Abstract
Nymphoides peltata has been widely used pharmacologically in traditional Chinese medicine to treat heat strangury and polyuria. The aim of this study was to isolate the bioactive components from N. peltata and evaluate their potential use as antioxidant and anti-wrinkle agents. Phytochemical investigation of the methanolic extract of N. peltata roots led to the isolation of 15 compounds (1–15), which were structurally determined as α-spinasterol (1), 3-O-β-D-glucopyranosyl-oleanolic acid 28-O-β-D-glucuronopyranoside (2), 4-hydroxybenzoic acid (3), protocatechuic acid (4), vanillic acid (5), p-coumaric acid (6), caffeic acid (7), ferulic acid (8), neochlorogenic acid (neo-CQA) (9), chlorogenic acid (CQA) (10), cryptochlorogenic acid (crypto-CQA) (11), isochlorogenic acid B (3,4-DCQA) (12), isochlorogenic acid A (3,5-DCQA) (13), isochlorogenic acid C (4,5-DCQA) (14), and 3,4,5-tri-O-caffeoylquinic acid (TCQA) (15). Of these 15 compounds, compound 2 was a new oleanane saponin, the chemical structure of which was characterized by 1D and 2D nuclear magnetic resonance (NMR) spectroscopic data and high-resolution electrospray ionization mass spectrometry (HRESIMS), as well as chemical reaction. Biological evaluation of the isolated compounds revealed that 3,4,5-tri-O-caffeoylquinic acid (TCQA) significantly improved Nrf2 levels in an Nrf2–ARE reporter HaCaT cell screening assay. TCQA was found to potently inhibit the Nrf2/HO-1 pathway and to possess strong anti-wrinkle activity by modulating the MAPK/NF-κB/AP-1 signaling pathway and thus inhibiting MMP-1 synthesis in HaCaT cells exposed to UVB. Our results suggest that TCQA isolated from N. peltata might be useful for developing effective antioxidant and anti-wrinkle agents.
Keywords:
Nymphoides peltata; 3,4,5-tri-O-caffeoylquinic acid (TCQA); anti-wrinkle; antioxidant; Nrf2; NF-κB; MAPK; AP-1; MMP-1 1. Introduction
The skin is the largest body organ, and it protects internal organs from physical harm and chemical irritants, and plays an important role in maintaining skin homeostasis [1]. However, complex interactions between multiple intrinsic and extrinsic factors cause skin aging [2]. Photoaging due to continuous ultraviolet (UV) exposure is a major contributor to skin aging and accounts for more than 80% of facial aging [3]. In particular, UVB irradiation can penetrate the epidermis and dermis and induce oxidative stress by generating reactive oxygen species (ROS) within cells and tissues, which can lead to skin inflammation, aging, and skin cancer [4]. Nuclear factor erythroid-2-related factor 2 (Nrf2) is activated by oxidative stress and regulates antioxidant activity and cellular redox levels by neutralizing ROS and reactive electrophiles [5,6,7,8]. In other words, when exposed to oxidative stress, Nrf2 dissociates from Keap1 and binds to antioxidant response element (ARE) to induce the expression of antioxidant-related enzymes, such as heme oxygenase-1 (HO-1) and NAD(P)H: quinone oxidoreductase 1 (NQO1), and thereby inhibits ROS-induced oxidative damage and skin aging [9,10].
Photoaging is characterized by the activation of key transcription factors, including mitogen-activated protein kinase (MAPK), nuclear factor-κB (NF-κB), and activator protein-1 (AP-1) [11]. These factors promote collagen degradation in skin tissue by inducing matrix metalloproteinase-1 (MMP-1), which leads to an abnormal skin structure [12,13]. MAPKs are phosphorylated and activated by ROS and oxidative stress, and they enter the nucleus and upregulate the transcription factors AP-1 and NF-κB [4,14]. AP-1 increases the expression of MMPs, including MMP-1, MMP-3, and MMP-9, which degrade the extracellular matrix (ECM) and basement membrane components [15,16,17,18]. In particular, MMP-1 is a collagen-degrading protease that promotes the degradation of procollagen type-I, a crucial constituent of ECM; thus, it contributes to skin aging and wrinkle formation [19,20]. NF-κB is another transcription factor that increases MMP-1 levels in the dermis and is activated by UVB-generated ROS [21]. Furthermore, activation of the NF-κB pathway results in the synthesis of proinflammatory enzymes and cytokines, leading to inflammation and tissue damage, enhanced MMP-1 production, and skin aging [22,23,24]. Therefore, agents with antioxidant properties that promote Nrf2 activation and inhibit the MAPK/NF-κB/AP-1 signaling pathway are considered potential anti-photoaging and anti-wrinkle agents. For this reason, research is being actively conducted to identify phytochemicals with antioxidant, anti-aging, and anti-wrinkle effects [25,26].
The genus Nymphoides is one of the five genera of the family Menyanthaceae and contains approximately fifty species worldwide [27]. Three species of this genus, that is, Nymphoides coreana (H. Lev.) H. Hara, Nymphoides indica (L.) Kuntze, and Nymphoides peltata (S. G. Gmel.) Kuntze, are endemic in Korea [28]. Nymphoides peltata (syn. Limnanthemum peltatum, Menyanthes nymphoides), known as Xing-Cai in China and Asaza in Japan, is a perennial aquatic plant that is widely distributed in temperate and subtropical regions of Eurasia, including Korea, Japan, and China [29]. In the Compendium of Materia Medica (Ben Cao Gang Mu), a compendium of traditional Chinese medicines, Nymphoides are mainly used to treat heat strangury and polyuria. Pharmacologically, N. peltata extracts have been reported to inhibit platelet-activating factor (PAF) and N-formyl methionyl-leucyl-phenylalanine (fMLP), which are both potent pro-inflammatory factors [30]. In our recent study, we found that a 95% EtOH extract of N. peltata roots ameliorated atopic dermatitis and activated the Nrf2/HO-1 signaling pathway [31]. As part of ongoing research for N. peltata, phytochemical investigation of the MeOH extract of N. peltata roots was conducted, which led to the isolation of 15 compounds (1–15) including a new oleanane saponin (2). The structures of the isolated compounds were determined by nuclear magnetic resonance (NMR) spectroscopic data and LC–MS analysis. Herein, we describe the isolation and structural determination of compounds 1–15, evaluating their antioxidant, anti-photoaging, and anti-wrinkle effects in UVB-irradiated HaCaT cells.
2. Materials and Methods
2.1. General
Column chromatography was conducted using Silica gel 60 (70–230 mesh, Merck, Darmstadt, Germany) and Sephadex LH-20 (25–100 μm; Pharmacia, Stockholm, Sweden). Thin-layer chromatography (TLC) analysis was performed on silica gel 60 F254 Art. 5715 plates (Merck, Germany). High-performance liquid chromatography (HPLC) preparative analysis was performed using a Gilson HPLC system equipped with two pumps (305 master pump, 307 slave pump) and a mixer (811C dynamic mixer), and a Shimadzu HPLC system with a UV/vis detector (SPD-20A), pump (LC-20AT), and system controller (CBM-20A). HPLC was conducted using a Watchers 120 ODS-BP column (S-10 μm, 150 mm × 10 mm, Isu Industry Corp., Seoul, Republic of Korea). High-resolution electrospray ionization mass spectrometry (HR-ESI-MS) was conducted using an Agilent 6530 Accurate-Mass Quadrupole Time-of-Flight (Q-TOF) LC/MS system, and a 6545 Accurate-Mass Q-TOF LC/MS (Agilent Technologies, Santa Clara, CA, USA). Nuclear magnetic resonance (NMR) spectra were recorded using JEOL 400 MHz (JNM-ECZ400S, JEOL, Tokyo, Japan), Bruker 500 MHz (Bruker, Billerica, MA, USA), and Agilent Technologies 600 MHz (Santa Clara, CA, USA) instruments. DMSO-d6 and chloroform-d (Cambridge Isotope Laboratories, Andover, MA, USA) were used as solvents for NMR studies.
2.2. Plant Material
N. peltata roots were collected at the Hantaek Botanical Garden Foundation (Yongin-si, Gyeonggi-do, Republic of Korea) and authenticated by Dr. Jung Hwa Kang. Voucher specimens (PNU-0040) were deposited at the Medicinal Herb Garden, Pusan National University.
2.3. Isolation of Compounds from N. peltata Extract
Dried roots of N. peltata (2.8 kg) were first ground to a powder, soaked in 100% methanol (MeOH), ultrasonicated for 90 min at room temperature twice, and immediately freeze-dried. The yield of MeOH extract obtained was 15.25% (427 g). This extract was then suspended in 2 L of H2O and sequentially extracted with n-hexane (Hex), ethyl acetate (EA), and n-butanol (n-BuOH) to obtain three fractions: NPH (80.3 g), NPE (24 g), and NPB (108 g), respectively.
The NPH fraction was subjected to silica gel column chromatography using Hex:EA (8:1 → 100% EA gradient) as the eluant to obtain 12 fractions. The NPH-6 (17.8996 g) subfraction was subjected to silica gel column chromatography using Hex:EA (5:1 → 100% EA gradient) as the eluant to obtain 9 fractions. The NPH-6-4 (2.1 g) subfraction was recrystallized using 100% methanol (MeOH) to obtain pure compound 1 (21.3 mg).
The NPE fraction was subjected to silica gel column chromatography using EA:MeOH (10:1 → 100% MeOH gradient) as the eluant to obtain 7 fractions. The NPE-1 (463.2 mg) subfraction was subjected to silica gel column chromatography using EA:MeOH (20:1 → 100% MeOH gradient) as the eluant to obtain 6 fractions. The NPE-1-4 (10.1 mg) subfraction was subjected to semi-preparative TLC using a methylene chloride (CH2Cl2):MeOH = 20:1 mobile phase to obtain 6 fractions. The NPE-1-4-6 (0.5 mg) subfraction was subjected to the Shimadzu prep HPLC system (UV wavelengths 250 and 330 nm; flow rate 2 mL/min) using an ODS column using 0.1% formic acid in acetonitrile (ACN):0.1% formic acid in H2O (25:75 isocratic) as the eluant to obtain pure compound 3 (retention time (tR) 19 min, 0.4 mg). The NPE-2 (1.025 g) fraction was subjected to silica gel column chromatography using chloroform (CHCl3):MeOH (20:1 → 100% MeOH gradient) as the eluant to obtain 9 fractions. The NPE-2-2,3 (73.7 mg) subfraction was subjected to the Shimadzu prep HPLC system (UV wavelengths 250 and 330 nm; flow rate 2 mL/min) using an ODS column and 0.1% formic acid in ACN:0.1% formic acid in H2O (27:73 isocratic) as the eluant to obtain pure compounds 5 (tR 13 min, 2.2 mg) and 8 (tR 19 min, 2.3 mg). The NPE-2-9 (206 mg) fraction was subjected to Sephadex LH-20 column chromatography using 100% MeOH as the eluant to obtain 3 fractions. The NPE-2-9-1 (115.7 mg) subfraction was subjected to Sephadex LH-20 column chromatography using 100% MeOH as the eluant to obtain 5 fractions. The NPE-2-9-1-4 (18.1 mg) subfraction was subjected to Shimadzu prep HPLC (UV wavelengths 250 and 330 nm; flow rate 2 mL/min) using an ODS column and 0.1% formic acid in ACN:0.1% formic acid in H2O (18:82 isocratic) as the eluant to obtain pure compounds 4 (tR 13 min, 0.5 mg), 7 (tR 22.2 min, 4.2 mg), and 6 (tR 39 min, 1.4 mg). The NPE-6 (5.9752 g) subfraction was subjected to Gilson prep HPLC (UV wavelength 250 nm; flow rate 2 mL/min) using an ODS column and MeOH:H2O (10:90 → 100% MeOH gradient) as the eluant to obtain 4 fractions. The NPE-6-3 (397.1 mg) subfraction was subjected to Shimadzu prep HPLC (UV wavelengths 250 and 330 nm; flow rate 2 mL/min) using an ODS column and 0.1% formic acid in ACN:0.1% formic acid in H2O (21:79 isocratic) as the eluant to obtain 7 fractions. The NPE-6-3-7 (147.4 mg) subfraction was subjected to the Shimadzu prep HPLC system (UV wavelengths 250 and 330 nm; flow rate 2 mL/min) using an ODS column and 0.1% formic acid in ACN:0.1% formic acid in H2O (30:70 isocratic) as the eluant to obtain 4 fractions. The NPE-6-3-7-4 (7.6 mg) subfraction was subjected to the Shimadzu prep HPLC system (UV wavelengths 250 and 330 nm; flow rate 2 mL/min) using an ODS column and 0.1% formic acid in ACN:0.1% formic acid in H2O (20:80 isocratic) as the eluant to obtain pure compound 15 (tR 48 min, 4.2 mg). The NPE-7 (1.893 g) subfraction was subjected to Shimadzu prep HPLC (UV wavelengths 250 and 330 nm; flow rate 2 mL/min) using an ODS column and 0.1% formic acid in ACN:0.1% formic acid in H2O (20:80 isocratic) as the eluant to obtain pure compounds 12 (tR 38 min, 13.2 mg), 13 (tR 48 min, 24.3 mg) and 14 (tR 60 min, 10.8 mg).
