Abstract
Selenocysteine (Sec), the 21st amino acid, is structurally similar to cysteine but with a sulfur to selenium replacement. This single change retains many of the chemical properties of cysteine but often with enhanced catalytic and redox activity. Incorporation of Sec into proteins is unique, requiring additional translation factors and multiple steps to insert Sec at stop (UGA) codons. These Sec-containing proteins (selenoproteins) are found in all three domains of life where they often are involved in cellular homeostasis (e.g., reducing reactive oxygen species). The essential role of selenoproteins in humans requires us to maintain appropriate levels of selenium, the precursor for Sec, in our diet. Too much selenium is also problematic due to its toxic effects. Deciphering the role of Sec in selenoproteins is challenging for many reasons, one of which is due to their complicated biosynthesis pathway. However, clever strategies are surfacing to overcome this and facilitate production of selenoproteins. Here, we focus on one of the 25 human selenoproteins, selenoprotein M (SELENOM), which has wide-spread expression throughout our tissues. Its thioredoxin motif suggests oxidoreductase function; however, its mechanism and functional role(s) are still being uncovered. Furthermore, the connection of both high and low expression levels of SELENOM to separate diseases emphasizes the medical application for studying the role of Sec in this protein. In this review, we aim to decipher the role of SELENOM through detailing and connecting current evidence. With multiple proposed functions in diverse tissues, continued research is still necessary to fully unveil the role of SELENOM.
1. Introduction
Selenium acts as a double-edged sword: an essential micronutrient for humans that becomes toxic in excess [1]. Selenium is found in both organic (selenomethionine, selenocysteine (Sec)) and inorganic (selenite, selenate) forms, of which the latter is found to be more toxic to humans [2,3]. Sec, the 21st natural amino acid, is biosynthesized on its tRNA to convert inorganic selenium to an organic form, readily used by cells for protein translation [3]. Humans have 25 selenoproteins (proteins containing Sec) that are responsible for cellular function (e.g., redox reactions, immune response, thyroid hormone metabolism). The inability to express these selenoproteins (due to selenium deficiency) have been associated with diseases including cancer, neurodegenerative diseases, Keshan disease, inflammatory bowel diseases, and diabetes [1,4,5]. Selenium supplementation has been proposed to prevent and treat some of these diseases; however, there are also data that show that excess selenium or overexpression of selenoproteins is connected to disease (e.g., diabetes, neurodegenerative diseases) [6,7,8,9,10]. These conflicting results suggest that maximizing selenoprotein production through selenium supplementation is not always a solution and we are missing key information to properly prescribe selenium [11]. Part of this gap is due to lack of understanding on the role that Sec has in the cellular function of selenoproteins [12].
The enhanced chemical reactivity of Sec has been an advantage when studying selenoproteins in vivo. Probes have been designed to specifically recognize Sec over cysteine, and radioactive selenium can be fed to cultures to visualize incorporation [13]. Studying selenoproteins in vitro, however, is more challenging. This is because they follow a translation path that is unique to all other proteins. Sec is encoded by a nonsense codon (UGA), but only in the presence of specific regulatory elements to designate Sec insertion instead of termination. Downstream of the specific UGA codon, in the 3′-untranslated region (UTR) of eukaryotes, is an mRNA hairpin known as the Sec insertion sequence (SECIS) element. This hairpin structure is highly conserved [14] for interaction with SECIS-binding protein 2 (SBP2) and a specialized elongation factor (eEFSec). eEFSec can discriminate the unique structure of tRNASec from tRNAs for the other 20 amino acids, promoting elongation instead of termination [15,16]. Details of the translation mechanism involving these additional factors: SECIS element, SBP2, and eEFSec, are still not fully understood [17]. Moreover, the mechanism differs in each domain of life, adding to the complexity of feasibly overexpressing selenoproteins for detailed analysis of their cellular function [18].
Nevertheless, researchers have used clever strategies to unveil this looming question about selenoprotein function [13]. In this review, we have combined the studies on selenoprotein M (SELENOM) in an effort to connect what has been determined thus far and propose new directions that should be investigated. The extensive expression of SELENOM in the brain directed focus to this region initially; however, it is also found in other tissues. Therefore, researchers have started questioning the role of SELENOM throughout the body. The evidence presented suggests that SELENOM supports cellular growth and, specifically in the brain, has a neuroprotective role. Advancement in the technology to produce selenoproteins [19,20,21] opens the ability to further characterize SELENOM function.
2. Expression of SELENOM Is Widespread
SELENOM is widely expressed throughout the body (e.g., heart, lung, kidney, stomach, small intestine, skin, testis, uterus, ovary, and brain), but not expressed in all tissue types (e.g., muscle and thymus) [22,23] (Figure 1). In the brain, SELENOM expression is extensive, detectable in multiple brain regions including the olfactory bulb, cortex, hippocampus, hypothalamus, brain stem, cerebellum, and cerebellar cortex lysates [23,24]. Immunohistochemistry staining for SELENOM distribution in mice coronal brain sections revealed SELENOM localization in multiple brain regions including the periventricular and arcuate nuclei of the hypothalamus; the ventral tegmental area; red nucleus; the CA1, CA2, and CA3 regions of the hippocampus; the medial septum; and the granular, Purkinje, and molecular layers of the cerebellum [24]. This high expression level of SELENOM in multiple brain regions suggests an important role in brain function.
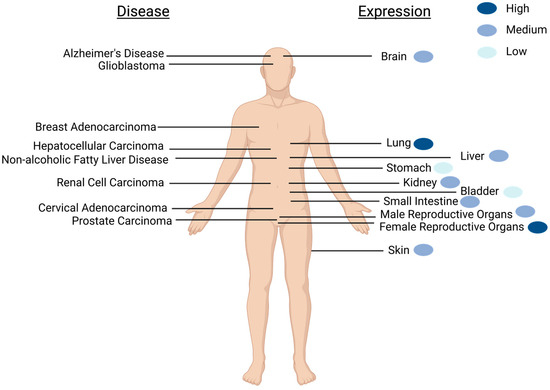
Figure 1.
Schematic of SELENOM expression and SELENOM-related diseases as found in humans. Diseases that correlate with aberrant SELENOM expression are shown on the left, while the location of major organs with detectable SELENOM expression is on the right. Relative expression levels of healthy individuals are portrayed with blue-colored dots. The widespread expression of SELENOM is not limited to what is depicted. Created with BioRender.com.
A global knockout of SELENOM in mice did not result in adverse cognitive or motor defects as one may have expected. Instead, metabolic dysregulation caused by diminished hypothalamic leptin signaling was observed [24,25]. Given that most of SELENOM is found in GABAergic cells, this observation made sense. However, one thing to consider when studying SELENOM in mice is that human expression levels are much lower and the distribution of selenoproteins differ between the organisms [26]. For example, mice have high expression levels of GPx1, GPx4, SELENOF, SELENOK, SELENOM, SELENOS, and SELENOW, while SELENOW and SELENOF are the highest expressed in humans [26]. Since the mechanisms by which many selenoproteins function are still not clear, we cannot rule out whether another selenoprotein in the brain compensates for cognitive and motor function in the absence of SELENOM.
Beyond the brain and examining the entirety of the human body, the Human Protein Atlas (https://www.proteinatlas.org/ENSG00000198832-SELENOM, accessed 14 March 2023) shows that generally SELENOM is expressed in the cytoplasm but localizes to the perinuclear region and nucleoplasm. SELENOM is also highly expressed in the thyroid gland, lungs, and female reproductive organs. The glandular system is most prominent for mRNA expression, with exocrine glandular cells expressing the highest levels. There is low cancer specificity for SELENOM; however, its expression in renal cell carcinoma (RCC) is unfavorable and used as a prognostic marker, discussed in a later section.
3. Elucidating SELENOM Function from Structure
3.1. SELENOM Is Structurally Close to Thioredoxin
As with elucidating the function of a newly discovered protein, when studying a new selenoprotein, an initial step is to scan the structure space for similar proteins that have been previously characterized. Structurally, the closest relative to SELENOM is thioredoxin [27]. Thioredoxins are oxidoreductases with a defined thioredoxin (TXN)-fold identified by a two-layer α/β/α sandwich with a βαβββα secondary structure. Moreover, they harbor a CXXC active-site motif, where X can be any amino acid [28]. Through this CXXC motif, TXNs catalyze the reduction of disulfide bonds as part of a catalytic cycle involving thioredoxin reductase (TXNRD) or through activation by reaction oxygen species (ROS) [29,30] (Figure 2a). In SELENOM, the CXXC motif is found as CXXU, where U refers to Sec [22]. The similar chemistry between C and U suggests that this motif also serves as a redox center and SELENOM participates as an oxidoreductase.
