Spatial and Temporal Protein Modules Signatures Associated with Alzheimer Disease in 3xTg-AD Mice Are Restored by Early Ubiquinol Supplementation
Abstract
1. Introduction
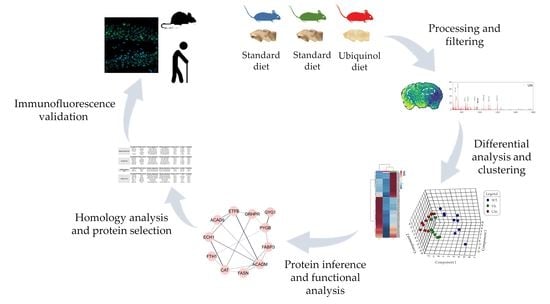
2. Materials and Methods
2.1. Animals
2.2. Behaviourial Tests
2.3. Processing of Samples
2.3.1. Murine Samples
2.3.2. Human Samples
2.4. MALDI Mass Spectrometry Imaging (MALDI–MSI)
2.4.1. Sample Preparation
2.4.2. On-Tissue Digestion and Matrix Deposition
2.4.3. Data Acquisition
2.4.4. Pre-Processing Analysis of MALDI–MSI Data
2.5. Protein Identification
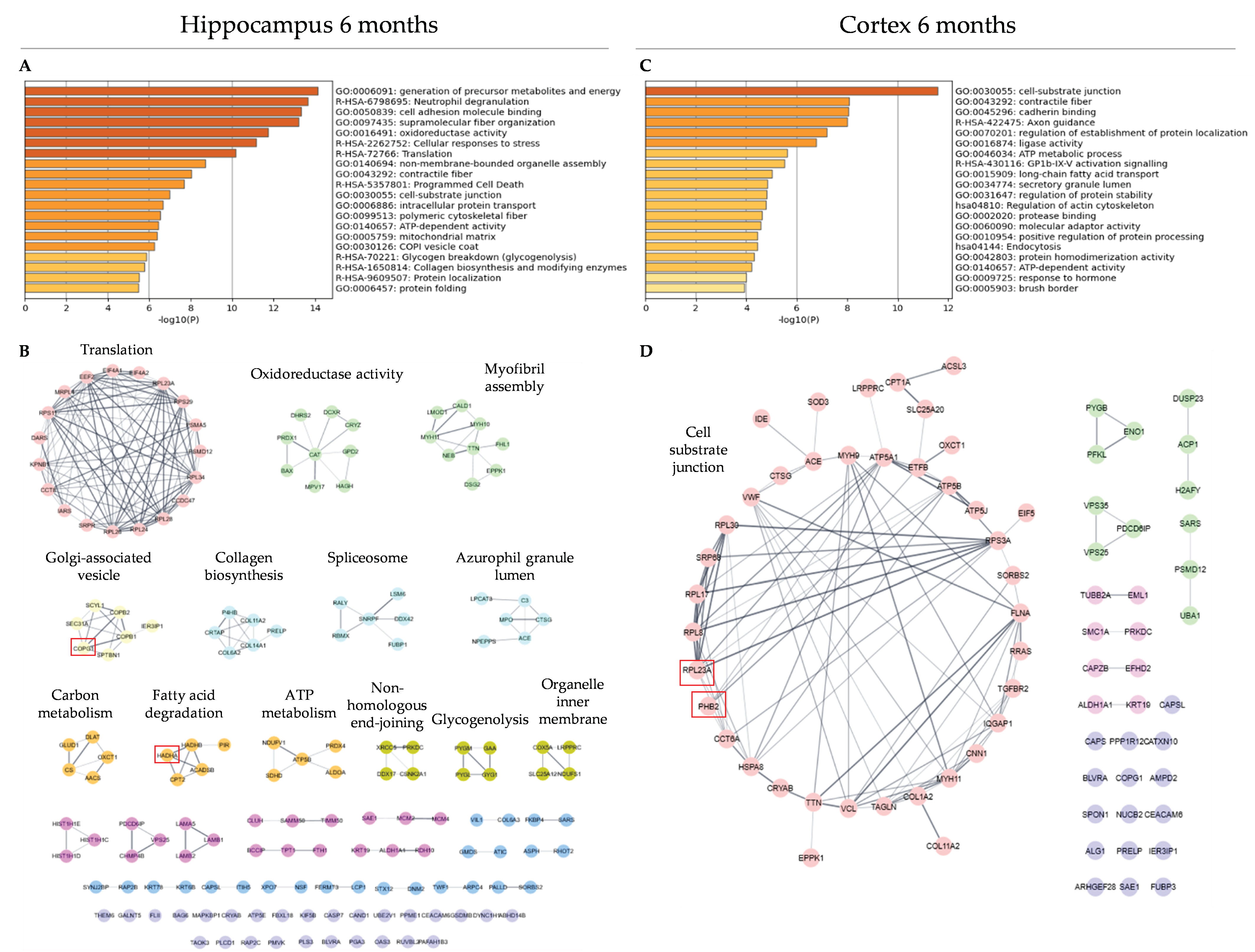
2.6. Functional Analysis and Protein–Protein Interaction Networks
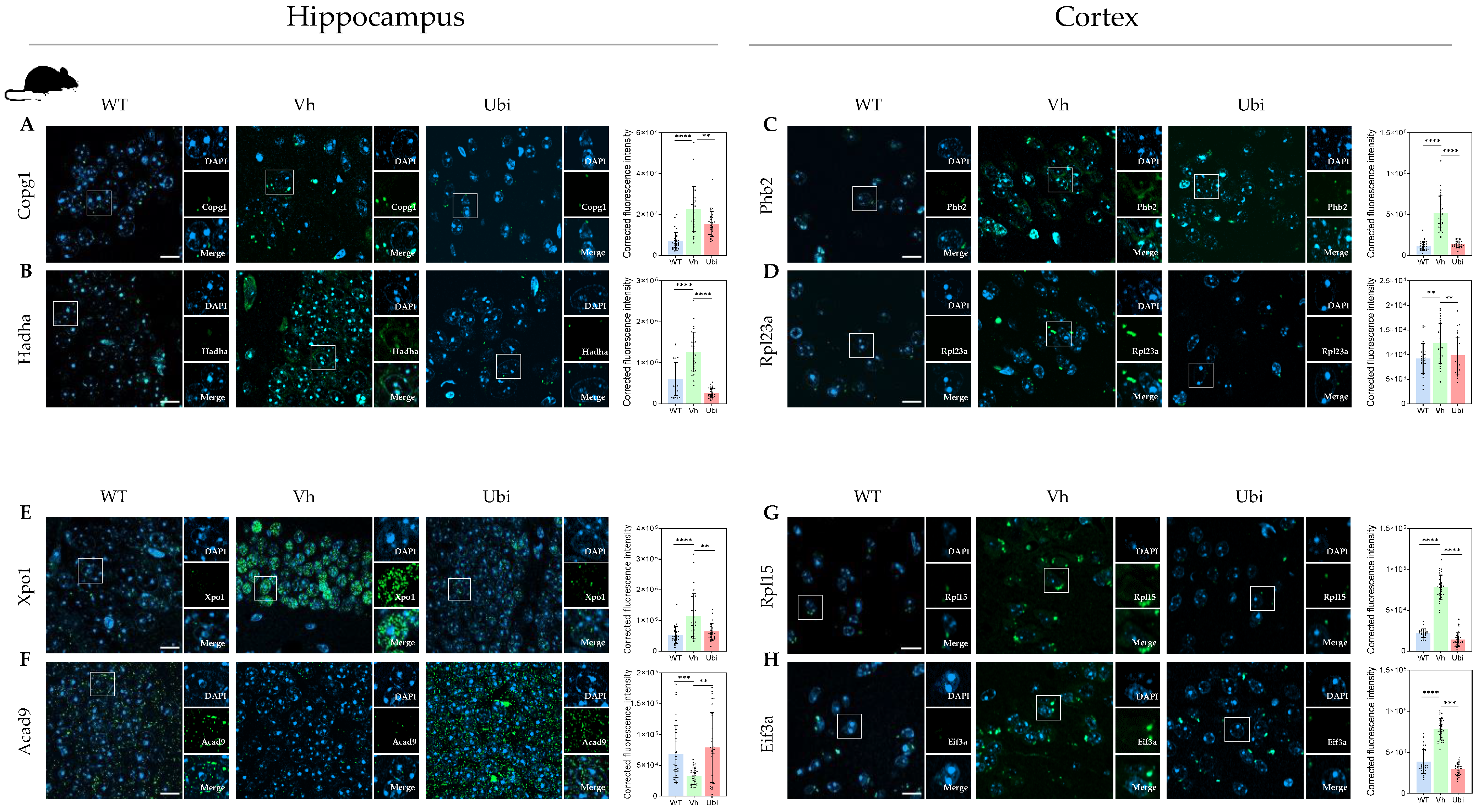
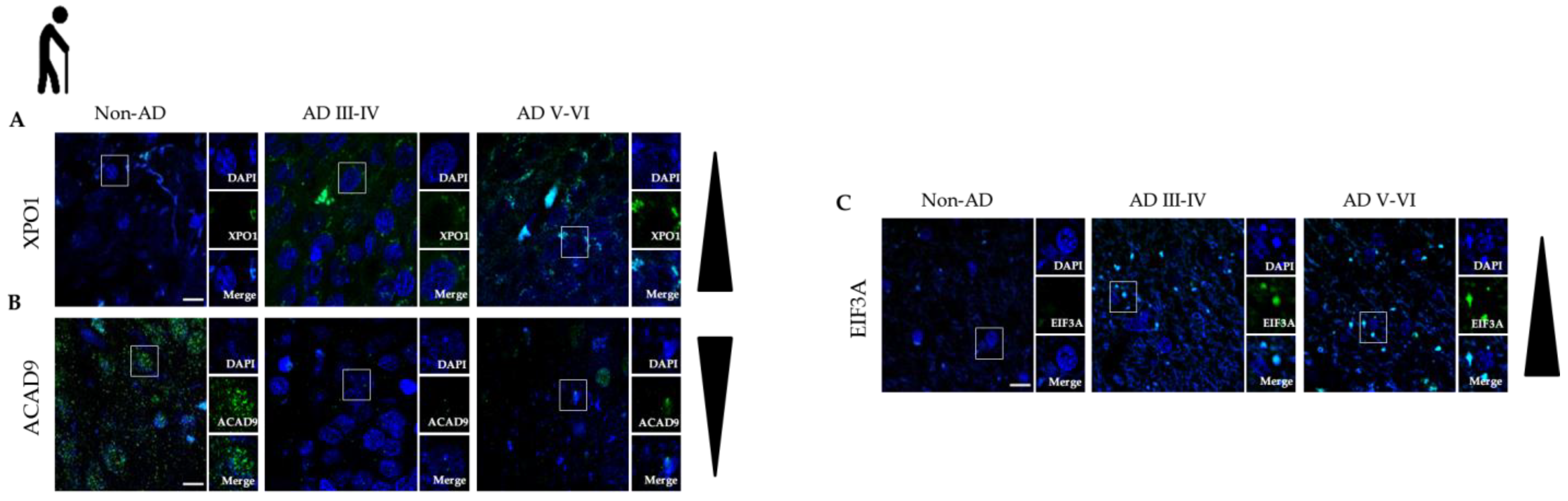
2.7. Validation by Immunofluorescence
2.7.1. Selection Criteria for Validation
2.7.2. Immuno-Histochemistry and Imaging Protocol
2.8. Statistical Analysis
3. Results
3.1. Behavioural Tests
3.2. MALDI–MSI Methodology
3.3. Functional Analysis and Protein–Protein Interaction Analysis
3.4. Protein Modules Validation by Immunofluorescence