The NPB fraction was subjected to silica gel column chromatography using EA:MeOH (20:1 → 100% MeOH) as the eluant to obtain 5 fractions. The NPB-4 (28.7062 g) subfraction was subjected to silica gel column chromatography using Hex:EA (10:1 → 100% EA → 100% MeOH) as the eluant to obtain 5 fractions. The NPB-4-5 (19.4128 g) subfraction was subjected to silica gel column chromatography using CHCl3:MeOH (20:1 → 100% MeOH) as the eluant to obtain 7 fractions. The NPB-4-5-7 (8.5607 g) subfraction was subjected to Gilson prep HPLC (UV wavelength at 250 nm; flow rate 2 mL/min) using an ODS column and MeOH:H2O (10:90 → 100% MeOH gradient) as the eluant to obtain 6 fractions. The NPB-4-5-7-1 (200.8 mg) subfraction was subjected to the Shimadzu prep HPLC system (UV wavelengths 250 and 330 nm; flow rate 2 mL/min) using an ODS column and 0.1% formic acid in ACN:0.1% formic acid in H2O (10:90 isocratic) as the eluant to obtain pure compounds 9 (tR 21 min, 7.8 mg), 10 (tR 38 min, 15.4 mg), and 11 (tR 46 min, 9.6 mg). The NPB-4-5-7-5 (512.9 mg) subfraction was recrystallized using 100% MeOH to obtain pure compound 2 (258.1 mg).
3-O-β-D-Glucopyranosyl-Oleanolic Acid 28-O-β-D-Glucuronopyranoside (2)
White amorphous powder; + 15.9 (c 0.2, MeOH); UV (MeOH) λmax (log ) 217 (3.8) nm; IR (neat) max: 3450, 1742, 1725, 1075 cm−1; 1H (400 MHz) and 13C (100 MHz) NMR data (Table 1); HR-ESI-MS (positive ion mode) m/z 817.4332 [M + Na]+ (calcd. for C42H66NaO14, 817.4350).

Table 1.
1H (400 MHz) and 13C NMR (100 MHz) data of compound 2 in DMSO-d6.
2.4. Cell Culture and UVB Irradiation
HaCaT cells (a spontaneously immortalized human keratinocyte cell line) were purchased from N.E. Fusenig (Deutsches Krebsforschungszentrum, Heidelberg, Germany) and cultured in Dulbecco’s modified Eagle’s medium (DMEM; HyClone, Logan, UT, USA) supplemented with 10% fetal bovine serum (FBS) and 100 units/mL of penicillin and 100 mg/mL streptomycin (HyClone) at 37 °C in a 5% CO2 humidified atmosphere. Seeded HaCaT cells were pretreated with various concentrations of TCQA (3,4,5-tri-O-caffeoylquinic acid, compound 15) for one hour and then exposed to 15 mJ/cm2 of UVB using a 280–360 mm light source (G15T8E UV-B lamp; Tokyo, Japan). UV strength was measured using a photo/radiometer (HD2102.1; Delta OHM, Padoba, Italy). After UVB exposure, cells were treated with various concentrations of TCQA (15) in 1% FBS.
2.5. Cell Viability
Cell viabilities were determined using a CCK-8 kit (WST-8; Abcam, Cambridge, MA, USA). HaCaT cells seeded in 96-well plates (2 × 104 cells/well) were pretreated with the indicated concentrations of TCQA (15) for 4 h, washed with PBS, and then exposed to UVB (15 mJ/cm2) and TCQA (15) for 3, 6, 12, or 24 h in 1% FBS. After UVB exposure, CCK-8 assay reagent was added to each well, and the cells were incubated for 30 min at 37 °C. Formazan absorbances were measured at 450 nm using a multifunctional plate reader (Tecan Infinite M1000 Microplate Reader, Tecan, Männedorf, Zürich, Switzerland), and compared to untreated cells.
2.6. ROS Measurement (DCFDA Assay)
Intracellular ROS formation was assessed using dichlorofluorescin diacetate (DCFDA) as the substrate. HaCaT cells were seeded at 2 × 104 cells/well in black 96-well plates and, 24 h later, pretreated with the indicated concentrations of TCQA (15) for 4 h. Then, they were washed with PBS and exposed to UVB (15 mJ/cm2) and TCQA (15) for 3, 6, 12, or 24 h in 1% FBS. After UVB irradiation, cells were incubated with DCFDA (20 μM in 1% PBS) for 30 min. DCF concentrations in media caused by the ROS-induced oxidation of DCFDA were measured at excitation/emission wavelengths of 485/525 nm using a multifunctional plate reader (Tecan Infinite M1000 Microplate Reader, Tecan).
2.7. Luciferase Reporter Gene Assay
HaCaT cells were seeded into 24-well plates, cultured for 24 h, and co-transfected with ARE-Luc reporter plasmid (300 ng/well) and internal control plasmid pRL-SV-40 (5 ng/well) using the FuGENE® 4K Transfection Reagent (Fugent LLC, Madison, WI, USA). Twenty-four hours after transfection, cells were treated with the indicated concentrations of TCQA (15) for an additional 24 h. Luciferase activities of cell lysates were then measured using the Dual-Luciferase® Reporter Assay System according to the manufacturer’s instructions (Promega, Madison, WI, USA). To determine transfection efficiencies, luciferase activities were expressed as ratios of Renilla luciferase activity (SV40).
Transactivation of AP-1 or NF-κB was evaluated using pNF-κB-Luc plasmid or pAP-1-luc plasmid (300 ng/well; Stratagene, San Diego, CA, USA) with pRL-SV40 (5 ng/well) in HaCaT cells. Twenty-four hours after transfection, the cells were pretreated with a TCQA (15) for 1 h. The cells were then treated with the proinflammatory cytokine [NF-κB (TNF-α and IFN-γ 10 ng/mL), AP-1 (PMA 2 μM)] for 20 h. Transactivation of NF-κB or AP-1 was determined as described above.
2.8. Real-Time Quantitative PCR (qPCR)
HaCaT cells were seeded at 2 × 105 cells/well in 12-well plates. Then, 24 h later, they were pretreated with the indicated concentrations of TCQA (15) for 4 h, washed with PBS, and treated with UVB (15 mJ/cm2) and TCQA (15) for 24 h when total RNA was isolated. This was carried out using an RNeasy mini kit (Qiagen, Hilden, Germany) and reverse transcribed using a RevertAid first-strand cDNA synthesis kit (Thermo Fisher Scientific, Waltham, MA, USA). Real-time PCR was conducted using QuantaStudio 6 pro (Thermo Fisher Scientific) using SYBR1 Green (Power SYBR Green PCR Master Mix; Applied Biosystems, Foster City, CA, USA). PCR settings were as follows: initial incubation for 2 min at 50 °C, denaturing for 10 min at 95 °C, followed by 40 cycles of PCR (15 s at 95 °C and 60 s at 60 °C). The primer sets used were MMP-1 (accession no. DQ399597), forward 5′-GCC CAG ATG TGG AGT GCC TG-3′ and reverse 5′-GTT TGC TCC CAG CGA GGG TT-3′; and GAPDH (accession no. NM_001357943), forward 5′-ACA CCC ACT CCT CCA CCT TT-3′ and reverse 5′-TGC TGT AGC CAA ATT CGT TG-3′. The qPCR data were analyzed using the QuantaStudio 6 pro System (Thermo Fisher Scientific). Transcript levels were normalized versus GAPDH.
2.9. Western Blot Analysis
Protein levels were measured by Western blot. Briefly, HaCaT cells were seeded at 5 × 105 cells/well in 6-well plates. Then, 24 h later, they were pretreated with the indicated concentrations of TCQA (15) for 4 h, washed with PBS, and exposed to UVB (15 mJ/cm2). After 24 h, cells were lysed in appropriate amounts of RIPA buffer (Bioprince, Chuncheon, Republic of Korea) and Phosphatase Inhibitor Cocktail 2 (Sigma-Aldrich, St. Louis, MO, USA). Lysates were centrifuged at 13,000 rpm for 15 min at 4 °C, and supernatants were collected for further studies. Denatured protein lysates were separated by SDS-PAGE and transferred to PVDF membranes (Merck, Darmstadt, Germany). Blots were treated with primary antibodies against p-Nrf2 (1/1000; Invitrogen, Carlsbad, CA, USA), Nrf2 (1/1000; BioLegend, San Diego, CA, USA), HO-1 (1/1000; Abcam), p-extracellular signal-regulated kinase (ERK)1/2 (1/1000; Cell Signaling, Danvers, MA, USA), ERK1/2 (1/1000; Santa Cruz Biotechnology, Santa Cruz, CA, USA), p-c-Jun N-terminal kinase (JNK) (1/1000; Cell Signaling), JNK (1/1000; Santa Cruz Biotechnology), p-p38 (1/1000; Cell Signaling), p38 (1/1000; Santa Cruz Biotechnology), p-c-fos (1/1000; Cell Signaling), c-fos (1/1000; Santa Cruz Biotechnology), p-c-jun (1/1000; Santa Cruz Biotechnology), c-jun (1/1000; Santa Cruz Biotechnology), p-nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha (IκBα) (1/1000; Cell Signaling), IκBα (1/1000; Cell Signaling), p-p65 (1/1000; Cell Signaling), p65 (1/1000; Cell Signaling), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (1/3000; Cell Signaling). Then, they were treated with horseradish-peroxidase-conjugated secondary antibodies (rabbit; 1/3000; Cell Signaling, mouse; 1/3000, Santa Cruz Biotechnology) and visualized using an ECL kit (Thermo Fisher Scientific).
2.10. Determination of MMP-1 Secretions by ELISA
HaCaT cells were seeded at 2 × 105 cells/well in 12-well plates. Then, 24 h later, they were pretreated with TCQA (15) (5, 10, or 20 μM) for 4 h, washed with PBS, and then exposed to UVB (15 mJ/cm2) and TCQA (15) overnight in 1% FBS medium. After exposure to UVB, culture supernatants were collected and centrifuged at 13,000 rpm for 5 min. Concentrations of matrix metalloproteinase-1 secreted into culture media were determined using human total MMP-1 enzyme-linked immunosorbent assay (ELISA) kits (Human Total MMP-1 DuoSet ELISA kit; R&D Systems, Minneapolis, MN, USA). Absorbances at 450 nm were read using a microplate reader (Tecan Infinite M1000 Microplate Reader, Tecan).
2.11. Statistical Analysis
The statistical analysis was performed using GraphPad Prism software v4.0 (GraphPad, La Jolla, CA, USA), and results are presented as the means ± SDs of 2 to 3 independent experiments. One-way analysis of variance (ANOVA) followed by Tukey’s multiple comparisons test for independent samples was used to determine the significances of intergroup differences, and statistical significance was accepted for p-values < 0.05: # p < 0.05, ## p < 0.01, and ### p < 0.001; * p < 0.05, ** p < 0.01, and *** p < 0.001.