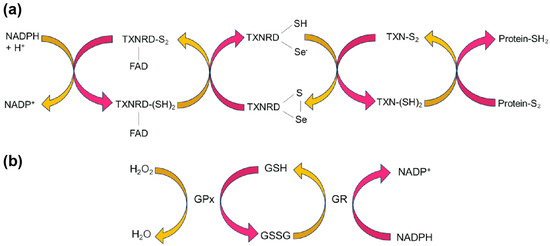
Figure 2.
Oxidoreductase pathways. (a) The thioredoxin (TXN) cycle illustrates how TXNRD, an essential selenoprotein, catalyzes the reduction of oxidized thioredoxin (TXN-S2) using NADPH as an electron donor. Reduced thioredoxin (TXN-(SH)2) plays a critical role in oxidizing proteins involved in cellular redox homeostasis, influencing processes like DNA synthesis, antioxidant defense, and apoptosis. (b) In the GPx pathway, GPx catalyzes the reduction of peroxides, including hydrogen peroxide (H2O2), using reduced glutathione (GSH) as an electron donor. The resulting oxidized glutathione (GSSG) can be converted back to GSH through the action of glutathione reductase (GR), ensuring a continuous supply for cellular antioxidant defense. From activity assays, SELENOM could play a role in reducing peroxides like GPx.
The redox active amino acids (C or U) in the CXXC/U motif are surface-accessible in SELENOM. This differs from TXN, but is seen in protein disulfide isomerases (PDIs). PDIs are also oxidoreductases that typically consist of two catalytic TXN-like domains (containing the CXXC motif), separated by two non-catalytic TXN-like domains. However, some PDIs only contain a single catalytic domain [31,32], which is analogous to what is observed in the structure of SELENOM. Furthermore, the position of the CXXU motif is located between the C-terminus of strand β1 and N-terminus of helix α1, which compares to what is found in both TXNs and PDIs [27].
3.2. SELENOM Defines the New Thioredoxin Family
Another structural homolog found for SELENOM is SELENOF (previously named Sep15) [22,33]. Although their sequence identity is only 31%, SELENOF shares multiple regions of significant sequence identity to SELENOM [34]. The major similarity that distinguishes SELENOM and SELENOF from other TXN families is its unique TXN-like fold. Its central α/β domain, composed of three α-helices (α1-α3) and a mixed parallel/anti-parallel four-stranded β-sheet (β1-β4), represents the most basic TXN-like fold [35] (Figure 3). The CXXU motif located within this unique TXN-like fold is also unlike other oxidoreductases. While X refers to any amino acid, only certain amino acids are typically found in nature. These include GP, GH, and PH, found in TXNs, PDIs, and disulfide oxidases, respectively [27]. SELENOM has the sequence motif CGGU, which has only been observed thus far in SELENOF as CGU [27].
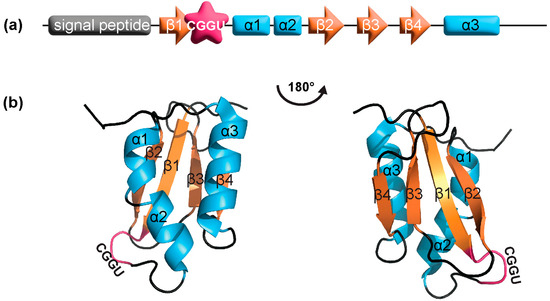
Figure 3.
SELENOM (a) domain structure and (b) NMR structure (PDB: 2A2P). The first 25 residues are the endoplasmic reticulum signal peptide, which was removed for protein expression. Residues 25–34 and 121–145 are not shown in the structure due to high flexibility. The CXXU motif is shown in magenta, α-helices in cyan, and β-strands in orange.
In addition to a defining TXN-like fold, both SELENOM and SELENOF have other structural features that group them into a separate subfamily of TXNs. The conserved proline at the N-terminus of strand β3 is typically found in the cis-conformation [29], while in SELENOM and SELENOF, it is in the trans-conformation [28]. Furthermore, these proteins are missing a charge pair that in TXNs and PDIs are involved in proton transfer [30]. The functional importance of these structural differences remains unclear and is still under study.
3.3. SELENOM Does Not Bind UGGT
Among the many similarities between SELENOF and SELENOM, there are differences that potentially separate their functions. These differences lie at the termini. Both selenoproteins have an N-terminus that contains an ER-signaling sequence, which is subsequently cleaved during protein maturation. In SELENOF, an elongated cysteine-rich extension follows the signaling sequence prior to strand β1, while in SELENOM, this is a short extension. Furthermore, the C-terminus of SELENOM is a flexible extension, but it is short and unstructured in SELENOF [27]. The cysteine-rich extension at the N-terminus of SELENOF is known to mediate a high-affinity complex with the folding sensor of the calnexin cycle-UDP-glucose:glycoprotein glucosyltransferase (UGGT) [34,36]. The function of this binding interaction is not fully investigated, though it is suggested to be a PDI co-factor, assisting UGGT in assessing misfolded glycoproteins [27]. The absence of this cysteine-rich region in SELENOM confirmed that UGGT is not an interacting partner for SELENOM [34].
4. Elucidating SELENOM Function from Observed Activity
While structurally SELENOM resembles other oxidoreductases, some of the defining functional features from these oxidoreductases are not shared with SELENOM. Therefore, experimental studies have been employed to investigate this further. Similar to the structural comparisons above, functional comparisons with predefined oxidoreductase assays were investigated.
4.1. Glutathione Peroxidase Activity
Glutathione peroxidase (GPx), a member of the selenoprotein family, serves as a critical antioxidant enzyme, and its functionality is intricately linked to its molecular structure. The enzyme’s catalytic efficiency is largely attributed to a Sec residue at its active site, which enables GPx to rapidly reduce hydrogen peroxides and lipid peroxides to water and alcohols, respectively. Peroxides oxidize GPx, which reacts with reduced glutathione (GSH). This oxidizes glutathione (GSSG), converting GPx back to its active (reduced) state for reaction with another peroxide. Glutathione reductase subsequently regenerates GSH from GSSG using NADPH as the reducing agent to complete the cycle (Figure 2b). This integrated antioxidant system, facilitated by the high reactivity of selenium and efficient enzyme kinetics, allows for rapid detoxification and maintenance of cellular redox balance [37]. Furthermore, the enzyme’s multimeric architecture—existing as either a dimer or tetramer—enhances its kinetics and enables substrate-specific channels that guide peroxides toward the Sec active site for catalysis [38].
To test for GPx activity, a GPx assay is established, which monitors the conversion of reduced glutathione (GSH) to oxidized glutathione (GSSG) while reducing hydrogen peroxide (H2O2) or other peroxides to water or alcohol. This enzymatic reaction cycle involves GPx and glutathione reductase, which helps convert GSSG back to GSH, thereby maintaining a pool of available GSH for the GPx reaction. The assay is designed to monitor the rate of NADPH oxidation to NADP+ as a direct measure of GPx activity. SELENOM has been shown to have GPx activity in vitro [39,40] and also shown to reduce ROS in HEK293T cells. Replacing Sec in the CXXU motif with a Cys, significantly increased the ROS present, presumably decreasing the activity of the enzyme. This shows that efficient reduction of ROS relies on the presence of Sec, due to the difference in chemistry between the two amino acids [37].
4.2. Thioredoxin Activity
TXN is a small redox-active protein known for its pivotal role in cellular redox homeostasis and other cellular processes, such as DNA synthesis and signal transduction [41]. It acts through the reduction of disulfide bonds in target proteins, often initiated by reduction of TXN by other proteins like TXNRD [42]. This catalytic cycle of reducing disulfide bonds with TXN, TXNRD, and NADPH is known as the TXN system (Figure 2b). Moreover, in the presence of NADPH and TXNRD, TXN activity can be measured on a target protein such as insulin. Based on the proposed similarity in disulfide bond formation function of TXN and SELENOM, this assay was performed on hypothalamic tissues and cells that are SELENOM deficient. Through this, a decrease in the TXN activity was observed, although levels of TXN and TXNRD were not affected. Further evidence showed TXN activity in SELENOM-immunoprecipitated samples suggesting that indeed SELENOM has intrinsic TXN activity and plays a crucial role in enhancing the TXN system’s antioxidant activity. This highlights SELENOM’s potential influence on regulating energy balance in hypothalamic tissues and defending cells from oxidative stress [25].
4.3. Absence of Thioredoxin Reductase Activity
Another oxidoreductase assay that has been established is the TXNRD assay. TXNRD is a selenoprotein involved in reducing the oxidized form of TXN. This occurs through the Sec-containing active site located at the C-terminus of the enzyme. Sec is critical to the enzyme’s efficiency because of its higher nucleophilicity compared to a sulfur-containing Cys. Thus, the selenium atom in Sec is directly involved in the enzyme’s catalytic mechanism, contributing to rapid and efficient reduction reactions. In addition to the Sec requirement for TXNRD activity, the enzyme also relies on its FAD cofactor to reduce TXN. In TXNRD, FAD is tightly, yet non-covalently, anchored within a specific segment of the protein, commonly referred to as the FAD-binding domain [43]. This domain is pivotal for enabling redox reactions by initially accepting electrons from NADPH. FAD serves as a redox-active cofactor, facilitating the transfer of electrons, pivotal for TXNRD’s role in reducing oxidized TXN (Figure 2b). In assays that assess TXNRD activity, a common approach is to measure the increase in DTNB (5,5′-dithiobis-(2-nitrobenzoic acid) levels, indicative of the enzyme’s capability to reduce TXN, which acts as an intermediary substrate in this process [44].