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tahami Monfared, A.A.; Byrnes, M.J.; White, L.A.; Zhang, Q. Alzheimer’s Disease: Epidemiology and Clinical Progression. Neurol. Ther. 2022, 11, 553–569. [Google Scholar] [CrossRef] [PubMed]
- Hampel, H.; Hardy, J.; Blennow, K.; Chen, C.; Perry, G.; Kim, S.H.; Villemagne, V.L.; Aisen, P.; Vendruscolo, M.; Iwatsubo, T.; et al. The Amyloid-β Pathway in Alzheimer’s Disease. Mol. Psychiatry 2021, 26, 5481–5503. [Google Scholar] [CrossRef] [PubMed]
- Javaid, S.F.; Giebel, C.; Khan, M.A.; Hashim, M.J. Epidemiology of Alzheimer’s disease and other dementias: Rising global burden and forecasted trends. F1000Research 2021, 10, 425. [Google Scholar] [CrossRef]
- Vyas, Y.; Montgomery, J.M.; Cheyne, J.E. Hippocampal Deficits in Amyloid-β-Related Rodent Models of Alzheimer’s Disease. Front. Neurosci. 2020, 14, 266. [Google Scholar] [CrossRef]
- Collins, A.E.; Saleh, T.M.; Kalisch, B.E. Naturally Occurring Antioxidant Therapy in Alzheimer’s Disease. Antioxidants 2022, 11, 213. [Google Scholar] [CrossRef]