3. Results
3.1. Isolation and Structural Identification of Compounds from N. peltata
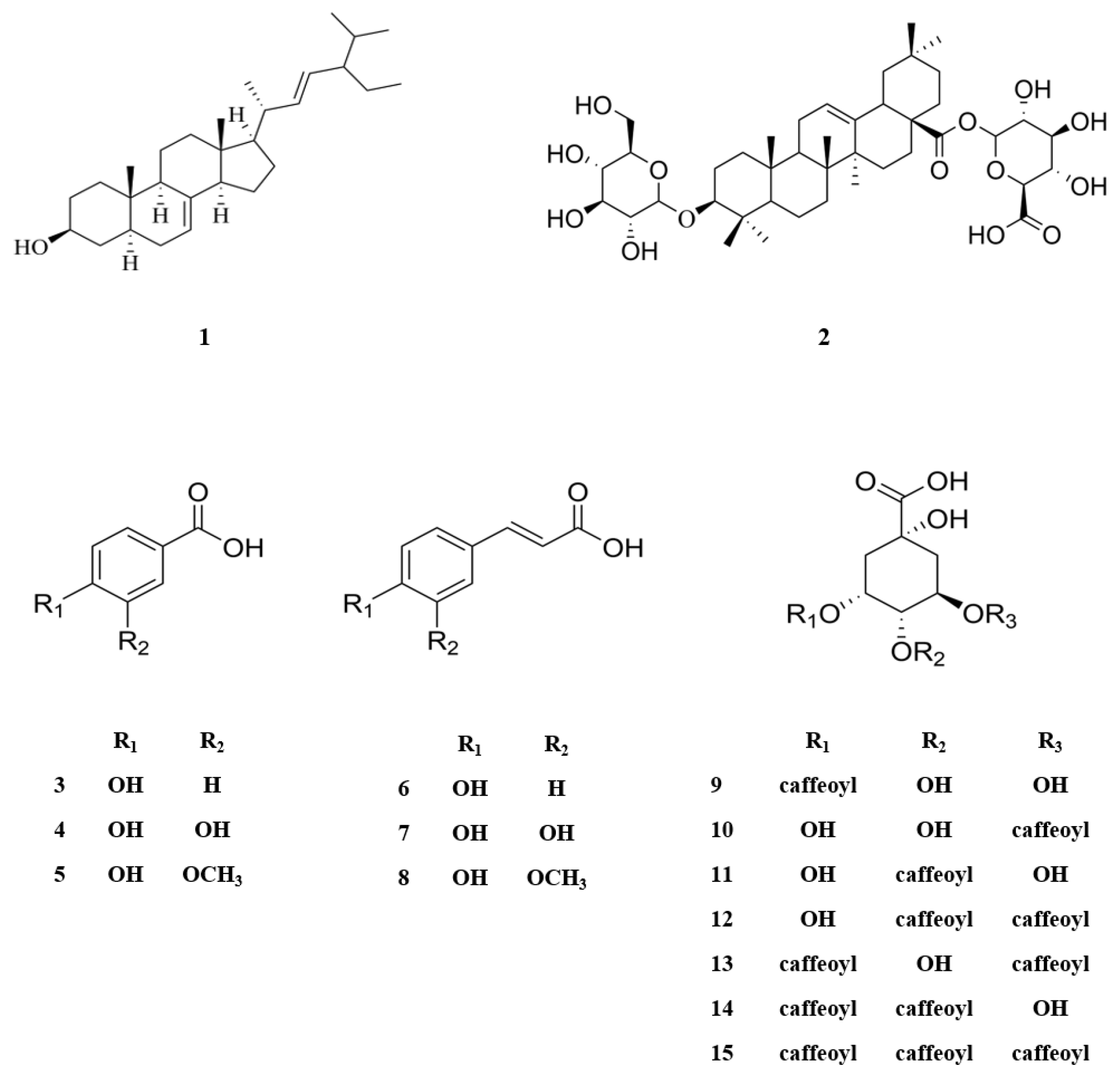
The MeOH extract of N. peltata roots was subjected to phytochemical investigation through liquid–liquid partitioning, column chromatography, and HPLC, leading to the isolation of one steroid derivative, one terpenoid saponin, six phenolic acid derivatives, and seven caffeoylquinic acid derivatives. The isolated compounds (1–15) were structurally identified as follows: α-spinasterol (1) (Figure S1) [32], 3-O-β-D-glucopyranosyl-oleanolic acid 28-O-β-D-glucuronopyranoside (2) (Figures S2–S6), 4-hydroxybenzoic acid (3) (Figure S7) [33], protocatechuic acid (4) (Figure S8) [34], vanillic acid (5) (Figure S9) [34], p-coumaric acid (6) (Figure S10) [35], caffeic acid (7) (Figure S11) [36], ferulic acid (8) (Figure S12) [37], neochlorogenic acid (neo-CQA) (9) (Figure S13) [38], chlorogenic acid (CQA) (10) (Figure S14) [38], cryptochlorogenic acid (crypto-CQA) (11) (Figure S15) [38], isochlorogenic acid B (3,4-DCQA) (12) (Figure S16) [39], isochlorogenic acid A (3,5-DCQA) (13) (Figure S17) [39], isochlorogenic acid C (4,5-DCQA) (14) (Figure S18) [39], and 3,4,5-tri-O-caffeoylquinic acid (TCQA) (Figure S19) (15) [40] (Figure 1). These identifications were based on their LC/MS data and comparison of their NMR spectroscopic data with those previously reported. A literature survey revealed that compound 2 is a new oleanane saponin, the chemical structure of which was characterized by 1D and 2D NMR spectroscopic data and high-resolution electrospray ionization mass spectrometry (HRESIMS), as well as chemical reaction.
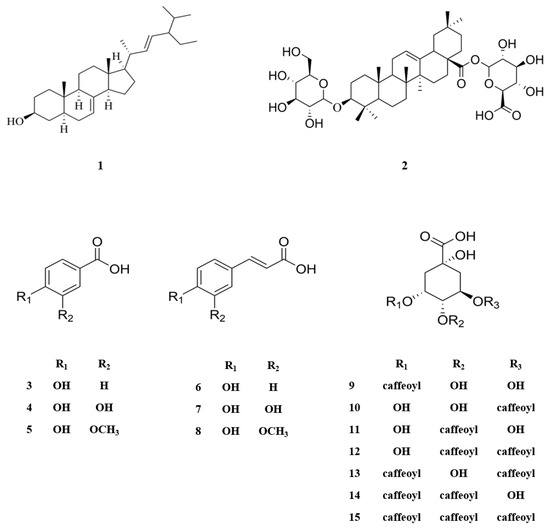
Figure 1.
Chemical structures of compounds 1–15 isolated from N. peltata.
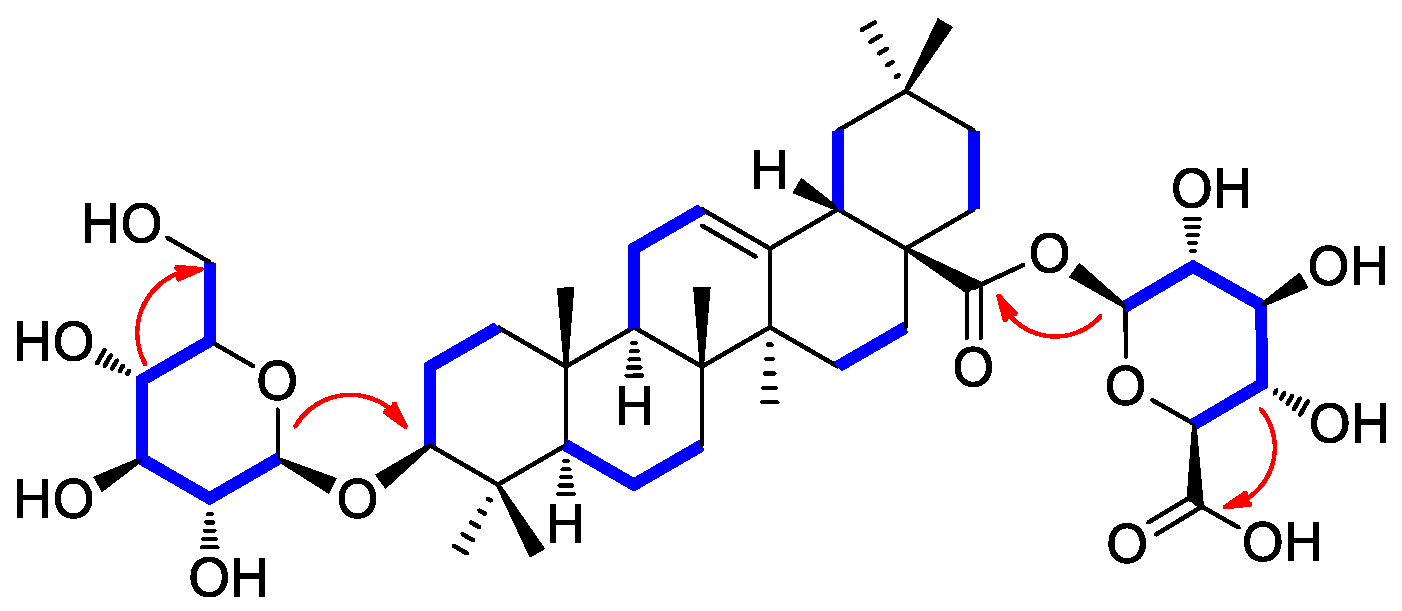
Compound 2 was isolated as a white amorphous powder with a molecular formula of C42H66O14, as deduced from the molecular ion peak [M + Na]+ at m/z 817.4332 (calcd. for C42H66NaO14, 817.4350) in the positive-ion mode of HRESIMS. The 1H NMR data (Table 1) of compound 2 showed proton NMR resonances corresponding to seven methyl groups [δH 0.67 (3H, s), 0.74 (3H, s), 0.86 (3H, s), 0.86 (3H, s), 0.87 (3H, s), 0.96 (3H, s), and 1.07 (3H, s)], one oxymethine proton [δH 3.01 (1H, dd, J = 10.5, 3.5 Hz)], and one olefinic proton [δH 5.16 (1H, br s)]. This was in addition to proton signals for sugar units, including two anomeric protons at δH 4.13 (1H, d, J = 7.5 Hz) and δH 5.23 (1H, d, J = 8.0 Hz), and other oxygenated protons at δH 3.43 (1H, dd, J = 11.0, 5.5 Hz), δH 3.61 (1H, dd, J = 10.0, 5.0 Hz), and δH 3.28–2.95. The 13C NMR data (Table 1) of compound 2 showed 42 carbon resonances, including seven methyl groups (δC 15.7, 17.0, 17.1, 23.8, 26.0, 28.1, and 33.2), one highly deshielded oxygenated methine (δC 88.4), two olefinic methines (δC 122.2 and 143.9), and one carboxyl carbon (δC 175.7) for an aglycone part of oleanolic acid. Among the remaining carbon signals attributable to sugar units, anomeric carbon signals at δC 94.6 and δC 101.7, and characteristic oxygenated carbon at δC 61.1 and carboxyl carbon at δ 174.3 indicated the presence of glucose and glucuronic acid, respectively [41,42]. Overall, 1H and 13C NMR data and mass spectral data implied that compound 2 is an oleanane-type triterpenoid saponin, and the NMR data were revealed to be very similar to those of chikusetsusaponin IVa [41,42]. However, there were some noticeable differences in the 13C NMR chemical shifts observed for the sugar groups, which suggested that compound 2 may have the different linkages of glucose and glucuronic acid from chikusetsusaponin IVa. The accurate structure of compound 2 was confirmed by 1H−1H COSY, HSQC, and HMBC experiments. The 1H−1H COSY correlations starting at H-1′ via H-2′/H-3′/H-4′/H-5′ and ending at H2–6′ as well as the HMBC correlations of H-1′/C-3 (δC 88.4) and H-4′/C-6′ (δC 61.1) determined the linkage of glucose at C-3 (Figure 2). In addition, the key HMBC correlations of H-1″/C-28 (δC 175.7) and H-4″/C-6″ (δC 174.3) and 1H−1H COSY correlations of H-1″/H-2″/H-3″/H-4″/H-5″ confirmed the glucuronic acid moiety connected at C-28 (Figure 2). The completed gross structure of compound 2 was finally established via cross-peaks in the HMBC and 1H−1H COSY spectra (Figure 2). For the absolute configuration of sugar units, sugar moieties of compound 2 were gained through acid hydrolysis and then determined by LC/MS–UV as described in the previous paper [42], which verified the absolute configuration for both glucose and glucuronic acid as the D-form. The anomeric coupling constants (J = 7.5 Hz for glucose and J = 8.0 Hz for glucuronic acid) were indicative of the β-form, confirming the sugar units as β-D-glucose and β-D-glucuronic acid, respectively [41,42]. Finally, the aglycone, derived from acid hydrolysis, was identified as oleanolic acid by comparison of the optical rotation and NMR data [41,42]. Accordingly, the chemical structure of compound 2 was elucidated as 3-O-β-D-glucopyranosyl-oleanolic acid 28-O-β-D-glucuronopyranoside, and named peltatasaponin A.
