaling (summarized in Figure 10 and Figure S4).
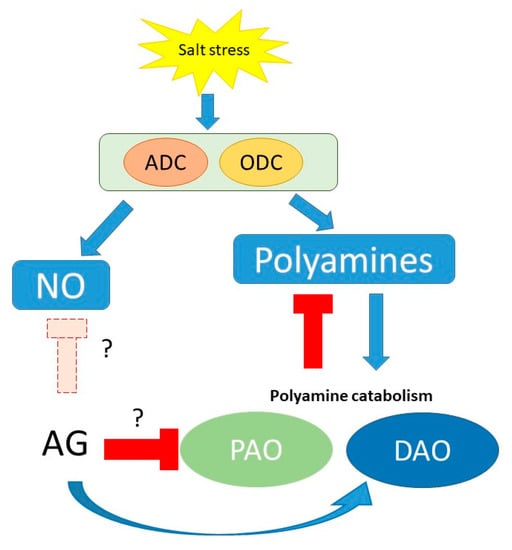
Figure 10.
Schematic presentation of potential mechanisms of the effect of AG on PA metabolism and related pathways during short-term salt-stress response to different NaCl concentrations (100 and 250 mM). Question marks indicate the questionable functions of AG during stress, as it can act as an inhibitor of NO synthesis or could induce the expression of PAO genes; however, these functions need to be studied.
Understanding the role of PA catabolism during the early responses of plants to salinity stress could fill the gap in our knowledge about the connection between salt sensing and PA homeostasis, focusing on the interplay between other signal pathways [72]. Precise modes of action of PA catabolism and the related signal pathways need to be deciphered by further studies.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/antiox12081614/s1, Table S1: Primer list of used primers in this study; Figure S1: Expressions of ODC1 gene of control and AG-treated tomato roots with and without NaCl treatment (100 and 250 mM); Figure S2: Expressions of PAO4 and PAO5 genes of control and AG-treated tomato roots with and without NaCl treatment (100 and 250 mM); Figure S3: Heat map of some important TCA cycle metabolite contents of control and AG-treated tomato roots with and without NaCl treatment (100 and 250 mM); Table S2: Levels of some important TCA cycle metabolite contents of control and AG-treated tomato roots with and without NaCl treatment (100 and 250 mM); Table S3: P values for AG and NaCl treatments in case of different parameters; Figure S4: Representative summary of AG treatment and salt stress on tomato roots exposed to NaCl at 100 and 250 mM concentration. References [21,73,74,75,76] are cited in the supplementary materials.
Author Contributions
Conceptualization, Á.S. and L.Z.; methodology, Á.S.; software, L.B.; validation, T.J., G.S. and O.K.G.; investigation, Á.S., L.Z., P.P. (Péter Pálfi) and H.K.; microscopy, P.P. (Péter Poór) and R.S.; molecular studies, L.S. and L.Z.; statistical analysis, Á.S. and L.B.; writing—original draft preparation, Á.S. and L.Z.; writing—review and editing, A.F., L.S. and L.Z.; nitrosothiol measurement, C.L.; funding acquisition, Á.S. and L.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Research, Development and Innovation (NRDI) Office of the Hungarian Ministry, with grant number FK129061 for Á.S and grant number FK128920 for L.Z. Additionally, Á.S. gratefully acknowledges the European Union and the State of Hungary for co-financing through the European Social Fund in the framework of TÁMOP 4.2.4.A/2-11-1-2012-0001 ‘National Excellence Program’ and thanks the Institute of Balassi for the Campus Hungary Scholarships.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
The authors are thankful for the excellent help of Etelka Bécsné Kozma and Elke Mattens for their laboratory assistance.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Munns, R.; Gilliham, M. Salinity tolerance of crops—What is the cost? New Phytol. 2015, 208, 668–673. [Google Scholar] [CrossRef]