TXNRD activity was also assessed for SELENOM in hypothalamic tissues and cells. Initial observations found a decrease in TXNRD activity in SELENOM-deficient cells but not tissues. However, upon further investigation with SELENOM-enriched samples, no increase in TXNRD activity was observed [25]. This result is not completely surprising given that structural similarities connect SELENOM to TXN rather than TXNRD. Therefore, as a parallel to the TXN system, SELENOM may also be part of a catalytic cycle for the reduction of disulfide bonds in proteins.
5. Elucidating SELENOM Function from Binding Partners
5.1. Thioredoxin Interacting Protein
Thioredoxin interacting protein (TXNIP), also known as thioredoxin-binding protein 2, is a major mediator in the TXN antioxidant system. TXNIP interacts with the CXXC motif of TXN, blocking its potential to scavenge ROS. This in turn increases the ROS levels in the cell and induces apoptosis [45]. TXNIP is also known to be a negative regulator of mTOR-dependent signaling [46], with implications that it modulates hypothalamic leptin signaling [47,48]. Micro-array analyses of SELENOM-deficient mHypoE-44 cells and hypothalamic tissue identified a downregulation of TXNIP but no change in expression levels for TXN and TXNRD [25]. This suggests that the reduction in TXNIP is a compensatory response adapting to the absence of SELENOM. Furthermore, an observed decrease in the TXN activity (but not the TXNRD activity) suggests that SELENOM contributes to the hypothalamic TXN system [25]. The CXXU motif in SELENOM and corresponding TXN activity implies an interaction with TXNIP similar to TXN. It follows that SELENOM and TXNIP work together to maintain cellular homeostasis.
This has been observed in neurons where SELENOM reduced neuronal apoptosis in response to oxidative stress through regulating cytosolic Ca2+ release from the ER [40]. TXNIP, on the other hand, is induced in high-oxidative environments, activating cellular apoptosis [49]. Independent observations found increased levels of TXNIP in neurodegenerative disease [49] and low levels of SELENOM [50,51]. In the absence of SELENOM, ER stress increases, promoting the expression of TXNIP and neuronal apoptosis and inflammation (Figure 4a) [49,52,53]. In cancers, the opposite has been observed, wherein TXNIP levels are lowered and SELENOM levels are higher [49]. The high levels of SELENOM reduce the ROS species, lowering the expression of TXNIP and promoting cellular growth (Figure 4b).
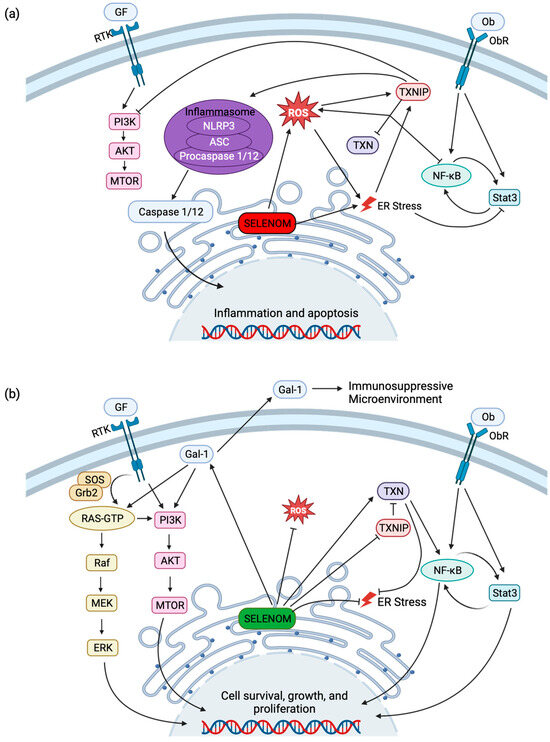
Figure 4.
SELENOM and related signaling pathways. (a) Low levels of SELENOM (red) are correlated with an increase in ROS and may induce ER stress. Elevated ROS and ER stress increase TXNIP activity, downregulating TXN activity, thereby resulting in the suppression of NF-κB and increase in ROS. TXNIP suppresses cellular growth and survival by inhibition of PI3K/AKT/mTOR signaling and promotes the caspase-mediated induction of inflammatory and apoptotic genes by activating the NLRP3 inflammasome. (b) High levels (green) of SELENOM decrease ROS generation and promote TXN activity by the suppression of TXNIP. Promotion of transcription factors NF-κB and Stat3 by leptin signaling and TXN activity as well as SELENOM/Gal-1 activation of PI3K/AKT/mTOR and Ras/Raf/MEK/ERK signaling results in the transcription of pro-survival, growth, and proliferation genes. Gal-1 also is secreted in the extracellular space as an anti-inflammatory signaling molecule. Created with BioRender.com.
5.2. Galectin-1
To identify other protein partners that could be involved in neuroprotection, a yeast two-hybrid system was used to screen a human fetal brain cDNA library. This resulted in the identification of galectin-1 (Gal-1) as an interactive binding partner of SELENOM [54]. Gal-1 is differentially expressed across many tissues and is suggested to have a wide range of biological activity both intra- and extracellularly. With respect to the brain, Gal-1 is found across the central and peripheral nervous systems, involved in nerve development and axon regeneration. Specifically, oxidized Gal-1 has been shown to promote neurite outgrowth and enhance axonal regeneration in nerves [55]. The detailed interaction of SELENOM and Gal-1 is not yet described; however, we know that Sec is not required for binding since the pull-down assay was performed with a Sec-to-Cys mutation in SELENOM. It is possible, however, that if SELENOM is involved in the oxidation of Gal-1, the presence of a Sec would be more efficient in the oxidoreductase activity than with Cys.
5.3. Cytoplasmic Actins
The presence of the CXXU motif in SELENOM, as mentioned above, suggests oxidoreductase activity like TXN. TXN activity functions through the formation of disulfide bonds using its CXXC motif. To search for proteins that interact with the proposed catalytic site of SELENOM, purified protein was incubated with cancer cell lysates (HT-1080 and MCF-7), followed by reduction to release proteins interacting through a disulfide bond. The multi-step translation path for Sec insertion challenges recombinant selenoprotein production [13]. Therefore, the search for interacting proteins was performed with a Sec-to-Cys mutation in SELENOM. A similar strategy has been used previously to screen for protein partners with other selenoproteins (SELENOV and SELENOW) [56,57]. This experiment led to the detection of cytoplasmic actin 1 and 2 (β- and γ-actin), key proteins in adhesion, migration, polarization, and mitosis of cells [58]. Interaction via a disulfide suggests that SELENOM is involved in reducing oxidized actin, playing a role in regulating the actin cytoskeleton. These cellular processes are involved in the metastasis of tumors and further enhance the explanation for the role of increased SELENOM that is observed in some cancers [12].
5.4. Osteoblast Colocalization
SELENOM has been shown to have a role in bone development as well as in cartilage formation. In situ hybridization studies on chickens detected SELENOM mRNA expression in the condensed mesenchyme, a cluster of cells that later differentiate into bone [59]. Furthermore, in situ hybridization on embryonic day 16.5 mice revealed SELENOM expression in the vertebrae, maxilla, mandibula, trabecular and periosteum long bones, and the olfactory epithelium. SELENOM expression was also observed in the teeth and bones of postnatal day 4 mice [60]. To further the involvement of SELENOM in bone formation, SELENOM expression was found to colocalize with markers for osteoblasts, such as the early osteoblast marker Bglap [59,60]. Deficiency in selenium leads to a reduction in SELENOM expression by almost 70% in chicken cartilage tissues [60]. While the evidence suggests that SELENOM functions in bone development, the understanding of mechanisms and key players involved in this process is still a mystery. Knowledge of this can bring new insights into healing fractured or broken bones and to combat developmental or adult bone disorders.