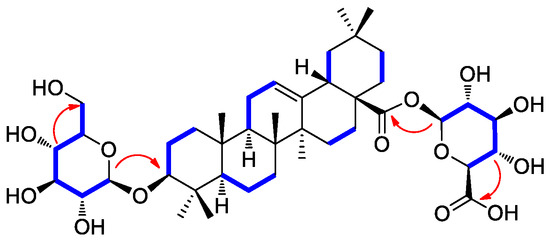
Figure 2.
1H-1H COSY (blue lines) and key HMBC correlations (red arrows) of compound 2.
3.2. Effects of Compounds Isolated from N. peltata on Nrf2–ARE Luciferase Activity in HaCaT Cells
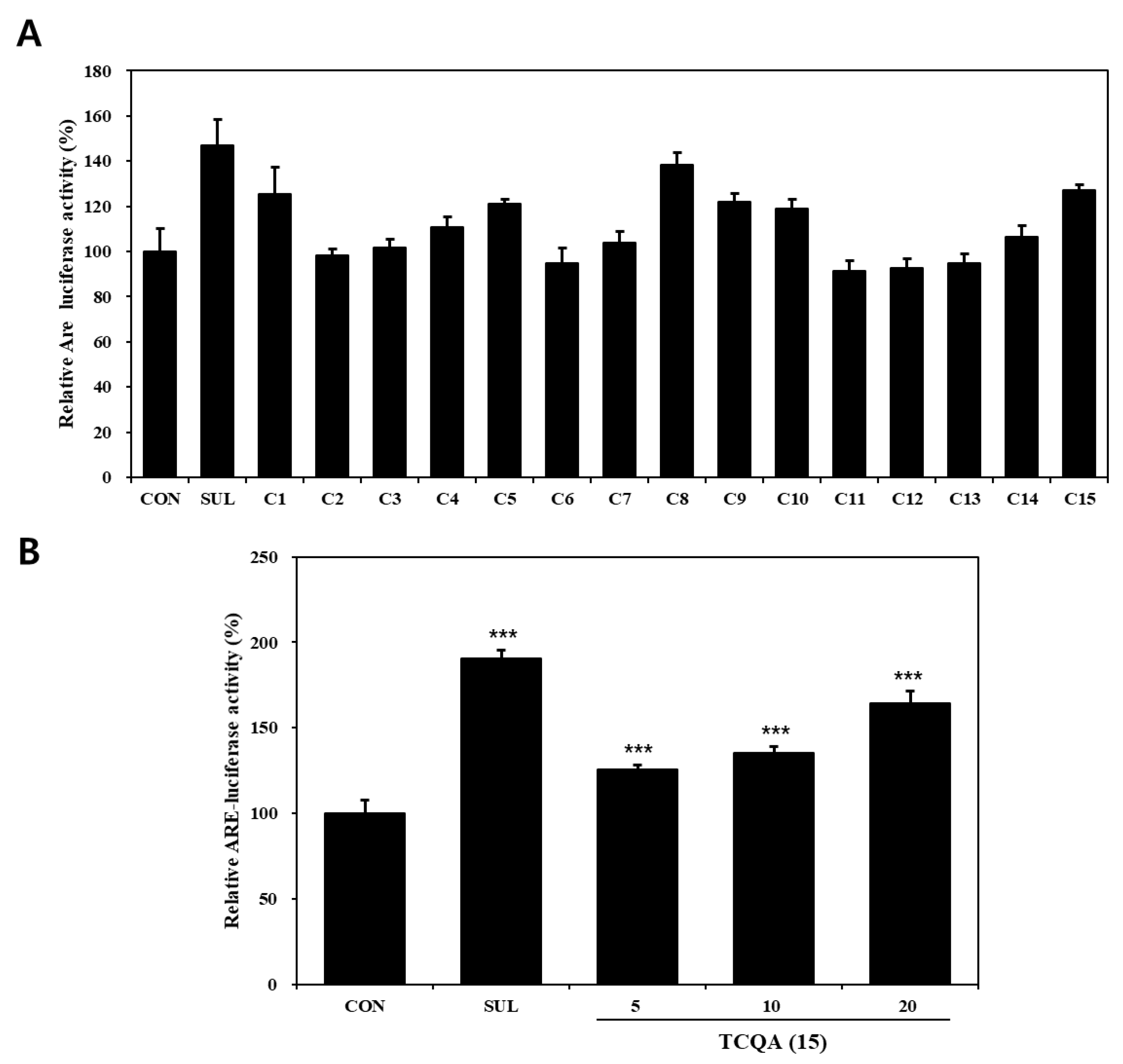
The effects of compounds (1–15) on the Nrf2–ARE pathway, as determined using Nrf2–ARE reporter, in HaCaT cells are shown in Figure 3A. Regarding activation of the Nrf2–ARE pathway, compounds 1 (125%), 8 (138%), 10 (119%), and 15 (127%) significantly increased Nrf2–ARE luciferase activity compared to the untreated control (CON) group (100%). Out of the four, compounds 1, 8, and 10 have already been reported to have potential use as skin anti-aging treatments due to their ability to activate Nrf2 [43,44,45]. TCQA (15) treatment of Nrf2–ARE reporter HaCaT cells resulted in a significant concentration-dependent increase in Nrf2 activity, that is, it enhanced activity by 25% at 5 μM, 35% at 10 μM, and 54% at 20 μM, compared to the CON group (Figure 3B).
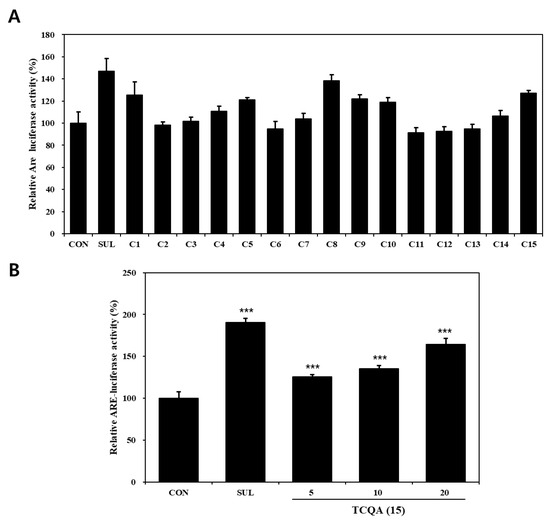
Figure 3.
Nrf2–ARE luciferase activities of compounds isolated from N. peltata. Transfected HaCaT cells were treated with (A) each of the fifteen compounds isolated N. peltata (10 μM) or (B) TCQA (5 μM, 10 μM, and 20 μM), and sulforaphane (2 μM) for 24 h. Results are presented as means ± SDs (n = 3). *** p < 0.001 versus CON group; CON: untreated control group; SUL: sulforaphane-treated group; TCQA: TCQA-treated group.
3.3. Effects of TCQA (15) on Antioxidant Activity in HaCaT Cells
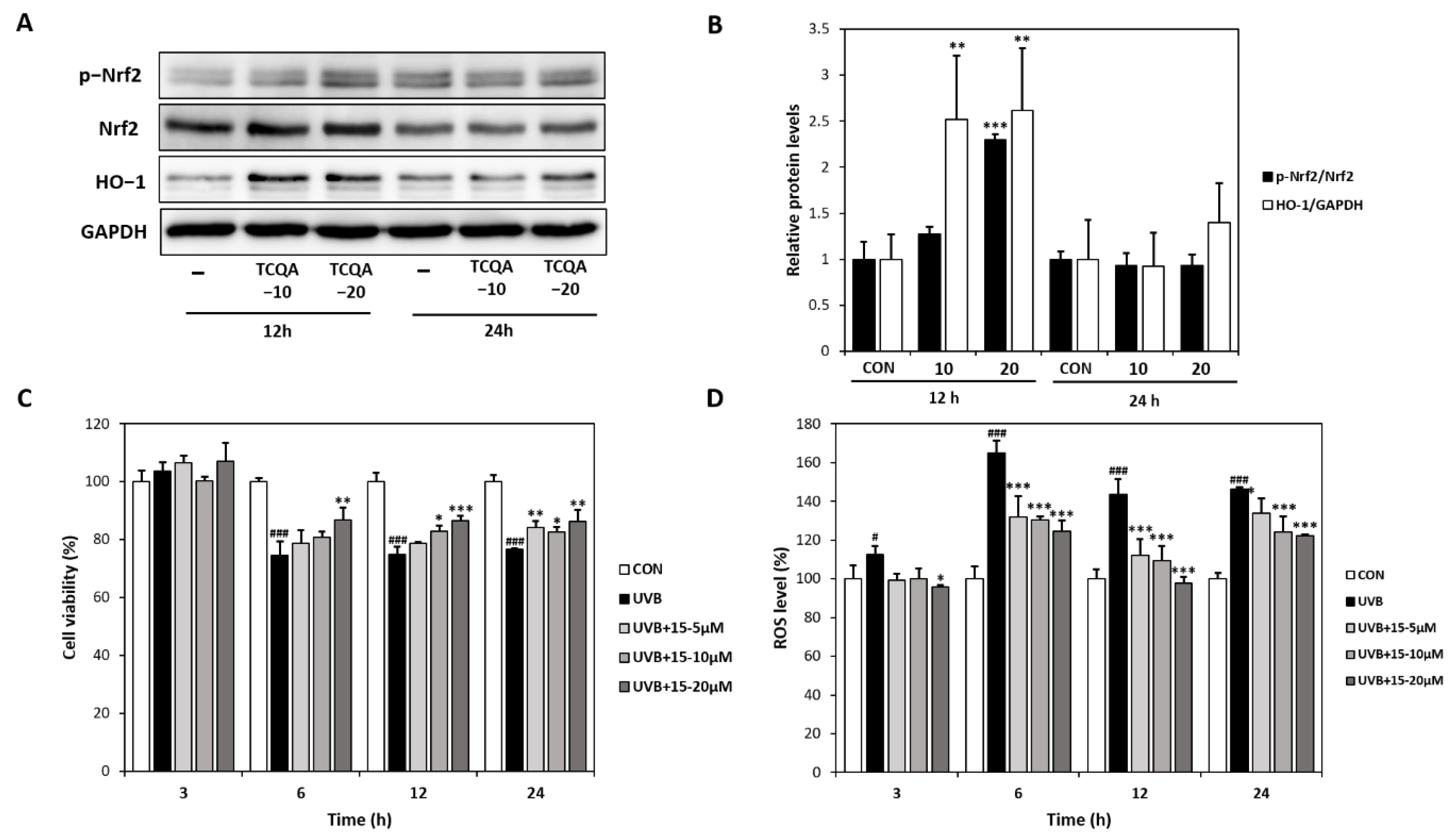
Treatment of HaCaT cells with TCQA for 12 h dose-dependently increased Nrf2 and HO-1 protein expressions by 1.3- and 2.3-fold at 10 μM and by 2.5- and 2.6-fold at 20 μM, respectively, compared to the CON group (Figure 4A,B). The CCK-8 assay showed that cultured HaCaT cells exhibited high ROS levels after exposure to UVB (6, 12, or 24 h). However, treatment with TCQA dose-dependently inhibited apoptosis and maintained cell viability for 24 h (Figure 4C). Furthermore, DCFDA analysis showed ROS levels were concentration-dependently reduced by TCQA at 5, 10, or 20 μM, compared to the UVB-treated (UVB) group. ROS levels were reduced by 20% (5 μM), 21% (10 μM), and 25% (20 μM) after 6 h of TCQA treatment and by 21% (5 μM), 24% (10 μM), and 31% (20 μM) after 12 h of TCQA treatment (Figure 4D).