- Isayenkov, S.V.; Maathuis, F.J.M. Plant Salinity Stress: Many Unanswered Questions Remain. Front. Plant Sci. 2019, 10, 80. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Zhang, H.; Song, C.; Zhu, J.-K.; Shabala, S. Mechanisms of Plant Responses and Adaptation to Soil Salinity. Innovation 2020, 1, 100017. [Google Scholar] [CrossRef]
- Sequera-Mutiozabal, M.; Antoniou, C.; Tiburcio, A.F.; Alcázar, R.; Fotopoulos, V. Polyamines: Emerging Hubs Promoting Drought and Salt Stress Tolerance in Plants. Curr. Mol. Biol. Rep. 2017, 3, 28–36. [Google Scholar] [CrossRef]
- Szepesi, Á. Chapter 22 Molecular Mechanisms of Polyamines-Induced Abiotic Stress Tolerance in Plants. In Book Approaches for Enhancing Abiotic Stress Tolerance in Plants, 1st ed.; Hasanuzzaman, M., Nahar, K., Fujita, M., Oku, H., Islam, T., Eds.; CRC Press: Boca Raton, FL, USA, 2019; p. 18. [Google Scholar]
- Alcázar, R.; Marco, F.; Cuevas, J.C.; Patron, M.; Ferrando, A.; Carrasco, P.; Tiburcio, A.F.; Altabella, T. Involvement of polyamines in plant response to abiotic stress. Biotechnol. Lett. 2006, 28, 1867–1876. [Google Scholar] [CrossRef]
- Alcázar, R.; Bueno, M.; Tiburcio, A.F. Polyamines: Small Amines with Large Effects on Plant Abiotic Stress Tolerance. Cells 2020, 9, 2373. [Google Scholar] [CrossRef]
- Szepesi, Á.; Csiszár, J.; Gémes, K.; Horváth, E.; Horváth, F.; Simon, M.L.; Tari, I. Salicylic acid improves acclimation to salt stress by stimulating abscisic aldehyde oxidase activity and abscisic acid accumulation, and increases Na+ content in leaves without toxicity symptoms in Solanum lycopersicum L. J. Plant Physiol. 2009, 166, 914–925. [Google Scholar] [CrossRef] [PubMed]
- Minocha, R.; Majumdar, R.; Minocha, S.C. Polyamines and abiotic stress in plants: A complex relationship. Front. Plant Sci. 2014, 5, 175. [Google Scholar] [CrossRef]
- Liu, J.-H.; Wang, W.; Wu, H.; Gong, X.; Moriguchi, T. Polyamines function in stress tolerance: From synthesis to regulation. Front. Plant Sci. 2015, 6, 827. [Google Scholar] [CrossRef]
- Zapata, P.J.; Serrano, M.; García-Legaz, M.F.; Pretel, M.T.; Botella, M.A. Short Term Effect of Salt Shock on Ethylene and Polyamines Depends on Plant Salt Sensitivity. Front. Plant Sci. 2017, 8, 855. [Google Scholar] [CrossRef]
- Tanou, G.; Ziogas, V.; Belghazi, M.; Christou, A.; Filippou, P.; Job, D.; Fotopoulos, V.; Molassiotis, A. Polyamines reprogram oxidative and nitrosative status and the proteome of citrus plants exposed to salinity stress. Plant Cell Environ. 2014, 37, 864–885. [Google Scholar] [CrossRef] [PubMed]
- Pál, M.; Szalai, G.; Janda, T. Speculation: Polyamines are important in abiotic stress signaling. Plant Sci. 2015, 237, 16–23. [Google Scholar] [CrossRef]
- Corpas, F.J.; González-Gordo, S.; Muñoz-Vargas, M.A.; Rodríguez-Ruiz, M.; Palma, J.M. The Modus Operandi of Hydrogen Sulfide(H2S)-Dependent Protein Persulfidation in Higher Plants. Antioxidants 2021, 10, 1686. [Google Scholar] [CrossRef] [PubMed]