- Frontiñán-Rubio, J.; Rabanal-Ruiz, Y.; Durán-Prado, M.; Alcain, F.J. The Protective Effect of Ubiquinone against the Amyloid Peptide in Endothelial Cells Is Isoprenoid Chain Length-Dependent. Antioxidants 2021, 10, 1806. [Google Scholar] [CrossRef]
- Cummings, J.; Aisen, P.S.; DuBois, B.; Frolich, L.; Jack, C.R., Jr.; Jones, R.W.; Morris, J.C.; Raskin, J.; Dowsett, S.A.; Scheltens, P. Drug development in Alzheimer’s disease: The path to 2025. Alz. Res. Therapy. 2016, 8, 39. [Google Scholar] [CrossRef]
- Frontiñán-Rubio, J.; Sancho-Bielsa, F.J.; Peinado, J.R.; LaFerla, F.M.; Giménez-Llort, L.; Durán-Prado, M.; Alcain, F.J. Sex-dependent co-occurrence of hypoxia and β-amyloid plaques in hippocampus and entorhinal cortex is reversed by long-term treatment with ubiquinol and ascorbic acid in the 3xTg-AD mouse model of Alzheimer’s disease. Mol. Cell. Neurosci. 2018, 92, 67–81. [Google Scholar] [CrossRef]
- Johnson, E.C.B.; Carter, E.K.; Dammer, E.B.; Duong, D.M.; Gerasimov, E.S.; Liu, Y.; Liu, J.; Betarbet, R.; Ping, L.; Yin, L.; et al. Large-scale deep multi-layer analysis of Alzheimer’s disease brain reveals strong proteomic disease-related changes not observed at the RNA level. Nat. Neurosci. 2022, 25, 213–225. [Google Scholar] [CrossRef]