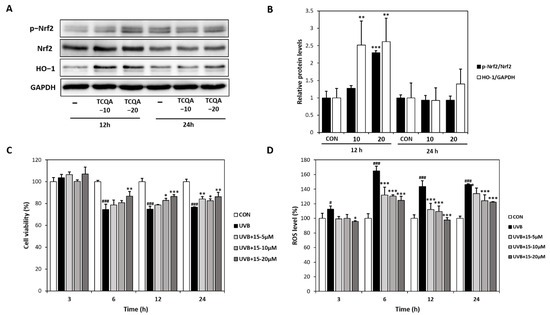
Figure 4.
Effects of TCQA on Nrf2 expression, UVB-induced cell viability, and ROS levels in HaCaT cells. (A,B) pNrf2, Nrf2, and HO-1 protein expressions were analyzed by Western blot (normalized vs. GAPDH). (C) Cell viabilities were determined using a CCK-8 assay, and (D) ROS levels were assessed using a DCFDA assay. HaCaT cells were treated with TCQA at 5, 10, or 20 μM and cultured in the absence or presence of UVB for 3, 6, 12, and 24 h. Data are expressed as means ± SDs (n = 2 to 3). # p < 0.05 and ### p < 0.001 versus CON group; * p < 0.05, ** p < 0.01, *** p < 0.001, versus CON or UVB-treated group (UVB). CON: untreated control group; UVB: UVB-treated group; TCQA: TCQA-treated group.
3.4. Effects of TCQA on MAPK-, AP-1-, and NF-κB-Related Transcription Factors in HaCaT Cells Exposed to UVB
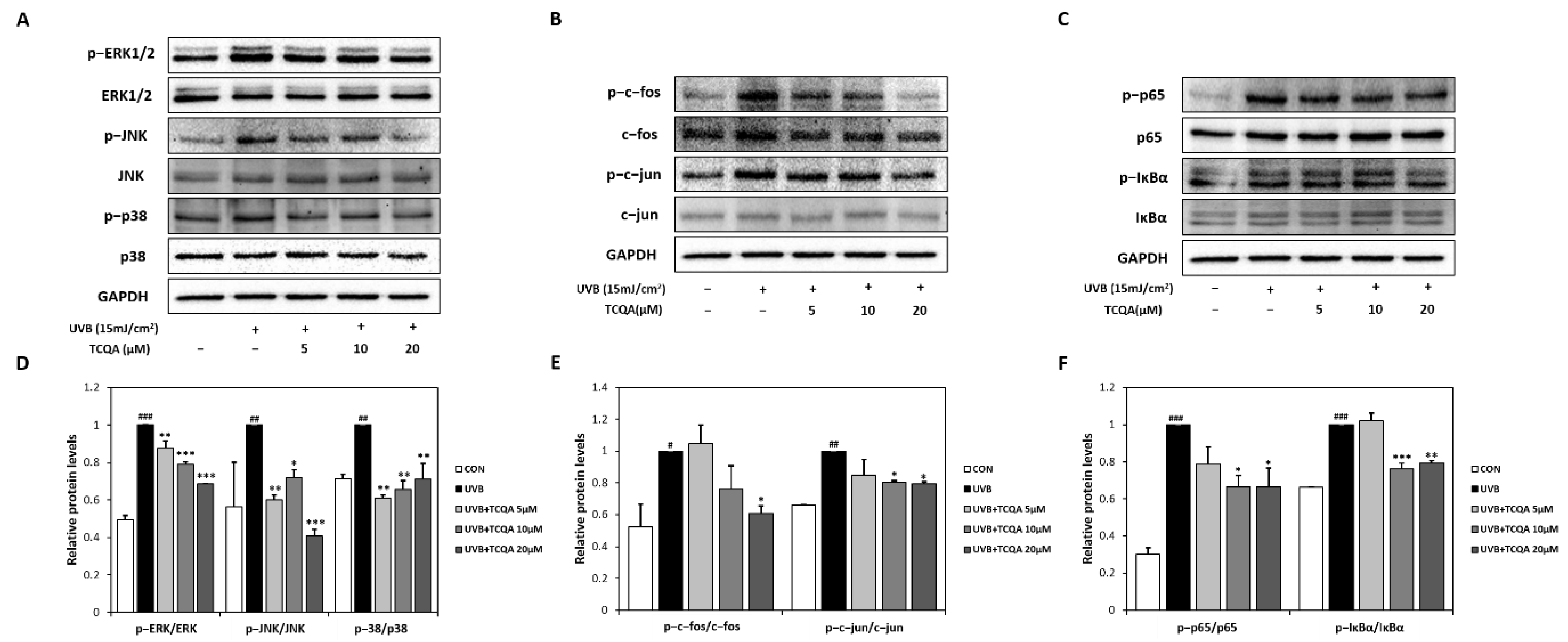
UVB exposure significantly increased the expression of MAPK-related transcription factors, such as p-ERK/ERK, p-JNK/JNK, and p-p38/p38. Treatment with TCQA significantly inhibited the expression levels of p-ERK/ERK (10% at 5 μM, 20% at 10 μM, and 30% at 20 μM), p-JNK/JNK (40% at 5 μM, 30% at 10 μM, and 60% at 20 μM), and p-p38/p38 (40% at 5 μM, 34% at 10 μM, and 30% at 20 μM), compared to the UVB group (Figure 5A,D). TCQA also decreased the expressions of AP-1-related transcription factors, such as p-c-fos/c-fos and p-c-jun/c-jun, in HaCaT cells exposed to UVB. Significant reductions were observed after treatment with 20 μM of TCQA, such as a 40% decrease in p-c-fos/c-fos expression and an 80% decrease in the p-c-jun/c-jun expression (Figure 5B,E). Furthermore, TCQA treatment dose-dependently inhibited the expressions of NF-κB-related transcription factors, such as p-p65/p65 and p-IκBα/IκBα. Significant inhibition was observed in the TCQA 10 μM group, with reductions of 33% and 25% in p-p65/p65 and p-IκBα/IκBα, respectively, compared to the UVB group (Figure 5C,F).
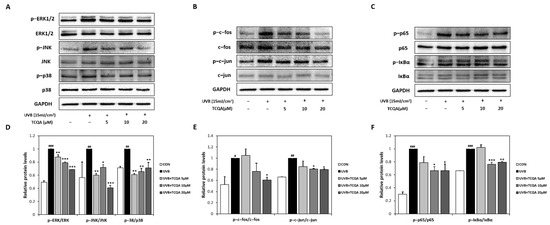
Figure 5.
Effect of TCQA on ERK, p38 and JNK/MAPK, c-fos and c-jun/AP-1, and p65 and IκBα/NF-κB transcription activities. After treating cells with the indicated concentrations of TCQA for 24 h. (A,D) ERK, p38, and JNK; (B,E), c-fos and c-jun; and (C,F) p65 and IκBα. The expressions of subunits gene were analyzed by Western blot (normalized vs. GAPDH) in HaCaT cells exposed to UVB. Data are expressed as means ± SDs (n = 2 to 3). # p < 0.05, ## p < 0.01 and ### p < 0.001, versus CON group; * p < 0.05, ** p < 0.01, and *** p < 0.001 versus UVB group. CON: untreated control group; UVB: UVB-treated group; TCQA: TCQA-treated group.
3.5. Effects of TCQA on AP-1 Activity in PMA-Induced HaCaT Cells and NF-κb Activity in TNF-α/IFN-γ-Induced HaCaT Cells
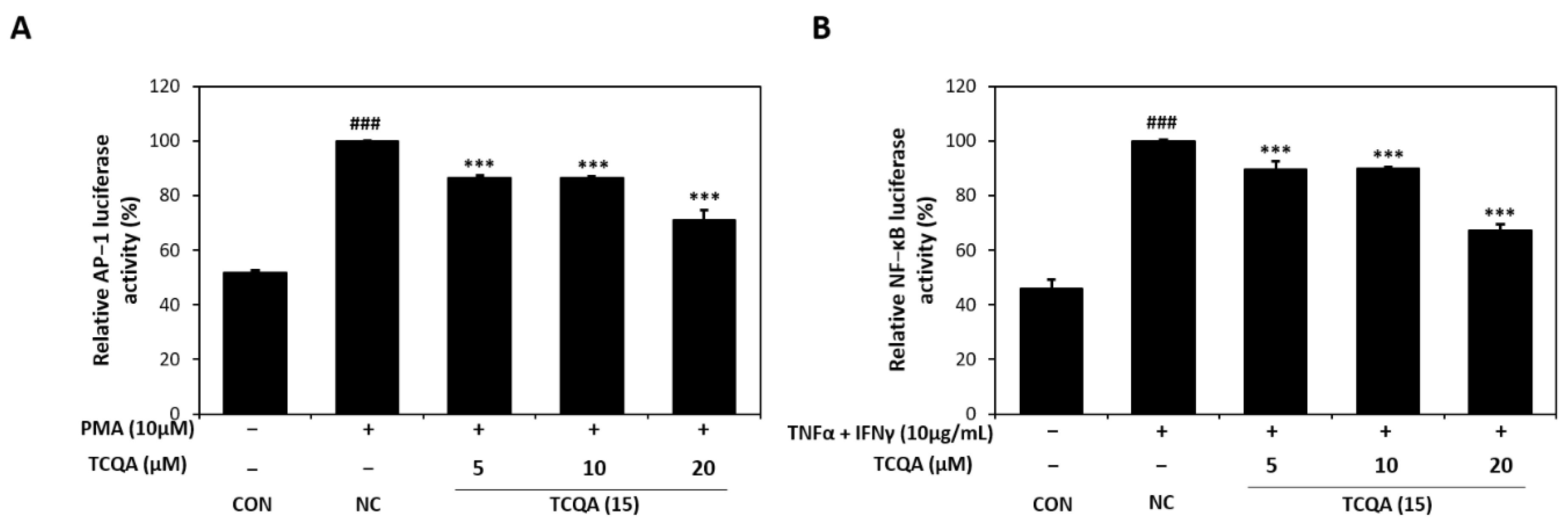
As id shown in Figure 6A, AP-1 luciferase activity increased to 100% in PMA (10 μM)-induced HaCaT cells. On the other hand, AP-1 activity significantly decreased to 86%, 86%, and 71% in the 5, 10, and 20 μM TCQA groups, respectively, vs. the NC group. In addition, exposure to TNF-α/IFN-γ increased NF-κB luciferase activity to approximately 100% of the CON group (46%). However, TCQA at 5, 10, and 20 μM significantly reduced the activation of NF-κB by 12%, 11%, and 33%, respectively (Figure 6B).
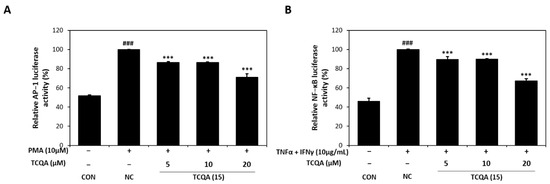
Figure 6.
Effect of TCQA on AP-1 and NF-κB luciferase activities. (A) Transactivation of AP-1 or (B) NF-κB was evaluated using luciferase reporter gene assay in PMA or TNF-α/IFN-γ-induced HaCaT cells. Data are expressed as mean ± SD (n = 3). ### p < 0.001, compared with the CON group; *** p < 0.001, versus NC group. CON: untreated control group; NC: PMA or TNF-α and IFN-γ-treated group; TCQA: TCQA-treated group.