- Biondi, S.; Antognoni, F.; Marincich, L.; Lianza, M.; Tejos, R.; Ruiz, K.B. The polyamine “multiverse” and stress mitigation in crops: A case study with seed priming in quinoa. Sci. Hortic. 2022, 304, 111292. [Google Scholar] [CrossRef]
- Wang, W.; Paschalidis, K.; Feng, J.-C.; Song, J.; Liu, J.-H. Polyamine Catabolism in Plants: A Universal Process with Diverse Functions. Front. Plant Sci. 2019, 10, 561. [Google Scholar] [CrossRef]
- Planas-Portell, J.; Gallart, M.; Tiburcio, A.F.; Altabella, T. Copper-containing amine oxidases contribute to terminal polyamine oxidation in peroxisomes and apoplast of Arabidopsis thaliana. BMC Plant Biol. 2013, 13, 109. [Google Scholar] [CrossRef]
- Tavladoraki, P.; Cona, A.; Angelini, R. Copper-Containing Amine Oxidases and FAD-Dependent Polyamine Oxidases Are Key Players in Plant Tissue Differentiation and Organ Development. Front. Plant Sci. 2016, 7, 141–160. [Google Scholar] [CrossRef]
- Fraudentali, I.; Ghuge, S.A.; Carucci, A.; Tavladoraki, P.; Angelini, R.; Rodrigues-Pousada, R.A.; Cona, A. Developmental, hormone- and stress-modulated expression profiles of four members of the Arabidopsis copper-amine oxidase gene family. Plant Physiol. Biochem. 2020, 147, 141–160. [Google Scholar] [CrossRef]
- Fraudentali, I.; Rodrigues-Pousada, R.A.; Angelini, R.; Ghuge, S.A.; Cona, A. Plant Copper Amine Oxidases: Key Players in Hormone Signaling Leading to Stress-Induced Phenotypic Plasticity. Int. J. Mol. Sci. 2021, 22, 5136. [Google Scholar] [CrossRef]
- Hao, Y.; Huang, B.; Jia, D.; Mann, T.; Jiang, X.; Qiu, Y.; Niitsu, M.; Berberich, T.; Kusano, K.; Liu, T. Identification of seven polyamine oxidase genes in tomato (Solanum lycopersicum L.) and their expression profiles under physiological and various stress conditions. J. Plant Physiol. 2018, 228, 1–11. [Google Scholar]
- Sagor, G.H.M.; Zhang, S.; Kojima, S.; Simm, S.; Berberich, T.; Kusano, T. Reducing Cytoplasmic Polyamine Oxidase Activity in Arabidopsis Increases Salt and Drought Tolerance by Reducing Reactive Oxygen Species Production and Increasing Defense Gene Expression. Front. Plant Sci. 2016, 7, 214. [Google Scholar] [CrossRef]
- Gémes, K.; Mellidou, I.; Karamanoli, K.; Beris, D.; Park, K.Y.; Matsi, T.; Haralampidis, K.; Constantinidou, H.-I.; Roubelakis-Angelakis, K.A. Deregulation of apoplastic polyamine oxidase affects development and salt response of tobacco plants. J. Plant Physiol. 2017, 211, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Mittler, R.; Zandalinas, S.I.; Fichman, Y.; Van Breusegem, F. Reactive oxygen species signalling in plant stress responses. Nat. Rev. Mol. Cell Biol. 2022, 23, 663–679. [Google Scholar] [CrossRef] [PubMed]
- Saha, J.; Brauer, E.K.; Sengupta, A.; Popescu, S.C.; Gupta, K.; Gupta, B. Polyamines as redox homeostasis regulators during salt stress in plants. Front. Environ. Sci. 2015, 3, 21. [Google Scholar] [CrossRef]
- Moschou, P.N.; Paschalidis, K.A.; Delis, I.D.; Andriopoulou, A.H.; Lagiotis, G.D.; Yakoumakis, D.I.; Roubelakis-Angelakis, K.A. Spermidine Exodus and Oxidation in the Apoplast Induced by Abiotic Stress Is Responsible for H2O2 Signatures That Direct Tolerance Responses in Tobacco. Plant Cell 2008, 20, 1708–1724. [Google Scholar] [CrossRef]