- Kusaka, S.; Miyake, Y.; Tokumaru, Y.; Morizane, Y.; Tamaki, S.; Akiyama, Y.; Sato, F.; Murata, I. Boron Delivery to Brain Cells via Cerebrospinal Fluid (CSF) Circulation in BNCT of Brain-Tumor-Model Rats—Ex Vivo Imaging of BPA Using MALDI Mass Spectrometry Imaging. Life 2022, 12, 1786. [Google Scholar] [CrossRef] [PubMed]
- Kwon, H.J.; Oh, J.Y.; Lee, K.S.; Lim, H.K.; Lee, J.; Yoon, H.-R.; Jung, J. Lipid Profiles Obtained from MALDI Mass Spectrometric Imaging in Liver Cancer Metastasis Model. Int. J. Anal. Chem. 2022, 2022, 6007158. [Google Scholar] [CrossRef]
- Strnad, Š.; Strnadová, V.; Sýkora, D.; Cvačka, J.; Maletínská, L.; Vrkoslav, V. MALDI Mass Spectrometry Imaging of Lipids on Free-Floating Brain Sections and Immunohistochemically Colocalized Markers of Neurodegeneration; Lee, Y.-J., Ed.; Mass Spectrometry Imaging of Small Molecules; Springer: New York, NY, USA, 2022; pp. 229–239. Available online: https://link.springer.com/10.1007/978-1-0716-2030-4_16 (accessed on 13 November 2022).