5.5. Hypothalamic Leptin Signalling
SELENOM is found to be highly expressed in several regions of the brain. In efforts to identify its role, a global knockout of the SELENOM gene (SelenoM−/−) in mice was generated. As these mice developed, they were found to have increased body weight, elevated white adipose tissue levels, and reduced leptin response [24]. Leptin is a hormone that acts within the hypothalamus to regulate energy balance via signaling pathways to suppress appetite or increase energy expenditure. When the hypothalamus does not respond normally to these leptin signals (i.e., leptin resistance), this results in obesity. One established cause of leptin resistance is continual stress in the ER, which can be caused by either accumulation of misfolded proteins, Ca2+ depletion, or a combination of the two [61,62]. The proposed oxidoreductase activity of SELENOM from its CXXU motif suggests its function in the maturation of proteins through disulfide formation. Therefore, without SELENOM present in the mouse, misfolded proteins could accumulate, inducing ER stress. Moreover, in vitro studies found SELENOM to be involved in Ca2+ homeostasis. Overexpression of SELENOM in HT22 and C8-D1A cell lines reduced changes in cytosolic Ca2+ levels that would otherwise increase during oxidative stress. The increased basal levels observed in the knock-down suggest that Ca2+ regulation is impaired preventing any response to increased Ca2+ levels [40]. This was further corroborated with SELENOM shRNA-expressing hypothalamic cells (silencing expression of the protein), where Ca2+ levels remained unchanged under leptin treatment [25]. These data propose that SELENOM facilitates communication between the leptin receptor and ER proteins involved in Ca2+ signaling. A hypothesis based off the mechanism of other selenoproteins [63,64], is that this is accomplished by indirectly reducing the thiol groups on ER Ca2+ channels or pumps.
6. SELENOM Implications in Disease
To further interrogate the localization and function of SELENOM, we investigated the diseases associated with aberrations in normal SELENOM expression. In this section, we summarize key findings connecting SELENOM to diseases such as Alzheimer’s dementia (AD), non-alcoholic fatty liver disease (NAFLD), and cancers. These are the most studied and well-known diseases that SELENOM has been associated with.
6.1. Alzheimer’s Disease
Alzheimer’s dementia (AD) is a neurodegenerative disease that results in progressive loss of cognition and ultimately dementia. There are three known genes wherein mutations cause the development of AD: amyloid precursor protein (APP), presenilin-1 (PSEN-1), and presenilin-2 (PSEN-2) [65]. PSEN-1 and -2 are components of γ-secretase, which is responsible for the cleavage of APP to produce amyloid-β (Aβ). Mutations in human PSEN genes alter the cleavage of APP by γ-secretase, resulting in a higher ratio of Aβ isoform Aβ42 and the development of early onset familial AD (eFAD) [66,67]. eFAD makes up a small percentage (less than 5%) of AD in humans caused by a single genetic mutation [68]. Although single gene mutation mouse models of AD typically present with just one hallmark of AD and lack other pathologies routinely observed in patients with AD, they still provide a useful tool to study such a complex, typically multigenetic and multifactorial disorder whose etiology remains largely unclear.
The connection of selenoproteins to AD was found through a specific knockout of Trsp (the gene for tRNASec) in mouse neurons. This disrupts the ability to express selenoproteins (including SELENOM) and induces a neurodevelopmental and neurodegenerative phenotype affecting the cerebral cortex and hippocampus [50]. The similar yet milder phenotype found in Gpx4-deficient mice suggests that GPx4 could be one of the major selenoproteins involved [69]. However, there must be at least one additional selenoprotein participating in neuroprotection and neural development to reach the extent of the phenotype observed through Trsp knockout. In a mouse model of AD that expresses human mutant PSEN-2 (N141I), SELENOM expression was found to be downregulated [51]. These data propose that SELENOM may be one of the missing selenoproteins involved in neurodegeneration. The pathogenicity of AD has been correlated to the secretase-mediated generation of Aβ42 peptides deposited at neuritic plaques [70]. Selenium has been found to decrease the γ-secretase activity in mice through activation of the extracellular-signaling-regulated kinase (ERK) pathway [71]. Overexpression of human SELENOM in addition to selenium supplementation further decreased the γ-secretase activity, while a decrease in α-secretase activity and an increase in β-secretase activity was also observed. The combination of these changes contributed to a decrease in Aβ42 production, lowering the progression of AD [72]. Being that SELENOM is a selenoprotein, the natural question to ask is whether the Sec residue is involved in protein function. In a study using HEK293 cells, cotransfection of Aβ42 with either SELENOM or SELENOM’ (a SELENOM variant in which the Sec is replaced with Cys) resulted in a significant reduction of Aβ42 aggregates in vitro [73]. This suggests that SELENOM prevention of Aβ42 aggregates is not influenced by the presence of Sec. Additionally, cotransfection of HEK293 cells with Aβ42 and an empty vector produces abnormal mitochondrial localization near Aβ plaques and swollen morphology. Addition of either SELENOM or SELENOM’ restores mitochondrial localization and morphology by preventing an increase in intracellular ROS [73]. Therefore, SELENOM may serve a neuroprotective role through exerting antioxidant activity in the brain.
Another hallmark of AD is the development of neurofibril tangles (NFT) due to the aggregation of hyperphosphorylated tau. NFTs purified from AD brains are enriched with hyperphosphorylated tau [74]. It has been shown that this hyperphosphorylation reduces tau’s ability to bind to microtubules, promoting self-aggregation and formation of NFTs in the cytoplasm of neurodegenerative disease cells [75,76,77]. Thus, uncovering mechanisms that reduce tau phosphorylation is attractive for the purposes of preventing NFT formation. ERK pathway activation via SELENOM overexpression and selenium supplementation led to decreased tau phosphorylation at three sites, Ser404, Ser202, and Thr231 [72]. Taken together, the data in this section indicate a connection of SELENOM expression with neuronal processes.
6.2. Cancer
SELENOM has also been studied in the realm of cancer biology with altered SELENOM expression observed in human tumor tissues. In some tumor types such as lymphoma, breast, fallopian tube, and ovarian cancers, SELENOM expression is decreased, while in others such as in parotid and uterine tumors, it is increased [1]. Human renal cell carcinoma (RCC) is a tumor type that has increased levels of SELENOM expression compared to normal kidney tissue. SELENOM can be used for prognosis with higher levels correlated to shorter overall patient survival [78]. This is suggested to be due to the role that SELENOM plays in cell survival; in vitro SELENOM silencing resulted in lower cell viability, and SELENOM overexpression produced increased cell viability. Moreover, SELENOM knockdown reduced the migratory capacity of two cell lines (CAKI-1 and 786O) derived from RCC [78]. For tumor cells to incur metastatic ability, they must achieve epithelial–mesenchymal transition (EMT). In the absence of SELENOM in CAKI-1 and 786O cells, key EMT-related genes were significantly downregulated, namely, N-cadherin, vimentin, β-catenin, and matrix metalloproteinase (MMP)-2 and MMP-9. Furthermore, the pathways involved in tumor growth and progression (PI3K/AKT/mTOR signaling) were also downregulated in the absence of SELENOM [78]. This demonstrates that SELENOM is positively correlated with the metastatic ability of RCC cells. RCC is one type of cancer that has been specifically investigated as a prognostic marker; however, other cell carcinomas require further studies [79].
The mitogen-activated protein kinase (MAPK) pathway is activated by selenium supplementation and SELENOM overexpression [72]. Specifically, the Raf/MEK/ERK pathway has been shown to be correlated with multiple cancer types such as in RCC, hepatocellular carcinoma (HCC), non-small cell lung cancer (NSCLC), melanoma, and papillary thyroid carcinoma [80]. ERK has been shown to promote cell migration, proliferation, and viability and has been associated with HCC progression. P38 and Jun N-terminal kinases, members of the two other MAPK pathways, have been associated with HCC development [81]. Moreover, higher SELENOM expression was discovered in hepatoma cell lines when compared to normal hepatocytes [82], and its overexpression in HCC liver tissues warranted its proposal as a putative marker for HCC [83]. Together, these reports elucidate another correlation between SELENOM and cancer progression.
A study in 2016 investigated the expression of six selenoproteins (SELENOH, SELENOK, SELENOM, SELENOS, SELENOV, and GPx6) in a variety of tumor cell lines: HT 1080 (fibrosarcoma), HepG2 (hepatocellular carcinoma), MCF7 (breast adenocarcinoma), A-172 (glioblastoma), HeLa (cervical adenocarcinoma), and DU-145 (prostate carcinoma). All six of the tumor cell lines investigated were found to express SELENOM mRNA at a relatively consistent level, while SELENOH and SELENOK levels varied more. In some cell lines they were expressed significantly more than SELENOM (HT-1080, HepG2, and HeLa), while in others, their expression levels were similar (MCF7, A-172, and DU-145). SELENOS, SELENOV, and GPx6 on the other hand were not detected in any of these six tumor cell lines [84]. Given the pro-growth attributes of SELENOM, its dysregulation may promote tumor growth and metastasis in multiple tissue types and forms of cancer.