- Wimalasekera, R.; Villar, C.; Begum, T.; Scherer, G.F. COPPER AMINE OXIDASE1 (CuAO1) of Arabidopsis thaliana Contributes to Abscisic Acid-and Polyamine-Induced Nitric Oxide Biosynthesis and Abscisic Acid Signal Transduction. Mol. Plant 2011, 4, 663–678. [Google Scholar] [CrossRef]
- Wimalasekera, R.; Tebartz, F.; Scherer, G.F. Polyamines, polyamine oxidases and nitric oxide in development, abiotic and biotic stresses. Plant Sci. 2011, 181, 593–603. [Google Scholar] [CrossRef]
- Groß, F.; Rudolf, E.-E.; Thiele, B.; Durner, J.; Astier, J. Copper amine oxidase 8 regulates arginine-dependent nitric oxide production in Arabidopsis thaliana. J. Exp. Bot. 2017, 68, 2149–2162. [Google Scholar] [CrossRef]
- Ni Ni Tun, N.; Santa-Catarina, C.; Begum, T.; Silveira, V.; Handro, W.; Floh, E.I.S.; Scherer, G.F.E. Polyamines Induce Rapid Biosynthesis of Nitric Oxide (NO) in Arabidopsis thaliana Seedlings. Plant Cell Physiol. 2006, 47, 346–354. [Google Scholar] [CrossRef]
- Yu, M.; Lamattina, L.; Spoel, S.H.; Loake, G.J. Nitric oxide function in plant biology: A redox cue in deconvolution. New Phytol. 2014, 202, 1142–1156. [Google Scholar] [CrossRef] [PubMed]
- Fancy, N.N.; Bahlmann, A.; Loake, G.J. Nitric oxide function in plant abiotic stress. Plant Cell Environ. 2017, 40, 462–472. [Google Scholar] [CrossRef] [PubMed]
- Romero-Puertas, M.C.; Sandalio, L.M. Nitric Oxide Level Is Self-Regulating and Also Regulates Its ROS Partners. Front. Plant Sci. 2016, 7, 316. [Google Scholar] [CrossRef] [PubMed]
- Köhler, Z.M.; Szepesi, Á. More Than a Diamine Oxidase Inhibitor: L-Aminoguanidine Modulates Polyamine-Related Abiotic Stress Responses of Plants. Life 2023, 13, 747. [Google Scholar] [CrossRef]
- Takács, Z.; Poór, P.; Szepesi, Á.; Tari, I. In vivo inhibition of polyamine oxidase by a spermine analogue, MDL-72527, in tomato exposed to sublethal and lethal salt stress. Funct. Plant Biol. 2017, 44, 480–492. [Google Scholar] [CrossRef]
- Yang, R.; Guo, Y.; Wang, S.; Gu, Z. Ca2+ and aminoguanidine on γ-aminobutyric acid accumulation in germinating soybean under hypoxia-NaCl stress. J. Food Drug Anal. 2015, 23, 287–293. [Google Scholar] [CrossRef]
- Szepesi, Á.; Bakacsy, L.; Kovács, H.; Szilágyi, Á.; Köhler, Z.M. Inhibiting Copper Amine Oxidase Using L-Aminoguanidine Induces Cultivar and Age-Dependent Alterations of Polyamine Catabolism in Tomato Seedlings. Agriculture 2022, 12, 274. [Google Scholar] [CrossRef]
- Cona, A.; Rea, G.; Angelini, R.; Federico, R.; Tavladoraki, P. Functions of amine oxidases in plant development and defence. Trends Plant Sci. 2006, 11, 80–88. [Google Scholar] [CrossRef]
- Paschalidis, K.; Tsaniklidis, G.; Wang, B.-Q.; Delis, C.; Trantas, E.; Loulakakis, K.; Makky, M.; Sarris, P.F.; Ververidis, F.; Liu, J.-H. The Interplay among Polyamines and Nitrogen in Plant Stress Responses. Plants 2019, 8, 315. [Google Scholar] [CrossRef] [PubMed]