- Chen, Y.; Hu, D.; Zhao, L.; Tang, W.; Li, B. Unraveling metabolic alterations in transgenic mouse model of Alzheimer’s disease using MALDI MS imaging with 4-aminocinnoline-3-carboxamide matrix. Anal. Chim. Acta 2022, 1192, 339337. [Google Scholar] [CrossRef] [PubMed]
- Fornasiero, E.F.; Mandad, S.; Wildhagen, H.; Alevra, M.; Rammner, B.; Keihani, S.; Opazo, F.; Urban, I.; Ischebeck, T.; Sakib, M.S.; et al. Precisely measured protein lifetimes in the mouse brain reveal differences across tissues and subcellular fractions. Nat. Commun. 2018, 9, 4230. [Google Scholar] [CrossRef]
- Sjölin, K.; Kultima, K.; Larsson, A.; Freyhult, E.; Zjukovskaja, C.; Alkass, K.; Burman, J. Distribution of five clinically important neuroglial proteins in the human brain. Mol. Brain 2022, 15, 52. [Google Scholar] [CrossRef] [PubMed]
- Boyle, P.A.; Yu, L.; Wilson, R.S.; Leurgans, S.E.; Schneider, J.A.; Bennett, D.A. Person-specific contribution of neuropathologies to cognitive loss in old age: Neuropathologies and Cognition. Ann. Neurol. 2018, 83, 74–83. [Google Scholar] [CrossRef]
- Giménez-Llort, L.; Arranz, L.; Maté, I.; De la Fuente, M. Gender-Specific Neuroimmunoendocrine Aging in a Triple-Transgenic 3xTg-AD Mouse Model for Alzheimer’s Disease and Its Relation with Longevity. Neuroimmunomodulation 2008, 15, 331–343. [Google Scholar] [CrossRef]
- Ubeda-Bañon, I.; Flores-Cuadrado, A.; Saiz-Sanchez, D.; Martinez-Marcos, A. Differential Effects of Parkinson’s Disease on Interneuron Subtypes within the Human Anterior Olfactory Nucleus. Front. Neuroanat. 2017, 11, 113. [Google Scholar] [CrossRef] [PubMed]
- Perez-Riverol, Y.; Bai, J.; Bandla, C.; García-Seisdedos, D.; Hewapathirana, S.; Kamatchinathan, S.; Kundu, D.J.; Prakash, A.; Frericks-Zipper, A.; Eisenacher, M.; et al. The PRIDE database resources in 2022: A hub for mass spectrometry-based proteomics evidences. Nucleic. Acids Res. 2022, 50, D543–D552. [Google Scholar] [CrossRef]
- Chong, J.; Soufan, O.; Li, C.; Caraus, I.; Li, S.; Bourque, G.; Wishart, D.S.; Xia, J. MetaboAnalyst 4.0: Towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018, 46, W486–W494. [Google Scholar] [CrossRef]
- Maier, S.K.; Hahne, H.; Gholami, A.M.; Balluff, B.; Meding, S.; Schoene, C.; Walch, A.K.; Kuster, B. Comprehensive identification of proteins from MALDI imaging. Mol. Cell. Proteom. 2013, 12, 2901–2910. [Google Scholar] [CrossRef]
- Doncheva, N.T.; Morris, J.H.; Gorodkin, J.; Jensen, L.J. Cytoscape StringApp: Network Analysis and Visualization of Proteomics Data. J. Proteome Res. 2019, 18, 623–632. [Google Scholar] [CrossRef]
- Morris, J.H.; Apeltsin, L.; Newman, A.M.; Baumbach, J.; Wittkop, T.; Su, G.; Bader, G.D.; Ferrin, T.E. clusterMaker: A multi-algorithm clustering plugin for Cytoscape. BMC Bioinform. 2011, 12, 436. [Google Scholar] [CrossRef] [PubMed]
- Flores-Cuadrado, A.; Saiz-Sanchez, D.; Mohedano-Moriano, A.; Lamas-Cenjor, E.; Leon-Olmo, V.; Martinez-Marcos, A.; Ubeda-Bañon, I. Astrogliosis and sexually dimorphic neurodegeneration and microgliosis in the olfactory bulb in Parkinson’s disease. npj Park. Dis. 2021, 7, 11. [Google Scholar] [CrossRef]
- Tiwari, S.; Atluri, V.; Kaushik, A.; Yndart, A.; Nair, M. Alzheimer’s disease: Pathogenesis, diagnostics, and therapeutics. IJN 2019, 14, 5541–5554. [Google Scholar] [CrossRef]