3.6. Effects of TCQA on MMP-1 Expression in HaCaT Cells Exposed to UVB
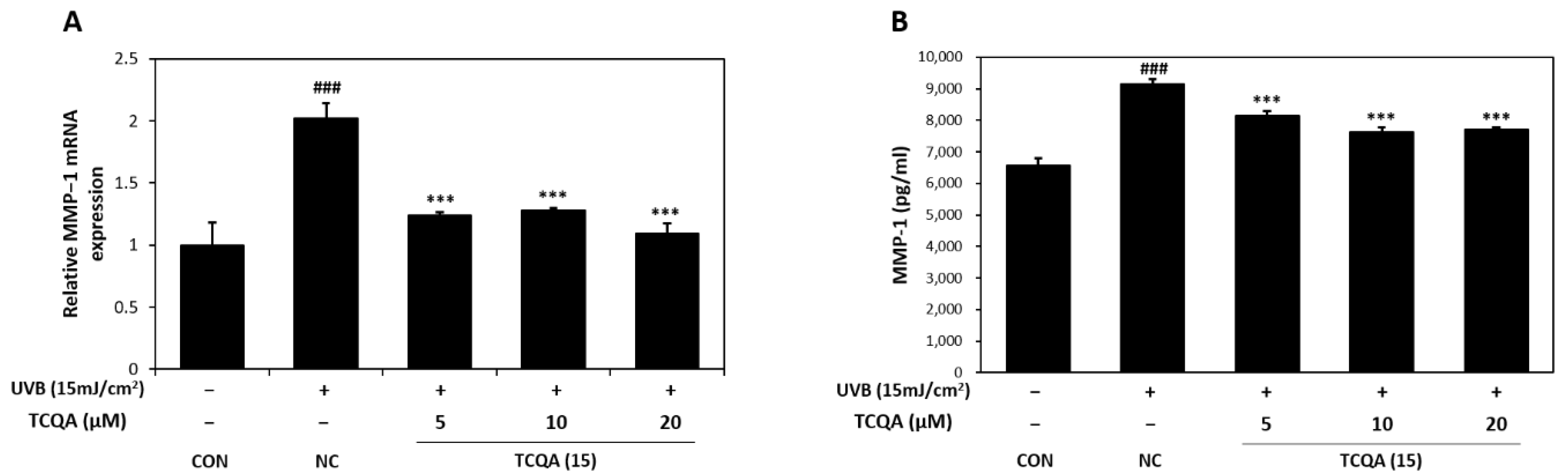
The relative MMP-1 mRNA expression levels in HaCaT cells were measured by qRT-PCR. In comparison to the CON group, the NC group showed a two-fold increase in MMP-1 mRNA expression. However, the MMP-1 mRNA expression level after treatment with TCQA was significantly decreased by 1.7-fold at 5 μM, 1.5-fold at 10 μM, and 2-fold at 20 μM, compared to the NC group (Figure 7A). After UVB treatment, MMP-1 concentration measured by ELISA increased to 9142 pg/mL and TCQA treatment after UVB exposure reduced MMP-1 concentrations to 8152 pg/mL at 5 μM, 7619 pg/mL at 10 μM, and 7700 pg/mL at 20 μM (Figure 7B).
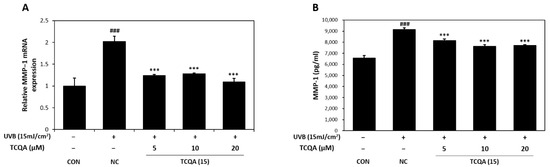
Figure 7.
Effect of TCQA on MMP-1 expression. (A) MMP-1 mRNA expression was analyzed in HaCaT cells exposed to UVB by qRT-PCR and (B) the concentration of MMP-1 was assessed using ELISA. Data are expressed as mean ± SD (n = 2 to 3). ### p < 0.001, versus CON group; *** p < 0.001, versus NC group. CON: untreated control group; NC: UVB-treated group; TCQA: TCQA-treated group.
4. Discussion
Photoaging is a significant extrinsic factor that causes skin aging, and the increase in ROS caused by UV exposure upregulates the MAPK/NF-κB/AP-1 pathway and Nrf2–ARE signaling pathway and causes skin aging and wrinkling [46,47]. Recently, to prevent photoaging, natural products with antioxidant and anti-aging activities have been actively studied [25,26]. N. peltata is a perennial aquatic plant in the family Menyanthaceae and has been used in Traditional Chinese Medicine to treat heat strangury and polyuria [27,48]. Extracts of this plant have been reported to have anti-inflammatory and anti-tumor activities [30,48]. Furthermore, we previously reported on the anti-atopic and antioxidant activities of the EtOH extract of N. peltata [31]. In this study, we isolated and identified 15 phytochemicals, including a new oleanane-type triterpenoid saponin, 3-O-β-D-glucopyranosyl-oleanolic acid 28-O-β-D-glucuronopyranoside, in a methanolic extract of N. peltata roots and evaluated their protective effects on UVB-induced cellular injuries in HaCaT cells. Of these 15 compounds, α-spinasterol, ferulic acid, CQA, and TCQA markedly enhanced Nrf2–ARE luciferase activity; the antioxidant effects of α-spinasterol, ferulic acid, and CQA have been previously reported [43,44,45]. Therefore, we conducted further experiments on TCQA.
UV exposure causes oxidative stress and accelerates skin aging [49], and activation of the Nrf2 signaling pathway inhibits oxidative stress by regulating various antioxidant enzymes, including HO-1 [50]. These help to maintain skin homeostasis by inhibiting DNA damage, cell membrane destruction, and lipid damage [50]. HO-1 regulates oxidative stress and immune responses [51], and the Nrf2 pathway has been demonstrated to suppress ROS increases in UVB-exposed HaCaT cells and, thus, to alleviate apoptosis and reduce skin damage [52,53]. In the present study, TCQA exhibited strong antioxidant activity by upregulating Nrf2 and HO-1 expressions and reducing UVB-induced ROS levels in HaCaT cells. Furthermore, UVB exposure and oxidative stress induce the activation of NF-κB [54], which is involved in cellular senescence and apoptosis by causing protein damage [55]. They also induce the nuclear translocation of p65, which stimulates various inflammatory cells and the expressions of various inflammatory cytokines, like TNF-α and IFN-γ [56,57]. In addition, inflammation mediated by TNF-α enhances ROS production and increases NF-κB activation and the expressions of other cytokines [58]. In addition, activated NF-κB has been demonstrated to be involved in the synthesis of proteins that inhibit collagen production in dermal fibroblasts and HaCaT cells [59], disrupting skin homeostasis and inflammatory cycles, and causing rapid skin aging [60]. We also observed that TCQA suppressed NF-κB, p65, and IκBα transcript levels in UVB- and TNF-α/IFN-γ-induced HaCaT cells, which indicated that TCQA inhibits oxidative stress and inflammatory pathways.
Caffeoylquinic acid (CQA) derivatives are secondary plant metabolites, and many studies have reported their strong antioxidant and anti-inflammatory effects [61,62]. These bioactivities have been attributed to the presence of an ortho-hydroxy group and esterification groups on caffeic acid and quinic acid moieties [63,64]. According to the research on the different bioactivities of dicaffeoylquinic acid (DCQA) isomers, DCQAs (4,5- or 3,4-) with caffeoyl groups substituted at C-4 exhibit stronger physiological activities than 3,5-DCQA [65,66,67], which suggests the importance of the presence of the cis-caffeoyl group in DCQA [68]. Nevertheless, TCQA has been reported to have more potent pharmacological activities than DCQA. 3, 4, 5-TCQA has a stronger antiradical ability than DCQA [69], induces ATP synthesis [70], and effectively inhibits proinflammatory substances such as TNF-α [71]. This enhanced bioactivity has been attributed to increasing the steric hindrance of TCQA due to the presence of three caffeoyl groups [69,72]. Our in vitro results obtained using Nrf2–ARE reporter HaCaT cells confirmed that TCQA better inhibits Nrf2 activation compared with several CQAs and DCQAs.
MAPKs are composed of extracellular signal-regulated kinase (ERK), c-Jun N-terminal kinase (JNK), and p38 kinase [73]. They are involved in a variety of cellular activities, including cell proliferation, differentiation, and death [74]. UV-induced oxidative stress increases ROS levels and promotes the phosphorylation of ERK, JNK, and p38, thereby activating the MAPK signaling pathway [75,76]. In this study, TCQA significantly decreased the protein expressions of p-ERK, p-JNK, and p38 in UVB-irradiated HaCaT cells. Thus, our results suggest that TCQA inhibits UVB-induced MAPK activation. AP-1 is a transcription factor composed of c-Jun and c-Fos and is mediated by an upstream MAPK signaling pathway consisting of ERK, JNK, and p38 [77]. In other words, the expression of c-Fos is induced by ERK activation, and the expression of c-Jun is induced by JNK and p38 [78,79]. Furthermore, c-Fos and c-Jun combine to form either a heterodimer (Fos–Jun) or a homodimer (Jun–Jun), which regulate the transcriptions of numerous genes, including AP-1 [80]. Moreover, the activation of AP-1 transcription can induce ceramide production, leading to the overproduction of proinflammatory cytokines [81].
In the present study, TCQA significantly and dose-dependently reduced the protein expressions of phosphorylated c-Fos and c-Jun in UVB-irradiated HaCaT cells and significantly inhibited the dose-dependent upregulation of AP-1 induced by PMA exposure. The ROS-induced MAPK, AP-1, and NF-κB signaling pathways activate MMPs, which leads to MMP secretion from keratinocytes and fibroblasts, and accelerates protein and collagen degradation [82]. In particular, MMP-1 is a key marker of skin photoaging and causes collagen breakdown in the dermis [83], which contributes to the aging responses of dermal cells, such as dermal layer atrophy, skin wrinkling, and loss of elasticity [84]. TCQA inhibited the UVB-induced activation of MMP-1 at all concentrations examined. Taken together, we conclude that TCQA protects against photoaging and skin wrinkling by modulating MAPK/NF-κB/AP-1 signaling, reducing MMP-1 activity, and activating Nrf2 in UVB-damaged HaCaT cells.
5. Conclusions
Phytochemical investigation of the MeOH extract of N. peltata roots led to the isolation of 15 compounds (1–15). Among these compounds was a new oleanane-type triterpenoid saponin, 3-O-β-D-glucopyranosyl-oleanolic acid 28-O-β-D-glucuronopyranoside (peltatasaponin A), the chemical structure of which was characterized by 1D and 2D NMR spectroscopic data and HRESIMS, as well as chemical reaction. Among the isolates, TCQA effectively inhibited ROS production by inducing the expression of Nrf2/HO-1 antioxidant enzyme in HaCaT cells. In addition, treatment with TCQA significantly inhibited MMP-1 expression by suppressing MAPK, AP-1, and NF-kB pathways and their respective subunits in HaCaT cells exposed to UVB. Thus, our study supports the potential use of TCQA as an antioxidant and anti-wrinkle treatment.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/antiox12101899/s1, Figure S1: The 1H NMR spectrum of compound 1; Figures S2–S6: The 1H, 13C, 1H-1H COSY, HSQC, and HMBC NMR spectrum of compound 2; Figures S7–S19: The 1H NMR spectrum of compound 3–15.