- Recalde, L.; Mansur, N.M.G.; Cabrera, A.V.; Matayoshi, C.L.; Gallego, S.M.; Groppa, M.D.; Benavides, M.P. Unravelling ties in the nitrogen network: Polyamines and nitric oxide emerging as essential players in signalling roadway. Ann. Appl. Biol. 2020, 178, 192–208. [Google Scholar] [CrossRef]
- Kasten, D.; Mithöfer, A.; Georgii, E.; Lang, H.; Durner, J.; Gaupels, F. Nitrite is the driver, phytohormones are modulators while NO and H2O2 act as promoters of NO2-induced cell death. J. Exp. Bot. 2016, 67, 6337–6349. [Google Scholar] [CrossRef]
- Astier, J.; Rasul, S.; Koen, E.; Manzoor, H.; Besson-Bard, A.; Lamotte, O.; Jeandroz, S.; Durner, J.; Lindermayr, C.; Wendehenne, D. S-nitrosylation: An emerging post-translational protein modification in plants. Plant Sci. 2011, 181, 527–533. [Google Scholar] [CrossRef]
- Fares, A.; Rossignol, M.; Peltier, J.-B. Proteomics investigation of endogenous S-nitrosylation in Arabidopsis. Biochem. Biophys. Res. Commun. 2011, 416, 331–336. [Google Scholar] [CrossRef]
- Barroso, J.B.; Valderrama, R.; Carreras, A.; Chaki, M.; Begara-Morales, J.C.; Sánchez-Calvo, B.; Corpas, F.J. Quantification and Localization of S-Nitrosothiols (SNOs) in Higher Plants. In Plant Nitric Oxide; Methods in Molecular Biology Book Series (MIMB); Humana Press: New York, NY, USA, 2016; Volume 1424, pp. 139–147. [Google Scholar] [CrossRef]
- Camejo, D.; Romero-Puertas, M.d.C.; Rodríguez-Serrano, M.; Sandalio, L.M.; Lázaro, J.J.; Jiménez, A.; Sevilla, F. Salinity-induced changes in S-nitrosylation of pea mitochondrial proteins. J. Proteom. 2013, 79, 87–99. [Google Scholar] [CrossRef] [PubMed]
- Jain, P.; von Toerne, C.; Lindermayr, C.; Bhatla, S.C. S-nitrosylation/denitrosylation as a regulatory mechanism of salt stress sensing in sunflower seedlings. Physiol. Plant. 2018, 162, 49–72. [Google Scholar] [CrossRef]
- Kollist, H.; Zandalinas, S.I.; Sengupta, S.; Nuhkat, M.; Kangasjärvi, J.; Mittler, R. Rapid Responses to Abiotic Stress: Priming the Landscape for the Signal Transduction Network. Trends Plant Sci. 2019, 24, 25–37. [Google Scholar] [PubMed]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, research0034.1. [Google Scholar] [CrossRef] [PubMed]
- Hellemans, J.; Mortier, G.; De Paepe, A.; Speleman, F.; Vandesompele, J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 2007, 8, R19. [Google Scholar] [CrossRef]
- Tari, I.; Csiszár, J.; Horváth, E.; Poór, P.; Takács, Z.; Szepesi, Á. The Alleviation of the Adverse Effects of Salt Stress in the Tomato Plant by Salicylic Acid Shows A Time- and Organ-Specific Antioxidant Response. Acta Biol. Cracoviensia Ser. Bot. 2015, 57, 21–30. [Google Scholar] [CrossRef]
- Gémes, K.; Poór, P.; Horváth, E.; Kolbert, Z.; Szopkó, D.; Szepesi, Á.; Tari, I. Cross-talk between salicylic acid and NaCl-generated reactive oxygen species and nitric oxide in tomato during acclimation to high salinity. Physiol. Plant. 2011, 142, 179–192. [Google Scholar] [CrossRef]