- Montal, V.; Vilaplana, E.; Pegueroles, J.; Bejanin, A.; Alcolea, D.; Carmona-Iragui, M.; Clarimón, J.; Levin, J.; Cruchaga, C.; Graff-Radford, N.; et al. Biphasic cortical macro- and microstructural changes in autosomal dominant Alzheimer’s disease. Alzheimers Dement 2021, 17, 618–628. [Google Scholar] [CrossRef]
- Signorile, A.; Sgaramella, G.; Bellomo, F.; De Rasmo, D. Prohibitins: A Critical Role in Mitochondrial Functions and Implication in Diseases. Cells 2019, 8, 71. [Google Scholar] [CrossRef] [PubMed]
- Rosenthal, S.L.; Wang, X.; Demirci, F.Y.; Barmada, M.M.; Ganguli, M.; Lopez, O.L.; Kamboh, M.I. Beta-amyloid toxicity modifier genes and the risk of Alzheimer’s disease. Am. J. Neurodegener. Dis. 2012, 1, 191–198. [Google Scholar] [PubMed]
- Speijer, D. Molecular characteristics of the multi-functional FAO enzyme ACAD9 illustrate the importance of FADH 2/NADH ratios for mitochondrial ROS formation. BioEssays 2022, 44, 2200056. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Li, Z.; Lin, E.; He, P.; Ru, G. Oxidative damage-induced hyperactive ribosome biogenesis participates in tumorigenesis of offspring by cross-interacting with the Wnt and TGF-β1 pathways in IVF embryos. Exp. Mol. Med. 2021, 53, 1792–1806. [Google Scholar] [CrossRef]
- Hao, W.; Dian, M.; Zhou, Y.; Zhong, Q.; Pang, W.; Li, Z.; Zhao, Y.; Ma, J.; Lin, X.; Luo, R.; et al. Autophagy induction promoted by m6A reader YTHDF3 through translation upregulation of FOXO3 mRNA. Nat. Commun. 2022, 13, 5845. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Llanos-González, E.; Sancho-Bielsa, F.J.; Frontiñán-Rubio, J.; Rabanal-Ruíz, Y.; García-Carpintero, S.; Chicano, E.; Úbeda-Banon, I.; Flores-Cuadrado, A.; Giménez-Llort, L.; Alcaín, F.J.; et al. Spatial and Temporal Protein Modules Signatures Associated with Alzheimer Disease in 3xTg-AD Mice Are Restored by Early Ubiquinol Supplementation. Antioxidants 2023, 12, 747. https://doi.org/10.3390/antiox12030747
Llanos-González E, Sancho-Bielsa FJ, Frontiñán-Rubio J, Rabanal-Ruíz Y, García-Carpintero S, Chicano E, Úbeda-Banon I, Flores-Cuadrado A, Giménez-Llort L, Alcaín FJ, et al. Spatial and Temporal Protein Modules Signatures Associated with Alzheimer Disease in 3xTg-AD Mice Are Restored by Early Ubiquinol Supplementation. Antioxidants. 2023; 12(3):747. https://doi.org/10.3390/antiox12030747
Chicago/Turabian StyleLlanos-González, Emilio, Francisco J. Sancho-Bielsa, Javier Frontiñán-Rubio, Yoana Rabanal-Ruíz, Sonia García-Carpintero, Eduardo Chicano, Isabel Úbeda-Banon, Alicia Flores-Cuadrado, Lydia Giménez-Llort, Francisco Javier Alcaín, and et al. 2023. "Spatial and Temporal Protein Modules Signatures Associated with Alzheimer Disease in 3xTg-AD Mice Are Restored by Early Ubiquinol Supplementation" Antioxidants 12, no. 3: 747. https://doi.org/10.3390/antiox12030747
APA StyleLlanos-González, E., Sancho-Bielsa, F. J., Frontiñán-Rubio, J., Rabanal-Ruíz, Y., García-Carpintero, S., Chicano, E., Úbeda-Banon, I., Flores-Cuadrado, A., Giménez-Llort, L., Alcaín, F. J., Peinado, J. R., & Durán-Prado, M. (2023). Spatial and Temporal Protein Modules Signatures Associated with Alzheimer Disease in 3xTg-AD Mice Are Restored by Early Ubiquinol Supplementation. Antioxidants, 12(3), 747. https://doi.org/10.3390/antiox12030747