6.3. Non-Alcoholic Fatty Liver Disease
The high prevalence of obesity worldwide poses a major risk for the development of NAFLD, the most common chronic liver disease type [85]. In NAFLD, patient health is complicated by abnormal lipid metabolism, mitochondrial dysfunction, oxidative stress, and lipid peroxidation [86]. NAFLD can develop from a high-fat diet (HFD) and is typically accompanied by an accumulation of lipid in the liver followed by lipotoxicity-induced hepatocyte inflammation and apoptosis [87]. SELENOM was recently connected to NAFLD utilizing an in vivo mouse model of HFD-induced NAFLD and an in vitro model of hepatocyte palmitic acid (PA)-mediated lipotoxicity [88]. In the in vivo model, a reduction in SELENOM mRNA and protein levels were accompanied by significantly higher body weight and increased levels of hepatic metabolism markers, liver weight, hepatic vacuolization, steatosis, nuclear atrophy, and overall liver degeneration including an increase in liver-fibrosis-related genes. Liver-specific knockout of SELENOM (Liv-SELENOM−/−) exacerbated these in vivo results. The in vitro model showed similar data, with reduced SELENOM mRNA and protein levels compared to non-PA-treated hepatocytes. Upon SELENOM overexpression, liver-fibrosis-related genes were restored [88]. These results demonstrate that HFD- and PA-mediated lipotoxicity downregulate SELENOM expression, and deletion of SELENOM worsens the development of HFD-induced NAFLD hepatic injury. Further laboratory and human studies have yet to be performed to confirm a connection between SELENOM and its potential to protect against hepatic injury.
Another observation in the NAFLD models was the higher levels of inflammatory markers such as TNF, interleukin (IL) 6, and IL1b inflammatory cytokines [88]. Chronic inflammation is a hallmark of NAFLD, which leads to cirrhosis, fibrosis, liver failure, and potentially hepatocellular carcinoma [89,90]. The connection to SELENOM was inferred when overexpression in the in vitro model restored expression levels of inflammatory markers and genes involved in oxidative stress and fatty acid oxidation (FAO) [88]. These have been shown to play a major role in hepatocyte lipid metabolism and the development of NAFLD [91]. A HFD results in reduced expression of antioxidant markers, increased lipid peroxidation, downregulated FAO genes, and increased lipogenic genes, which becomes further enhanced in Liv-SELENOM−/−. However, as with the inflammatory markers, overexpression of SELENOM restored all genes to their appropriate levels [88]. Together, these results suggest that SELENOM is also protective against oxidative stress and promotes lipid metabolism in hepatocytes. As we have discussed above, SELENOM is involved in the regulation of ROS, which is suggested to be important for neuroprotection in AD. Therefore, these antioxidant properties of SELENOM may be important in multiple tissues.
Further results indicated that SELENOM modulates mitochondrial stress by activating the Parkin-related mitophagy via the AMPKα1–MFN2 pathway. Mitochondrial apoptosis and mitophagy play central roles in lipotoxicity and in maintaining mitochondria quality in hepatocytes [92,93,94]. In HFD-fed mice, levels of pro-apoptotic genes were increased, and anti-apoptotic genes decreased. These mice also exhibited lower levels of mitophagy-related genes, which were further altered in HFD-fed Liv-SELENOM−/− mice. These results paralleled with the in vitro model, with SELENOM overexpression returning the pro- and anti-apoptotic and mitophagy related genes to their respective levels [88]. Together these findings allude to the role of SELENOM in promoting hepatocyte survival by regulating genes involved in apoptosis and inducing mitophagy. This supports the hypothesis that SELENOM is involved in cell survival.
7. Outlook
Selenoproteins are critical for human function, and of the 25 identified in humans, there is still much we do not know about their role in facilitating cellular homeostasis. A better understanding of the mechanism by which Sec facilitates the function of these proteins will help guide prescription of selenium supplementation for various diseases. In this review, we have outlined what has been established for SELENOM, a small oxidoreductase widely expressed throughout the body [22,23]. We first compared structural similarities to functions. The closest structural relative to SELENOM, SELENOF, was unable to provide any functional information aside from ruling out an interaction with UGGT. However, the next closest relative is the more well-known oxidoreductase, TXN. Given that TXN is involved in disulfide bond formation, oxidoreductase assays were tested with SELENOM and found to have both TXN and GPx activity, but not TXNRD activity [25,39,40]. This infers that SELENOM oxidoreductase activity can be activated by redox stress but may also play a role in a TXN-like catalytic cycle.
Further investigation into the TXN-like behavior of SELENOM found that SELENOM might interact with TXNIP through a disulfide or selenyl-sulfide bond, modulating cellular homeostasis. This correlation has been observed in cancer in which cell growth is promoted due to high levels of SELENOM and low levels of TXNIP [49]. The opposite is true in AD, where low levels of SELENOM and high levels of TXNIP promote neuronal apoptosis due to oxidative damage [50,51]. Other interacting proteins include cellular actin, which has been specifically identified to form a disulfide (or selenyl-sulfide) bond with SELENOM [58], and Gal-1, where the binding mechanism has not been identified [54]. An imbalance of SELENOM can have deleterious effects, but what is more apparent is that SELENOM strikes a balance with its binding partners such as TXNIP; too much can cause increased cell growth and cancers, too little can cause neurodegenerative diseases. As details emerge on the interactive partners of SELENOM, we can begin to connect these interactions to observed diseases through continued fusion of mouse models, human studies, and in vitro laboratory experiments.
Combining what is known thus far, we can conclude that SELENOM is heavily involved in cellular homeostasis, through its CXXU motif. With growing technology to enable selenoprotein overexpression, outstanding questions about SELENOM and the interaction with its protein partners can be tackled. While SELENOM plays a role in certain cancers, neither reducing daily selenium intake nor having an excess of it is a straightforward remedy. Both low and high levels of selenium can be detrimental, as selenium is essential for other selenoproteins in the body that contribute to various physiological processes [1,95]. Furthermore, a direct correlation between dietary selenium levels and selenoprotein-related cancers has yet to be conclusively established. Continued efforts to unveil the functional mechanism of this protein, along with the other 24 human selenoproteins, is imperative for effective treatment of diseases.
Author Contributions
Conceptualization, N.K.; writing—original draft preparation, L.G.A.N., C.C. and N.K.; writing—review and editing, A.C., L.G.A.N., P.R.H. and N.K.; visualization, A.C, L.G.A.N. and N.K.; supervision, P.R.H. and N.K.; funding acquisition, N.K. and P.R.H. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by the National Institute of Allergy and Infectious Diseases, grant number 5R01AI147496 to P.R.H., and the University of Georgia Provost’s Office and Franklin College of Arts and Sciences to N.K.
Data Availability Statement
No new data were created or analyzed in this study. Data sharing is not applicable to this article.