- Poór, P.; Kovács, J.; Borbély, P.; Takács, Z.; Szepesi, Á.; Tari, I. Salt stress-induced production of reactive oxygen- and nitrogen species and cell death in the ethylene receptor mutant Never ripe and wild type tomato roots. Plant Physiol. Biochem. 2015, 97, 313–322. [Google Scholar] [CrossRef]
- Molnár, Á.; Rónavári, A.; Bélteky, P.; Szőllősi, R.; Valyon, E.; Oláh, D.; Rázga, Z.; Ördög, A.; Kónya, Z.; Kolbert, Z. ZnO nanoparticles induce cell wall remodeling and modify ROS/ RNS signalling in roots of Brassica seedlings. Ecotoxicol. Environ. Saf. 2020, 206, 111158. [Google Scholar] [CrossRef] [PubMed]
- Ederli, L.; Pasqualini, S.; Batini, P.; Antonielli, M. Photoinhibition and oxidative stress: Effects on xanthophyll cycle, scavenger enzymes and abscisic acid content in tobacco plants. J. Plant Physiol. 1997, 151, 422–428. [Google Scholar] [CrossRef]
- Mishra, V.; Singh, P.; Tripathi, D.K.; Corpas, F.J.; Singh, V.P. Nitric oxide and hydrogen sulfide: An indispensable combination for plant functioning. Trends Plant Sci. 2021, 26, 1270–1285. [Google Scholar] [CrossRef] [PubMed]
- Michaeli, S.; Fromm, H. Closing the loop on the GABA shunt in plants: Are GABA metabolism and signaling entwined? Front. Plant Sci. 2015, 6, 419. [Google Scholar] [CrossRef]
- Kabała, K.; Reda, M.; Wdowikowska, A.; Janicka, M. Role of Plasma Membrane NADPH Oxidase in Response to Salt Stress in Cucumber Seedlings. Antioxidants 2022, 11, 1534. [Google Scholar] [CrossRef]
- Nilsson, B.-O. Biological effects of aminoguanidine: An update. Inflamm. Res. 1999, 48, 509–515. [Google Scholar] [CrossRef]
- Gémes, K.; Kim, Y.J.; Park, K.Y.; Moschou, P.N.; Andronis, E.; Valassaki, C.; Roussis, A.; Roubelakis-Angelakis, K.A. An NADPH-Oxidase/Polyamine Oxidase Feedback Loop Controls Oxidative Burst Under Salinity. Plant Physiol. 2016, 172, 1418–1431. [Google Scholar] [CrossRef]
- González-Hernández, A.I.; Scalschi, L.; Vicedo, B.; Marcos-Barbero, E.L.; Morcuende, R.; Camañes, G. Putrescine: A Key Metabolite Involved in Plant Development, Tolerance and Resistance Responses to Stress. Int. J. Mol. Sci. 2022, 23, 2971. [Google Scholar] [CrossRef]
- Wimalasekera, R.; Schaarschmidt, F.; Angelini, R.; Cona, A.; Tavladoraki, P.; Scherer, G.F. POLYAMINE OXIDASE2 of Arabidopsis contributes to ABA mediated plant developmental processes. Plant Physiol. Biochem. 2015, 96, 231–240. [Google Scholar] [CrossRef]
- Alabdallah, O.; Ahou, A.; Mancuso, N.; Pompili, V.; Macone, A.; Pashkoulov, D.; Stano, P.; Cona, A.; Angelini, R.; Tavladoraki, P. The Arabidopsis polyamine oxidase/dehydrogenase 5 interferes with cytokinin and auxin signaling pathways to control xylem differentiation. J. Exp. Bot. 2017, 68, 997–1012. [Google Scholar] [CrossRef]
- Astier, J.; Gross, I.; Durner, J. Nitric oxide production in plants: An update. J. Exp. Bot. 2018, 69, 3401–3411. [Google Scholar] [CrossRef]
- Hasanuzzaman, M.; Oku, H.; Nahar, K.; Bhuyan, M.H.M.B.; Al Mahmud, J.; Baluska, F.; Fujita, M. Nitric oxide-induced salt stress tolerance in plants: ROS metabolism, signaling, and molecular interactions. Plant Biotechnol. Rep. 2018, 12, 77–92. [Google Scholar] [CrossRef]