Author Contributions
K.H.K., S.-N.K. and M.H.Y. designed the study; T.-Y.K., N.-J.P., B.-G.J., M.-J.K., T.-H.K. and B.S.L. performed the experiments. M.-J.K., T.-H.K. and B.S.L. analyzed the data and interpreted the results; T.-Y.K., N.-J.P., B.-G.J., K.H.K., S.-N.K. and M.H.Y. drafted the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by a National Research Foundation of Korea (NRF) grant funded by the Ministry of Science, ICT, and Future Planning (NRF-2019M3A9I3080265, NRF-2019M3A9I3080266, and NRF-2022R1A2C1003088).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are contained in the article.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Mohd Zaid, N.A.; Sekar, M.; Bonam, S.R.; Gan, S.H.; Lum, P.T.; Begum, M.Y.; Mat Rani, N.N.I.; Vaijanathappa, J.; Wu, Y.S.; Subramaniyan, V.; et al. Promising Natural Products in New Drug Design, Development, and Therapy for Skin Disorders: An Overview of Scientific Evidence and Understanding Their Mechanism of Action. Drug Des. Dev. Ther. 2022, 16, 23–66. [Google Scholar] [CrossRef] [PubMed]
- Franco, A.C.; Aveleira, C.; Cavadas, C. Skin Senescence: Mechanisms and Impact on Whole-Body Aging. Trends Mol. Med. 2022, 28, 97–109. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; He, L.; Zhang, N.; Ma, L.; Yao, L. Photoprotective Effect of Artemisia sieversiana Ehrhart Essential Oil Against UVB-induced Photoaging in Mice. Photochem. Photobiol. 2022, 98, 958–968. [Google Scholar] [CrossRef]
- Tanveer, M.A.; Rashid, H.; Tasduq, S.A. Molecular Basis of Skin Photoaging and Therapeutic Interventions by Plant-Derived Natural Product Ingredients: A Comprehensive Review. Heliyon 2023, 9, e13580. [Google Scholar] [CrossRef] [PubMed]
- Bayo Jimenez, M.T.; Frenis, K.; Hahad, O.; Steven, S.; Cohen, G.; Cuadrado, A.; Münzel, T.; Daiber, A. Protective Actions of Nuclear Factor Erythroid 2-Related Factor 2 (NRF2) and Downstream Pathways against Environmental Stressors. Free Radic. Biol. Med. 2022, 187, 72–91. [Google Scholar] [CrossRef]
- Sharma, V.; Kaur, A.; Singh, T.G. Counteracting Role of Nuclear Factor Erythroid 2-Related Factor 2 Pathway in Alzheimer’s Disease. Biomed. Pharmacother. 2020, 129, 110373. [Google Scholar] [CrossRef]
- Kasai, S.; Shimizu, S.; Tatara, Y.; Mimura, J.; Itoh, K. Regulation of Nrf2 by Mitochondrial Reactive Oxygen Species in Physiology and Pathology. Biomolecules 2020, 10, 320. [Google Scholar] [CrossRef]
- Zhang, Q.; Pi, J.; Woods, C.G.; Andersen, M.E. A Systems Biology Perspective on Nrf2-Mediated Antioxidant Response. Toxicol. Appl. Pharmacol. 2010, 244, 84–97. [Google Scholar] [CrossRef]
- Liu, T.; Xia, Q.; Lv, Y.; Wang, Z.; Zhu, S.; Qin, W.; Yang, Y.; Liu, T.; Wang, X.; Zhao, Z.; et al. ErZhiFormula Prevents UV-Induced Skin Photoaging by Nrf2/HO-1/NQO1 Signaling: An in Vitro and in Vivo Studies. J. Ethnopharmacol. 2023, 309, 115935. [Google Scholar] [CrossRef]
- Chartoumpekis, D.; Ziros, P.G.; Psyrogiannis, A.; Kyriazopoulou, V.; Papavassiliou, A.G.; Habeos, I.G. Simvastatin Lowers Reactive Oxygen Species Level by Nrf2 Activation via PI3K/Akt Pathway. Biochem. Biophys. Res. Commun. 2010, 396, 463–466. [Google Scholar] [CrossRef]
- Papaccio, F.; D’Arino, A.; Caputo, S.; Bellei, B. Focus on the Contribution of Oxidative Stress in Skin Aging. Antioxidants 2022, 11, 1121. [Google Scholar] [CrossRef]
- Lee, C.S.; Bae, I.-H.; Han, J.; Choi, G.; Hwang, K.-H.; Kim, D.-H.; Yeom, M.-H.; Park, Y.-H.; Park, M. Compound K Inhibits MMP-1 Expression through Suppression of c-Src-Dependent ERK Activation in TNF-α-Stimulated Dermal Fibroblast. Exp. Dermatol. 2014, 23, 819–824. [Google Scholar] [CrossRef]
- Kirindage, K.G.I.S.; Jayasinghe, A.M.K.; Han, E.-J.; Jee, Y.; Kim, H.-J.; Do, S.G.; Fernando, I.P.S.; Ahn, G. Fucosterol Isolated from Dietary Brown Alga Sargassum horneri Protects TNF-α/IFN-γ-Stimulated Human Dermal Fibroblasts Via Regulating Nrf2/HO-1 and NF-κB/MAPK Pathways. Antioxidants 2022, 11, 1429. [Google Scholar] [CrossRef]
- Rahman, I. Oxidative Stress, Transcription Factors and Chromatin Remodelling in Lung Inflammation. Biochem. Pharmacol. 2002, 64, 935–942. [Google Scholar] [CrossRef]
- Jabłońska-Trypuć, A.; Matejczyk, M.; Rosochacki, S. Matrix Metalloproteinases (MMPs), the Main Extracellular Matrix (ECM) Enzymes in Collagen Degradation, as a Target for Anticancer Drugs. J. Enzym. Inhib. Med. Chem. 2016, 31, 177–183. [Google Scholar] [CrossRef] [PubMed]
- Pillai, S.; Oresajo, C.; Hayward, J. Ultraviolet Radiation and Skin Aging: Roles of Reactive Oxygen Species, Inflammation and Protease Activation, and Strategies for Prevention of Inflammation-Induced Matrix Degradation—A Review. Int. J. Cosmet. Sci. 2005, 27, 17–34. [Google Scholar] [CrossRef]
- Brenneisen, P.; Sies, H.; Scharffetter-Kochanek, K. Ultraviolet-B Irradiation and Matrix Metalloproteinases. Ann. N. Y. Acad. Sci. 2002, 973, 31–43. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.H.; Lee, M.J.; Lee, S.R.; Kim, K.H.; Cho, K.H.; Eun, H.C.; Chung, J.H. Augmentation of UV-Induced Skin Wrinkling by Infrared Irradiation in Hairless Mice. Mech. Ageing Dev. 2005, 126, 1170–1177. [Google Scholar] [CrossRef] [PubMed]
- Oh, J.H.; Joo, Y.H.; Karadeniz, F.; Ko, J.; Kong, C.-S. Syringaresinol Inhibits UVA-Induced MMP-1 Expression by Suppression of MAPK/AP-1 Signaling in HaCaT Keratinocytes and Human Dermal Fibroblasts. Int. J. Mol. Sci. 2020, 21, 3981. [Google Scholar] [CrossRef]
- Kim, M.-S.; Kim, Y.K.; Cho, K.H.; Chung, J.H. Regulation of Type I Procollagen and MMP-1 Expression after Single or Repeated Exposure to Infrared Radiation in Human Skin. Mech. Ageing Dev. 2006, 127, 875–882. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, L.; Wen, X.; Hao, D.; Zhang, N.; He, G.; Jiang, X. NF-κB Signaling in Skin Aging. Mech. Ageing Dev. 2019, 184, 111160. [Google Scholar] [CrossRef] [PubMed]
- Sunil, M.A.; Sunitha, V.S.; Santhakumaran, P.; Mohan, M.C.; Jose, M.S.; Radhakrishnan, E.K.; Mathew, J. Protective Effect of (+)–Catechin against Lipopolysaccharide-Induced Inflammatory Response in RAW 264.7 Cells through Downregulation of NF-κB and P38 MAPK. Inflammopharmacology 2021, 29, 1139–1155. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Oh, J.-Y.; Lee, W.; Jeon, Y.-J. Fucoidan Isolated from Hizikia fusiforme Suppresses Ultraviolet B-Induced Photodamage by down-Regulating the Expressions of Matrix Metalloproteinases and pro-Inflammatory Cytokines via Inhibiting NF-κB, AP-1, and MAPK Signaling Pathways. Int. J. Biol. Macromol. 2021, 166, 751–759. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Li, L.; Huang, G.; Zhou, T.; Zhang, X.; Leng, X.; Chen, Z.; Lin, J. Embelia laeta Aqueous Extract Suppresses Acute Inflammation via Decreasing COX-2/iNOS Expression and Inhibiting NF-κB Pathway. J. Ethnopharmacol. 2021, 281, 114575. [Google Scholar] [CrossRef]
- Cavinato, M.; Waltenberger, B.; Baraldo, G.; Grade, C.V.C.; Stuppner, H.; Jansen-Dürr, P. Plant Extracts and Natural Compounds Used against UVB-Induced Photoaging. Biogerontology 2017, 18, 499–516. [Google Scholar] [CrossRef]
- Mukherjee, P.K.; Maity, N.; Nema, N.K.; Sarkar, B.K. Bioactive Compounds from Natural Resources against Skin Aging. Phytomedicine 2011, 19, 64–73. [Google Scholar] [CrossRef]
- Tippery, N.P.; Sokolik, J.T. Phylogenetics and Seed Morphology of African Nymphoides (Menyanthaceae). Nord. J. Bot. 2020, 38, njb02545. [Google Scholar] [CrossRef]
- Lee, B.Y.; Nam, G.H.; Lee, J.Y.; Park, C.H.; Lim, C.E.; Kim, M.H.; Lee, S.J.; Roh, T.K.; Lim, J.A.; Han, J.E. National List of Species of Korea (Vascular Plants); National Institute of Biological Resources: Incheon, Republic of Korea, 2011; p. 633.
- Nishihiro, J.; Uesugi, R.; Takagawa, S.; Washitani, I. Toward the Restoration of a Sustainable Population of a Threatened Aquatic Plant, Nymphoides peltata: Integrated Genetic/Demographic Studies and Practices. Biol. Conserv. 2009, 142, 1906–1912. [Google Scholar] [CrossRef]
- Johansson, S.; Göransson, U.; Luijendijk, T.; Backlund, A.; Claeson, P.; Bohlin, L. A Neutrophil Multitarget Functional Bioassay to Detect Anti-Inflammatory Natural Products. J. Nat. Prod. 2002, 65, 32–41. [Google Scholar] [CrossRef]
- Kim, T.-Y.; Park, N.-J.; Jegal, H.; Paik, J.-H.; Choi, S.; Kim, S.-N.; Yang, M.H. Nymphoides peltata Root Extracts Improve Atopic Dermatitis by Regulating Skin Inflammatory and Anti-Oxidative Enzymes in 2,4-Dinitrochlorobenzene (DNCB)-Induced SKH-1 Hairless Mice. Antioxidants 2023, 12, 873. [Google Scholar] [CrossRef]
- Lee, D.; Kim, J.-Y.; Kwon, H.C.; Kwon, J.; Jang, D.S.; Kang, K.S. Dual Beneficial Effects of α-Spinasterol Isolated from Aster Pseudoglehnii on Glucose Uptake in Skeletal Muscle Cells and Glucose-Stimulated Insulin Secretion in Pancreatic β-Cells. Plants 2022, 11, 658. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.E.; Jeon, B.J.; Park, M.Y.; Kim, B.S. Inhibitory Activity of Sedum Middendorffianum-Derived 4-Hydroxybenzoic Acid and Vanillic Acid on the Type III Secretion System of Pseudomonas Syringae Pv. Tomato DC3000. Plant Pathol. J. 2020, 36, 608. [Google Scholar] [CrossRef]
- Colombo, R.; Batista, A.N.D.L.; Bomfim, G.C.; Burgos, R.C.; Cavalheiro, A.J.; Bolzani, V.D.S.; Silva, D.H.S.; Reimberg, M.C.H. Validated High-Performance Liquid Chromatographic Method for the Standardisation of Ptychopetalum olacoides Benth., Olacaceae, Commercial Extracts. Rev. Bras. Farmacogn. 2010, 20, 781–788. [Google Scholar] [CrossRef]
- Karthikeyan, R.; Devadasu, C.; Srinivasa Babu, P. Isolation, Characterization, and RP-HPLC Estimation of p-Coumaric Acid from Methanolic Extract of Durva Grass (Cynodon dactylon Linn.) (Pers.). Int. J. Anal. Chem. 2015, 2015, 201386. [Google Scholar] [CrossRef]