Acknowledgments
We thank Dieter Söll (Yale University) and Oscar Vargas-Rodriguez (University of Connecticut Health) for their insightful discussions.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Rayman, M.P. Selenium intake, status, and health: A complex relationship. Hormones 2020, 19, 9–14. [Google Scholar] [CrossRef] [PubMed]
- Marschall, T.A.; Bornhorst, J.; Kuehnelt, D.; Schwerdtle, T. Differing cytotoxicity and bioavailability of selenite, methylselenocysteine, selenomethionine, selenosugar 1 and trimethylselenonium ion and their underlying metabolic transformations in human cells. Mol. Nutr. Food Res. 2016, 60, 2622–2632. [Google Scholar] [CrossRef] [PubMed]
- Lazard, M.; Dauplais, M.; Blanquet, S.; Plateau, P. Recent advances in the mechanism of selenoamino acids toxicity in eukaryotic cells. Biomol. Concepts 2017, 8, 93–104. [Google Scholar] [CrossRef] [PubMed]
- Speckmann, B.; Steinbrenner, H. Selenium and selenoproteins in inflammatory bowel diseases and experimental colitis. Inflamm. Bowel Dis. 2014, 20, 1110–1119. [Google Scholar] [CrossRef]
- Avery, J.C.; Hoffmann, P.R. Selenium, selenoproteins, and immunity. Nutrients 2018, 10, 1203. [Google Scholar] [CrossRef]
- Fairweather-Tait, S.J.; Filippini, T.; Vinceti, M. Selenium status and immunity. Proc. Nutr. Soc. 2022, 82, 32–38. [Google Scholar] [CrossRef]
- Vinceti, M.; Filippini, T.; Jablonska, E.; Saito, Y.; Wise, L.A. Safety of selenium exposure and limitations of selenoprotein maximization: Molecular and epidemiologic perspectives. Environ. Res. 2022, 211, 113092. [Google Scholar] [CrossRef]
- Urbano, T.; Filippini, T.; Lasagni, D.; De Luca, T.; Sucato, S.; Polledri, E.; Bruzziches, F.; Malavolti, M.; Baraldi, C.; Santachiara, A.; et al. Associations between urinary and dietary selenium and blood metabolic parameters in a healthy northern italy population. Antioxidants 2021, 10, 1193. [Google Scholar] [CrossRef]
- Urbano, T.; Vinceti, M.; Mandrioli, J.; Chiari, A.; Filippini, T.; Bedin, R.; Tondelli, M.; Simonini, C.; Zamboni, G.; Shimizu, M.; et al. Selenoprotein P concentrations in the cerebrospinal fluid and serum of individuals affected by amyotrophic lateral sclerosis, mild cognitive impairment and Alzheimer’s Dementia. Int. J. Mol. Sci. 2022, 23, 9865. [Google Scholar] [CrossRef]
- Huang, Y.C.; Combs, G.F., Jr.; Wu, T.L.; Zeng, H.; Cheng, W.H. Selenium status and type 2 diabetes risk. Arch. Biochem. Biophys. 2022, 730, 109400. [Google Scholar] [CrossRef]
- Steinbrenner, H.; Duntas, L.H.; Rayman, M.P. The role of selenium in type-2 diabetes mellitus and its metabolic comorbidities. Redox Biol. 2022, 50, 102236. [Google Scholar] [CrossRef] [PubMed]
- Rayman, M.P. Selenium and human health. Lancet 2012, 379, 1256–1268. [Google Scholar] [CrossRef] [PubMed]
- Chung, C.Z.; Krahn, N. The selenocysteine toolbox: A guide to studying the 21st amino acid. Arch. Biochem. Biophys. 2022, 730, 109421. [Google Scholar] [CrossRef] [PubMed]
- Labunskyy, V.M.; Hatfield, D.L.; Gladyshev, V.N. Selenoproteins: Molecular pathways and physiological roles. Physiol. Rev. 2014, 94, 739–777. [Google Scholar] [CrossRef] [PubMed]
- Krahn, N.; Fischer, J.T.; Söll, D. Naturally occurring tRNAs with non-canonical structures. Front. Microbiol. 2020, 11, 596914. [Google Scholar] [CrossRef] [PubMed]
- Serrão, V.H.B.; Silva, I.R.; da Silva, M.T.A.; Scortecci, J.F.; de Freitas Fernandes, A.; Thiemann, O.H. The unique tRNASec and its role in selenocysteine biosynthesis. Amino Acids 2018, 50, 1145–1167. [Google Scholar] [CrossRef]
- Hilal, T.; Killam, B.Y.; Grozdanović, M.; Dobosz-Bartoszek, M.; Loerke, J.; Bürger, J.; Mielke, T.; Copeland, P.R.; Simonović, M.; Spahn, C.M.T. Structure of the mammalian ribosome as it decodes the selenocysteine UGA codon. Science 2022, 376, 1338–1343. [Google Scholar] [CrossRef]
- Peng, J.J.; Yue, S.Y.; Fang, Y.H.; Liu, X.L.; Wang, C.H. Mechanisms affecting the biosynthesis and incorporation rate of selenocysteine. Molecules 2021, 26, 7120. [Google Scholar] [CrossRef]
- Mukai, T.; Sevostyanova, A.; Suzuki, T.; Fu, X.; Söll, D. A facile method for producing selenocysteine-containing proteins. Angew. Chem. Int. Ed. Engl. 2018, 57, 7215–7219. [Google Scholar] [CrossRef]
- Cheng, Q.; Arnér, E.S. Selenocysteine insertion at a predefined UAG codon in a release factor 1 (RF1)-depleted Escherichia coli host strain bypasses species barriers in recombinant selenoprotein translation. J. Biol. Chem. 2017, 292, 5476–5487. [Google Scholar] [CrossRef]
- Novoselov, S.V.; Lobanov, A.V.; Hua, D.; Kasaikina, M.V.; Hatfield, D.L.; Gladyshev, V.N. A highly efficient form of the selenocysteine insertion sequence element in protozoan parasites and its use in mammalian cells. Proc. Natl. Acad. Sci. USA 2007, 104, 7857–7862. [Google Scholar] [CrossRef] [PubMed]
- Korotkov, K.V.; Novoselov, S.V.; Hatfield, D.L.; Gladyshev, V.N. Mammalian selenoprotein in which selenocysteine (Sec) incorporation is supported by a new form of Sec insertion sequence element. Mol. Cell. Biol. 2002, 22, 1402–1411. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhou, Y.; Schweizer, U.; Savaskan, N.E.; Hua, D.; Kipnis, J.; Hatfield, D.L.; Gladyshev, V.N. Comparative analysis of selenocysteine machinery and selenoproteome gene expression in mouse brain identifies neurons as key functional sites of selenium in mammals. J. Biol. Chem. 2008, 283, 2427–2438. [Google Scholar] [CrossRef]
- Pitts, M.W.; Reeves, M.A.; Hashimoto, A.C.; Ogawa, A.; Kremer, P.; Seale, L.A.; Berry, M.J. Deletion of selenoprotein M leads to obesity without cognitive deficits. J. Biol. Chem. 2013, 288, 26121–26134. [Google Scholar] [CrossRef] [PubMed]
- Gong, T.; Hashimoto, A.C.; Sasuclark, A.R.; Khadka, V.S.; Gurary, A.; Pitts, M.W. Selenoprotein M promotes hypothalamic leptin signaling and thioredoxin antioxidant activity. Antioxid. Redox Signal. 2021, 35, 775–787. [Google Scholar] [CrossRef] [PubMed]
- Sasuclark, A.R.; Khadka, V.S.; Pitts, M.W. Cell-type specific analysis of selenium-related genes in brain. Antioxidants 2019, 8, 120. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, A.D.; Labunskyy, V.M.; Fomenko, D.E.; Araç, D.; Chelliah, Y.; Amezcua, C.A.; Rizo, J.; Gladyshev, V.N.; Deisenhofer, J. NMR structures of the selenoproteins Sep15 and SelM reveal redox activity of a new thioredoxin-like family. J. Biol. Chem. 2006, 281, 3536–3543. [Google Scholar] [CrossRef]
- Oberacker, T.; Kraft, L.; Schanz, M.; Latus, J.; Schricker, S. The importance of thioredoxin-1 in health and disease. Antioxidants 2023, 12, 1078. [Google Scholar] [CrossRef]
- Lu, J.; Holmgren, A. The thioredoxin antioxidant system. Free Radic. Biol. Med. 2014, 66, 75–87. [Google Scholar] [CrossRef]
- Muri, J.; Kopf, M. The thioredoxin system: Balancing redox responses in immune cells and tumors. Eur. J. Immunol. 2023, 53, e2249948. [Google Scholar] [CrossRef]
- Alanen, H.I.; Williamson, R.A.; Howard, M.J.; Lappi, A.K.; Jantti, H.P.; Rautio, S.M.; Kellokumpu, S.; Ruddock, L.W. Functional characterization of ERp18, a new endoplasmic reticulum-located thioredoxin superfamily member. J. Biol. Chem. 2003, 278, 28912–28920. [Google Scholar] [CrossRef] [PubMed]
- Anelli, T.; Alessio, M.; Mezghrani, A.; Simmen, T.; Talamo, F.; Bachi, A.; Sitia, R. ERp44, a novel endoplasmic reticulum folding assistant of the thioredoxin family. EMBO J. 2002, 21, 835–844. [Google Scholar] [CrossRef] [PubMed]
- Flowers, B.; Bochnacka, O.; Poles, A.; Diamond, A.M.; Kastrati, I. Distinct roles of SELENOF in different human cancers. Biomolecules 2023, 13, 486. [Google Scholar] [CrossRef] [PubMed]