- Rosales, E.P.; Iannone, M.F.; Groppa, M.D.; Benavides, M.P. Polyamines modulate nitrate reductase activity in wheat leaves: Involvement of nitric oxide. Amino Acids 2012, 42, 857–865. [Google Scholar] [CrossRef] [PubMed]
- Lindermayr, C. Crosstalk between reactive oxygen species and nitric oxide in plants: Key role of S-nitrosoglutathione reductase. Free. Radic. Biol. Med. 2018, 122, 110–115. [Google Scholar] [CrossRef] [PubMed]
- Raza, A.; Tabassum, J.; Mubarik, M.S.; Anwar, S.; Zahra, N.; Sharif, Y.; Hafeez, M.B.; Zhang, C.; Corpas, F.J.; Chen, H. Hydrogen sulfide: An emerging component against abiotic stress in plants. Plant Biol. 2022, 24, 540–558. [Google Scholar] [CrossRef]
- Hasanuzzaman, M.; Bhuyan, M.B.; Zulfiqar, F.; Raza, A.; Mohsin, S.M.; Al Mahmud, J.; Fujita, M.; Fotopoulos, V. Reactive Oxygen Species and Antioxidant Defense in Plants under Abiotic Stress: Revisiting the Crucial Role of a Universal Defense Regulator. Antioxidants 2020, 9, 681. [Google Scholar] [CrossRef]
- Hasanuzzaman, M.; Bhuyan, M.H.M.B.; Parvin, K.; Bhuiyan, T.F.; Anee, T.I.; Nahar, K.; Hossen, S.; Zulfiqar, F.; Alam, M.; Fujita, M. Regulation of ROS Metabolism in Plants under Environmental Stress: A Review of Recent Experimental Evidence. Int. J. Mol. Sci. 2020, 21, 8695. [Google Scholar] [CrossRef]
- Goyal, V.; Jhanghel, D.; Mehrotra, S. Emerging warriors against salinity in plants: Nitric oxide and hydrogen sulphide. Physiol. Plant. 2021, 171, 896–908. [Google Scholar] [CrossRef]
- Van der Merwe, M.J.; Osorio, S.; Araújo, W.L.; Balbo, I.; Nunes-Nesi, A.; Maximova, E.; Carrari, F.; Bunik, V.I.; Persson, S.; Fernie, A.R. Tricarboxylic Acid Cycle Activity Regulates Tomato Root Growth via Effects on Secondary Cell Wall Production. Plant Physiol. 2010, 153, 611–621. [Google Scholar] [CrossRef]
- Szepesi, Á. Halotropism: Phytohormonal Aspects and Potential Applications. Front. Plant Sci. 2020, 11, 571025. [Google Scholar] [CrossRef]
- Liu, T.; Huang, B.; Chen, L.; Xian, Z.; Song, S.; Chen, R.; Hao, Y. Genome-wide identification, phylogenetic analysis, and expression profiling of polyamine synthesis gene family members in tomato. Gene 2018, 661, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Upadhyay, R.K.; Fatima, T.; Handa, A.K.; Mattoo, A.K. Polyamines and Their Biosynthesis/Catabolism Genes Are Differentially Modulated in Response to Heat Versus Cold Stress in Tomato Leaves (Solanum lycopersicum L.). Cells 2020, 9, 1749. [Google Scholar] [CrossRef] [PubMed]
- Ma, N.; Feng, H.; Meng, X.; Li, D.; Yang, D.; Wu, C.; Meng, Q. Overexpression of tomato SlNAC1transcription factor alters fruit pigmentation and softening. BMC Plant Biol. 2014, 14, 351. [Google Scholar] [CrossRef] [PubMed]
- Poyatos-Pertíñez, S.; Quinet, M.; Ortíz-Atienza, A.; Yuste-Lisbona, F.J.; Pons, C.; Giménez, E.; Angosto, T.; Granell, A.; Capel, J.; Lozano, R. A Factor Linking Floral Organ Identity and Growth Revealed by Characterization of the Tomato Mutant unfinished flower development (ufd). Front. Plant Sci. 2016, 7, 1648. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).