- Korotkov, K.V.; Kumaraswamy, E.; Zhou, Y.; Hatfield, D.L.; Gladyshev, V.N. Association between the 15-kDa selenoprotein and UDP-glucose:glycoprotein glucosyltransferase in the endoplasmic reticulum of mammalian cells. J. Biol. Chem. 2001, 276, 15330–15336. [Google Scholar] [CrossRef] [PubMed]
- Qi, Y.; Grishin, N.V. Structural classification of thioredoxin-like fold proteins. Proteins 2005, 58, 376–388. [Google Scholar] [CrossRef]
- Labunskyy, V.M.; Ferguson, A.D.; Fomenko, D.E.; Chelliah, Y.; Hatfield, D.L.; Gladyshev, V.N. A novel cysteine-rich domain of Sep15 mediates the interaction with UDP-glucose:glycoprotein glucosyltransferase. J. Biol. Chem. 2005, 280, 37839–37845. [Google Scholar] [CrossRef]
- Lubos, E.; Loscalzo, J.; Handy, D.E. Glutathione peroxidase-1 in health and disease: From molecular mechanisms to therapeutic opportunities. Antioxid. Redox Signal. 2011, 15, 1957–1997. [Google Scholar] [CrossRef]
- Margis, R.; Dunand, C.; Teixeira, F.K.; Margis-Pinheiro, M. Glutathione peroxidase family—An evolutionary overview. FEBS J. 2008, 275, 3959–3970. [Google Scholar] [CrossRef]
- Jerome-Morais, A.; Bera, S.; Rachidi, W.; Gann, P.H.; Diamond, A.M. The effects of selenium and the GPx-1 selenoprotein on the phosphorylation of H2AX. Biochim. Biophys. Acta 2013, 1830, 3399–3406. [Google Scholar] [CrossRef][Green Version]
- Reeves, M.A.; Bellinger, F.P.; Berry, M.J. The neuroprotective functions of selenoprotein M and its role in cytosolic calcium regulation. Antioxid. Redox Signal. 2010, 12, 809–818. [Google Scholar] [CrossRef]
- Ouyang, Y.; Peng, Y.; Li, J.; Holmgren, A.; Lu, J. Modulation of thiol-dependent redox system by metal ions via thioredoxin and glutaredoxin systems. Metallomics 2018, 10, 218–228. [Google Scholar] [CrossRef] [PubMed]
- Arnér, E.S.; Holmgren, A. Physiological functions of thioredoxin and thioredoxin reductase. Eur. J. Biochem. 2000, 267, 6102–6109. [Google Scholar] [CrossRef]
- Le, N.Q.; Ou, Y.Y. Prediction of FAD binding sites in electron transport proteins according to efficient radial basis function networks and significant amino acid pairs. BMC Bioinform. 2016, 17, 298. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z. Antioxidant activity of the thioredoxin system. Biophys. Rep. 2023, 9, 26–32. [Google Scholar] [CrossRef] [PubMed]
- Qayyum, N.; Haseeb, M.; Kim, M.S.; Choi, S. Role of thioredoxin-interacting protein in diseases and its therapeutic outlook. Int. J. Mol. Sci. 2021, 22, 2754. [Google Scholar] [CrossRef] [PubMed]
- Jin, H.O.; Seo, S.K.; Kim, Y.S.; Woo, S.H.; Lee, K.H.; Yi, J.Y.; Lee, S.J.; Choe, T.B.; Lee, J.H.; An, S.; et al. TXNIP potentiates Redd1-induced mTOR suppression through stabilization of Redd1. Oncogene 2011, 30, 3792–3801. [Google Scholar] [CrossRef]
- Blouet, C.; Liu, S.M.; Jo, Y.H.; Chua, S.; Schwartz, G.J. TXNIP in Agrp neurons regulates adiposity, energy expenditure, and central leptin sensitivity. J. Neurosci. 2012, 32, 9870–9877. [Google Scholar] [CrossRef]
- Blouet, C.; Schwartz, G.J. Nutrient-sensing hypothalamic TXNIP links nutrient excess to energy imbalance in mice. J. Neurosci. 2011, 31, 6019–6027. [Google Scholar] [CrossRef]
- Pan, M.; Zhang, F.; Qu, K.; Liu, C.; Zhang, J. TXNIP: A double-edged sword in disease and therapeutic outlook. Oxid. Med. Cell. Longev. 2022, 2022, 7805115. [Google Scholar] [CrossRef]
- Wirth, E.K.; Conrad, M.; Winterer, J.; Wozny, C.; Carlson, B.A.; Roth, S.; Schmitz, D.; Bornkamm, G.W.; Coppola, V.; Tessarollo, L.; et al. Neuronal selenoprotein expression is required for interneuron development and prevents seizures and neurodegeneration. FASEB J. 2010, 24, 844–852. [Google Scholar] [CrossRef]
- Hwang, D.Y.; Cho, J.S.; Oh, J.H.; Shim, S.B.; Jee, S.W.; Lee, S.H.; Seo, S.J.; Lee, S.K.; Lee, S.H.; Kim, Y.K. Differentially expressed genes in transgenic mice carrying human mutant presenilin-2 (N141I): Correlation of selenoprotein M with Alzheimer’s disease. Neurochem. Res. 2005, 30, 1009–1019. [Google Scholar] [CrossRef] [PubMed]
- Gong, T.; Torres, D.J.; Berry, M.J.; Pitts, M.W. Hypothalamic redox balance and leptin signaling—Emerging role of selenoproteins. Free. Radic. Biol. Med. 2018, 127, 172–181. [Google Scholar] [CrossRef] [PubMed]
- Lane, T.; Flam, B.; Lockey, R.; Kolliputi, N. TXNIP shuttling: Missing link between oxidative stress and inflammasome activation. Front. Physiol. 2013, 4, 50. [Google Scholar] [CrossRef]
- Qiao, X.; Tian, J.; Chen, P.; Wang, C.; Ni, J.; Liu, Q. Galectin-1 is an interactive protein of selenoprotein M in the brain. Int. J. Mol. Sci. 2013, 14, 22233–22245. [Google Scholar] [CrossRef]
- Camby, I.; Le Mercier, M.; Lefranc, F.; Kiss, R. Galectin-1: A small protein with major functions. Glycobiology 2006, 16, 137R–157R. [Google Scholar] [CrossRef] [PubMed]
- Dikiy, A.; Novoselov, S.V.; Fomenko, D.E.; Sengupta, A.; Carlson, B.A.; Cerny, R.L.; Ginalski, K.; Grishin, N.V.; Hatfield, D.L.; Gladyshev, V.N. SelT, SelW, SelH, and Rdx12: Genomics and molecular insights into the functions of selenoproteins of a novel thioredoxin-like family. Biochemistry 2007, 46, 6871–6882. [Google Scholar] [CrossRef] [PubMed]
- Varlamova, E.G.; Novoselov, S.V.; Novoselov, V.I.; Fesenko, E.E. New mammalian selenium-containing protein V: The search for protein partners. Dokl. Biochem. Biophys. 2011, 441, 255–257. [Google Scholar] [CrossRef]
- Varlamova, E.G.; Goltyaev, M.V.; Fesenko, E.E. Protein partners of selenoprotein SELM and the role of selenium compounds in regulation of its expression in human cancer cells. Dokl. Biochem. Biophys. 2019, 488, 300–303. [Google Scholar] [CrossRef]
- Chi, Q.; Luan, Y.; Zhang, Y.; Hu, X.; Li, S. The regulatory effects of miR-138-5p on selenium deficiency-induced chondrocyte apoptosis are mediated by targeting SelM. Metallomics 2019, 11, 845–857. [Google Scholar] [CrossRef]
- Grosch, M.; Fuchs, J.; Bosl, M.; Winterpacht, A.; Tagariello, A. Selenoprotein M is expressed during bone development. EXCLI J. 2013, 12, 967–979. [Google Scholar]
- Ozcan, L.; Ergin, A.S.; Lu, A.; Chung, J.; Sarkar, S.; Nie, D.; Myers, M.G., Jr.; Ozcan, U. Endoplasmic reticulum stress plays a central role in development of leptin resistance. Cell Metab. 2009, 9, 35–51. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhang, G.; Zhang, H.; Karin, M.; Bai, H.; Cai, D. Hypothalamic IKKβ/NF-κB and ER stress link overnutrition to energy imbalance and obesity. Cell 2008, 135, 61–73. [Google Scholar] [CrossRef] [PubMed]
- Marino, M.; Stoilova, T.; Giorgi, C.; Bachi, A.; Cattaneo, A.; Auricchio, A.; Pinton, P.; Zito, E. SEPN1, an endoplasmic reticulum-localized selenoprotein linked to skeletal muscle pathology, counteracts hyperoxidation by means of redox-regulating SERCA2 pump activity. Hum. Mol. Genet. 2015, 24, 1843–1855. [Google Scholar] [CrossRef] [PubMed]
- Fredericks, G.J.; Hoffmann, F.W.; Rose, A.H.; Osterheld, H.J.; Hess, F.M.; Mercier, F.; Hoffmann, P.R. Stable expression and function of the inositol 1,4,5-triphosphate receptor requires palmitoylation by a DHHC6/selenoprotein K complex. Proc. Natl. Acad. Sci. USA 2014, 111, 16478–16483. [Google Scholar] [CrossRef] [PubMed]
- Bateman, R.J.; Aisen, P.S.; De Strooper, B.; Fox, N.C.; Lemere, C.A.; Ringman, J.M.; Salloway, S.; Sperling, R.A.; Windisch, M.; Xiong, C. Autosomal-dominant Alzheimer’s disease: A review and proposal for the prevention of Alzheimer’s disease. Alzheimers Res. Ther. 2011, 3, 1. [Google Scholar] [CrossRef]
- Scheuner, D.; Eckman, C.; Jensen, M.; Song, X.; Citron, M.; Suzuki, N.; Bird, T.D.; Hardy, J.; Hutton, M.; Kukull, W.; et al. Secreted amyloid β-protein similar to that in the senile plaques of Alzheimer’s disease is increased in vivo by the presenilin 1 and 2 and APP mutations linked to familial Alzheimer’s disease. Nat. Med. 1996, 2, 864–870. [Google Scholar] [CrossRef]
- Kabir, M.T.; Uddin, M.S.; Setu, J.R.; Ashraf, G.M.; Bin-Jumah, M.N.; Abdel-Daim, M.M. Exploring the role of PSEN mutations in the pathogenesis of Alzheimer’s disease. Neurotox. Res. 2020, 38, 833–849. [Google Scholar] [CrossRef]
- Cacace, R.; Sleegers, K.; Van Broeckhoven, C. Molecular genetics of early-onset Alzheimer’s disease revisited. Alzheimers Dement. 2016, 12, 733–748. [Google Scholar] [CrossRef]
- Seiler, A.; Schneider, M.; Forster, H.; Roth, S.; Wirth, E.K.; Culmsee, C.; Plesnila, N.; Kremmer, E.; Radmark, O.; Wurst, W.; et al. Glutathione peroxidase 4 senses and translates oxidative stress into 12/15-lipoxygenase dependent- and AIF-mediated cell death. Cell Metab. 2008, 8, 237–248. [Google Scholar] [CrossRef]
- Selkoe, D.J. Alzheimer’s disease results from the cerebral accumulation and cytotoxicity of amyloid β-protein. J. Alzheimers Dis. 2001, 3, 75–80. [Google Scholar] [CrossRef]
- Tung, Y.T.; Hsu, W.M.; Wang, B.J.; Wu, S.Y.; Yen, C.T.; Hu, M.K.; Liao, Y.F. Sodium selenite inhibits γ-secretase activity through activation of ERK. Neurosci. Lett. 2008, 440, 38–43. [Google Scholar] [CrossRef] [PubMed]
- Yim, S.Y.; Chae, K.R.; Shim, S.B.; Hong, J.T.; Park, J.Y.; Lee, C.Y.; Son, H.J.; Sheen, Y.Y.; Hwang, D.Y. ERK activation induced by selenium treatment significantly downregulates β/γ-secretase activity and tau phosphorylation in the transgenic rat overexpressing human selenoprotein M. Int. J. Mol. Med. 2009, 24, 91–96. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chen, P.; Wang, R.R.; Ma, X.J.; Liu, Q.; Ni, J.Z. Different forms of selenoprotein M differentially affect Aβ aggregation and ROS generation. Int. J. Mol. Sci. 2013, 14, 4385–4399. [Google Scholar] [CrossRef] [PubMed]
- Naseri, N.N.; Wang, H.; Guo, J.; Sharma, M.; Luo, W. The complexity of tau in Alzheimer’s disease. Neurosci. Lett. 2019, 705, 183–194. [Google Scholar] [CrossRef]
- Bramblett, G.T.; Goedert, M.; Jakes, R.; Merrick, S.E.; Trojanowski, J.Q.; Lee, V.M. Abnormal tau phosphorylation at Ser396 in Alzheimer’s disease recapitulates development and contributes to reduced microtubule binding. Neuron 1993, 10, 1089–1099. [Google Scholar] [CrossRef]
- Alonso, A.; Zaidi, T.; Novak, M.; Grundke-Iqbal, I.; Iqbal, K. Hyperphosphorylation induces self-assembly of tau into tangles of paired helical filaments/straight filaments. Proc. Natl. Acad. Sci. USA 2001, 98, 6923–6928. [Google Scholar] [CrossRef]
- Shim, S.B.; Lim, H.J.; Chae, K.R.; Kim, C.K.; Hwang, D.Y.; Jee, S.W.; Lee, S.H.; Sin, J.S.; Leem, Y.H.; Lee, S.H.; et al. Tau overexpression in transgenic mice induces glycogen synthase kinase 3β and β-catenin phosphorylation. Neuroscience 2007, 146, 730–740. [Google Scholar] [CrossRef]
- Jiang, H.; Shi, Q.Q.; Ge, L.Y.; Zhuang, Q.F.; Xue, D.; Xu, H.Y.; He, X.Z. Selenoprotein M stimulates the proliferative and metastatic capacities of renal cell carcinoma through activating the PI3K/AKT/mTOR pathway. Cancer Med. 2019, 8, 4836–4844. [Google Scholar] [CrossRef]
- Jia, Y.; Dai, J.; Zeng, Z. Potential relationship between the selenoproteome and cancer. Mol. Clin. Oncol. 2020, 13, 83. [Google Scholar] [CrossRef]
- Gollob, J.A.; Wilhelm, S.; Carter, C.; Kelley, S.L. Role of Raf kinase in cancer: Therapeutic potential of targeting the Raf/MEK/ERK signal transduction pathway. Semin. Oncol. 2006, 33, 392–406. [Google Scholar] [CrossRef]
- Min, J.H.; Lee, M.W.; Rhim, H.; Choi, D.; Kim, Y.S.; Kim, Y.J.; Cha, D.I.; Lim, H.K. Recurrent hepatocellular carcinoma after transcatheter arterial chemoembolization: Planning sonography for radio frequency ablation. J. Ultrasound Med. 2011, 30, 617–624. [Google Scholar] [CrossRef] [PubMed]
- Guariniello, S.; Colonna, G.; Raucci, R.; Costantini, M.; Di Bernardo, G.; Bergantino, F.; Castello, G.; Costantini, S. Structure-function relationship and evolutionary history of the human selenoprotein M (SelM) found over-expressed in hepatocellular carcinoma. Biochim. Biophys. Acta 2014, 1844, 447–456. [Google Scholar] [CrossRef] [PubMed]
- Guerriero, E.; Accardo, M.; Capone, F.; Colonna, G.; Castello, G.; Costantini, S. Assessment of the Selenoprotein M (SELM) over-expression on human hepatocellular carcinoma tissues by immunohistochemistry. Eur. J. Histochem. 2014, 58, 2433. [Google Scholar] [CrossRef] [PubMed]
- Varlamova, E.G.; Goltyaev, M.V.; Fesenko, E.E. Expression of human selenoprotein genes selh, selk, selm, sels, selv, and gpx-6 in various tumor cell lines. Dokl. Biochem. Biophys. 2016, 468, 203–205. [Google Scholar] [CrossRef] [PubMed]
- Uppal, V.; Mansoor, S.; Furuya, K.N. Pediatric non-alcoholic fatty liver disease. Curr. Gastroenterol. Rep. 2016, 18, 24. [Google Scholar] [CrossRef] [PubMed]
- Musso, G.; Cassader, M.; Paschetta, E.; Gambino, R. Bioactive lipid species and metabolic pathways in progression and resolution of nonalcoholic steatohepatitis. Gastroenterology 2018, 155, 282–302. [Google Scholar] [CrossRef] [PubMed]
- Nageeb, M.M.; Khatab, M.I.; Abdel-Sameea, A.A.; Teleb, N.A. Adelmidrol protects against non-alcoholic steatohepatitis in mice. Naunyn-Schmiedeberg’s Arch. Pharmacol. 2020, 393, 777–784. [Google Scholar] [CrossRef]
- Cai, J.; Huang, J.; Yang, J.; Chen, X.; Zhang, H.; Zhu, Y.; Liu, Q.; Zhang, Z. The protective effect of selenoprotein M on non-alcoholic fatty liver disease: The role of the AMPKα1-MFN2 pathway and Parkin mitophagy. Cell. Mol. Life Sci. 2022, 79, 354. [Google Scholar] [CrossRef]
- Schuster, S.; Cabrera, D.; Arrese, M.; Feldstein, A.E. Triggering and resolution of inflammation in NASH. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 349–364. [Google Scholar] [CrossRef]
- Chen, Y.; Zhou, C.; Bian, Y.; Fu, F.; Zhu, B.; Zhao, X.; Zhang, M.; Zhou, C.; Yao, S.; Zhang, Z.; et al. Cadmium exposure promotes thyroid pyroptosis and endocrine dysfunction by inhibiting Nrf2/Keap1 signaling. Ecotoxicol. Environ. Saf. 2022, 249, 114376. [Google Scholar] [CrossRef]
- Sangineto, M.; Bukke, V.N.; Bellanti, F.; Tamborra, R.; Moola, A.; Duda, L.; Villani, R.; Romano, A.D.; Serviddio, G. A novel nutraceuticals mixture improves liver steatosis by preventing oxidative stress and mitochondrial dysfunction in a NAFLD model. Nutrients 2021, 13, 652. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhao, F.; Gai, X.; Cai, J.; Zhang, X.; Chen, X.; Zhu, Y.; Zhang, Z. Astilbin attenuates apoptosis induced by cadmium through oxidative stress in carp (Cyprinus carpio L.) head kidney lymphocyte. Fish Shellfish Immunol. 2022, 125, 230–237. [Google Scholar] [CrossRef] [PubMed]
- Paech, F.; Abegg, V.F.; Duthaler, U.; Terracciano, L.; Bouitbir, J.; Krähenbühl, S. Sunitinib induces hepatocyte mitochondrial damage and apoptosis in mice. Toxicology 2018, 409, 13–23. [Google Scholar] [CrossRef] [PubMed]
- Kruppa, A.J.; Buss, F. Actin cages isolate damaged mitochondria during mitophagy. Autophagy 2018, 14, 1644–1645. [Google Scholar] [CrossRef]
- Hatfield, D.L.; Tsuji, P.A.; Carlson, B.A.; Gladyshev, V.N. Selenium and selenocysteine: Roles in cancer, health, and development. Trends Biochem. Sci. 2014, 39, 112–120. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).