- Jeong, C.-H.; Jeong, H.R.; Choi, G.N.; Kim, D.-O.; Lee, U.K.; Heo, H.J. Neuroprotective and Anti-Oxidant Effects of Caffeic Acid Isolated from Erigeron annuus Leaf. Chin. Med. 2011, 6, 25. [Google Scholar] [CrossRef] [PubMed]
- Jain, P.G.; Surana, S.J. Isolation, Characterization and Hypolipidemic Activity of Ferulic Acid in High-Fat-Diet-Induced Hyperlipidemia in Laboratory Rats. EXCLI J. 2016, 15, 599. [Google Scholar] [CrossRef]
- Wong, S.K.; Lim, Y.Y.; Ling, S.K.; Chan, E.W.C. Caffeoylquinic Acids in Leaves of Selected Apocynaceae Species: Their Isolation and Content. Pharmacogn. Res. 2014, 6, 67. [Google Scholar] [CrossRef]
- Shi, S.; Huang, K.; Zhang, Y.; Zhao, Y.; Du, Q. Purification and Identification of Antiviral Components from Laggera pterodonta by High-Speed Counter-Current Chromatography. J. Chromatogr. B 2007, 859, 119–124. [Google Scholar] [CrossRef]
- Islam, M.S.; Yoshimoto, M.; Yahara, S.; Okuno, S.; Ishiguro, K.; Yamakawa, O. Identification and Characterization of Foliar Polyphenolic Composition in Sweetpotato (Ipomoea batatas L.) Genotypes. J. Agric. Food Chem. 2002, 50, 3718–3722. [Google Scholar] [CrossRef]
- Magalhaes, A.F.; Goulart de Azevedo Tozzi, A.M.; Santos, C.C.; Serrano, D.R.; Zanotti-Magalhaes, E.M.; Magalhaes, E.G.; Magalhaes, L.A. Saponins from Swartzia langsdorffii: Biological Activities. Mem. Inst. Oswaldo Cruz 2003, 98, 713–718. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mizui, F.; Kasai, R.; Ohtani, K.; Tanaka, O. Saponins from Bran of Quinoa, Chenopodium quinoa WILLD. II. Chem. Pharm. Bull. 1990, 38, 375–377. [Google Scholar] [CrossRef]
- Neopane, D.; Ansari, V.A.; Singh, A. Ferulic Acid: Signaling Pathways in Aging. Drug Res. 2023, 73, 318–324. [Google Scholar] [CrossRef]
- Feng, J.; Zhang, P.; Chen, X.; He, G. PI3K and ERK/Nrf2 Pathways Are Involved in Oleanolic Acid-Induced Heme Oxygenase-1 Expression in Rat Vascular Smooth Muscle Cells. J. Cell. Biochem. 2011, 112, 1524–1531. [Google Scholar] [CrossRef]
- Ha, J.W.; Boo, Y.C. Siegesbeckiae Herba Extract and Chlorogenic Acid Ameliorate the Death of HaCaT Keratinocytes Exposed to Airborne Particulate Matter by Mitigating Oxidative Stress. Antioxidants 2021, 10, 1762. [Google Scholar] [CrossRef]
- Hwang, B.-M.; Noh, E.-M.; Kim, J.-S.; Kim, J.-M.; You, Y.-O.; Hwang, J.-K.; Kwon, K.-B.; Lee, Y.-R. Curcumin Inhibits UVB-Induced Matrix Metalloproteinase-1/3 Expression by Suppressing the MAPK-P38/JNK Pathways in Human Dermal Fibroblasts. Exp. Dermatol. 2013, 22, 371–374. [Google Scholar] [CrossRef]
- Hwang, E.; Lin, P.; Ngo, H.T.T.; Gao, W.; Wang, Y.-S.; Yu, H.-S.; Yi, T.-H. Icariin and Icaritin Recover UVB-Induced Photoaging by Stimulating Nrf2/ARE and Reducing AP-1 and NF-κB Signaling Pathways: A Comparative Study on UVB-Irradiated Human Keratinocytes. Photochem. Photobiol. Sci. 2018, 17, 1396–1408. [Google Scholar] [CrossRef] [PubMed]
- Khan, Z.; Chowdhury, N.; Sharmin, S.; Sohrab, M.H. Medicinal Values of Aquatic Plant Genus Nymphoides Grown in Asia: A Review. Asian Pac. J. Trop. Biomed. 2018, 8, 113. [Google Scholar] [CrossRef]
- Nakai, K.; Tsuruta, D. What Are Reactive Oxygen Species, Free Radicals, and Oxidative Stress in Skin Diseases? Int. J. Mol. Sci. 2021, 22, 10799. [Google Scholar] [CrossRef]
- Ikehata, H.; Yamamoto, M. Roles of the KEAP1-NRF2 System in Mammalian Skin Exposed to UV Radiation. Toxicol. Appl. Pharmacol. 2018, 360, 69–77. [Google Scholar] [CrossRef]
- Campbell, N.K.; Fitzgerald, H.K.; Dunne, A. Regulation of Inflammation by the Antioxidant Haem Oxygenase 1. Nat. Rev. Immunol. 2021, 21, 411–425. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Chen, Z.; Wu, Z. Four-Octyl Itaconate Attenuates UVB-Induced Melanocytes and Keratinocytes Apoptosis by Nrf2 Activation-Dependent ROS Inhibition. Oxidative Med. Cell. Longev. 2022, 2022, e9897442. [Google Scholar] [CrossRef]
- Wang, S.-H.; Chen, Y.-S.; Lai, K.-H.; Lu, C.-K.; Chang, H.-S.; Wu, H.-C.; Yen, F.-L.; Chen, L.-Y.; Lee, J.-C.; Yen, C.-H. Prinsepiae Nux Extract Activates NRF2 Activity and Protects UVB-Induced Damage in Keratinocyte. Antioxidants 2022, 11, 1755. [Google Scholar] [CrossRef]
- Guo, C.; Ning, X.; Zhang, J.; Zhang, C.; Wang, J.; Su, L.; Han, J.; Ma, N. Ultraviolet B Radiation Induces Oxidative Stress and Apoptosis in Human Lens Epithelium Cells by Activating NF-κB Signaling to down-Regulate Sodium Vitamin C Transporter 2 (SVCT2) Expression. Cell Cycle 2023, 22, 1450–1462. [Google Scholar] [CrossRef] [PubMed]
- Kriete, A.; Mayo, K.L. Atypical Pathways of NF-κB Activation and Aging. Exp. Gerontol. 2009, 44, 250–255. [Google Scholar] [CrossRef]
- Giridharan, S.; Srinivasan, M. Mechanisms of NF-κB p65 and Strategies for Therapeutic Manipulation. J. Inflamm. Res. 2018, 11, 407–419. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Zhang, L.; Joo, D.; Sun, S.-C. NF-κB Signaling in Inflammation. Signal Transduct. Target. Ther. 2017, 2, 17023. [Google Scholar] [CrossRef]
- Young, C.N.; Koepke, J.I.; Terlecky, L.J.; Borkin, M.S.; Boyd, S.L.; Terlecky, S.R. Reactive Oxygen Species in Tumor Necrosis Factor-α-Activated Primary Human Keratinocytes: Implications for Psoriasis and Inflammatory Skin Disease. J. Investig. Dermatol. 2008, 128, 2606–2614. [Google Scholar] [CrossRef]
- Tanaka, K.; Asamitsu, K.; Uranishi, H.; Iddamalgoda, A.; Ito, K.; Kojima, H.; Okamoto, T. Protecting Skin Photoaging by NF-κB Inhibitor. Curr. Drug Metab. 2010, 11, 431–435. [Google Scholar] [CrossRef]
- García-García, V.A.; Alameda, J.P.; Page, A.; Casanova, M.L. Role of NF-κB in Ageing and Age-Related Diseases: Lessons from Genetically Modified Mouse Models. Cells 2021, 10, 1906. [Google Scholar] [CrossRef] [PubMed]
- Xue, N.; Liu, Y.; Jin, J.; Ji, M.; Chen, X. Chlorogenic Acid Prevents UVA-Induced Skin Photoaging through Regulating Collagen Metabolism and Apoptosis in Human Dermal Fibroblasts. Int. J. Mol. Sci. 2022, 23, 6941. [Google Scholar] [CrossRef]
- Li, R.; Tao, M.; Wu, T.; Zhou, Z.; Xu, T.; Pan, S.; Xu, X. A promising strategy for investigating the anti-aging effect of natural compounds: A case study of caffeoylquinic acids. Food Funct. 2021, 12, 8583. [Google Scholar] [CrossRef] [PubMed]
- Maurin, C.; Bailly, F.; Cotelle, P. Structure-Activity Relationships of HIV-1 Integrase Inhibitors-Enzyme-Ligand Interactions. Curr. Med. Chem. 2003, 10, 1795–1810. [Google Scholar] [CrossRef]
- Alcázar Magaña, A.; Kamimura, N.; Soumyanath, A.; Stevens, J.F.; Maier, C.S. Caffeoylquinic Acids: Chemistry, Biosynthesis, Occurrence, Analytical Challenges, and Bioactivity. Plant J. 2021, 107, 1299–1319. [Google Scholar] [CrossRef]
- Xu, J.-G.; Hu, Q.-P.; Liu, Y. Antioxidant and DNA-Protective Activities of Chlorogenic Acid Isomers. J. Agric. Food Chem. 2012, 60, 11625–11630. [Google Scholar] [CrossRef]
- Indy Tamayose, C.; dos Santos, E.A.; Roque, N.; Costa-Lotufo, L.V.; Pena Ferreira, M.J. Caffeoylquinic Acids: Separation Method, Antiradical Properties and Cytotoxicity. Chem. Biodivers. 2019, 16, e1900093. [Google Scholar] [CrossRef]
- Arulselvan, P.; Fard, M.T.; Tan, W.S.; Gothai, S.; Fakurazi, S.; Norhaizan, M.E.; Kumar, S.S. Role of Antioxidants and Natural Products in Inflammation. Oxidative Med. Cell. Longev. 2016, 2016, e5276130. [Google Scholar] [CrossRef]
- Ge, L.; Wan, H.; Tang, S.; Chen, H.; Li, J.; Zhang, K.; Zhou, B.; Fei, J.; Wu, S.; Zeng, X. Novel Caffeoylquinic Acid Derivatives from Lonicera japonica Thunb. Flower Buds Exert Pronounced Anti-HBV Activities. RSC Adv. 2018, 8, 35374–35385. [Google Scholar] [CrossRef]
- Liu, W.; Li, J.; Zhang, X.; Zu, Y.; Yang, Y.; Liu, W.; Xu, Z.; Gao, H.; Sun, X.; Jiang, X.; et al. Current Advances in Naturally Occurring Caffeoylquinic Acids: Structure, Bioactivity, and Synthesis. J. Agric. Food Chem. 2020, 68, 10489–10516. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, K.; Han, J.; Shigemori, H.; Isoda, H. Caffeoylquinic Acid Induces ATP Production and Energy Metabolism in Human Neurotypic SH-SY5Y Cells. Nutr. Aging 2012, 1, 141–150. [Google Scholar] [CrossRef]
- Abdel Motaal, A.; Ezzat, S.M.; Tadros, M.G.; El-Askary, H.I. In Vivo Anti-Inflammatory Activity of Caffeoylquinic Acid Derivatives from Solidago virgaurea in Rats. Pharm. Biol. 2016, 54, 2864–2870. [Google Scholar] [CrossRef]
- Tang, B.; Huang, Y.; Yang, H.; Tang, P.; Li, H. Molecular Mechanism of the Binding of 3,4,5-Tri-O-caffeoylquinic Acid to Human Serum Albumin: Saturation Transfer Difference NMR, Multi-Spectroscopy, and Docking Studies. J. Photochem. Photobiol. B 2016, 165, 24–33. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.K.; Choi, E.-J. Pathological Roles of MAPK Signaling Pathways in Human Diseases. Biochim. Biophys. Acta BBA Mol. Basis Dis. 2010, 1802, 396–405. [Google Scholar] [CrossRef]
- Wada, T.; Penninger, J.M. Mitogen-Activated Protein Kinases in Apoptosis Regulation. Oncogene 2004, 23, 2838–2849. [Google Scholar] [CrossRef] [PubMed]
- Long, Y.; Wang, W.; Zhang, Y.; Du, F.; Zhang, S.; Li, Z.; Deng, J.; Li, J. Photoprotective Effects of Dendrobium nobile Lindl. Polysaccharides against UVB-Induced Oxidative Stress and Apoptosis in HaCaT Cells. Int. J. Mol. Sci. 2023, 24, 6120. [Google Scholar] [CrossRef]
- Mantena, S.K.; Katiyar, S.K. Grape Seed Proanthocyanidins Inhibit UV-Radiation-Induced Oxidative Stress and Activation of MAPK and NF-κB Signaling in Human Epidermal Keratinocytes. Free Radic. Biol. Med. 2006, 40, 1603–1614. [Google Scholar] [CrossRef]
- Liou, J.-T.; Lin, C.-S.; Liao, Y.-C.; Ho, L.-J.; Yang, S.-P.; Lai, J.-H. JNK/AP-1 Activation Contributes to Tetrandrine Resistance in T-Cell Acute Lymphoblastic Leukaemia. Acta Pharmacol. Sin. 2017, 38, 1171–1183. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Hao, Z.; Xing, Z.; Tan, J.; Zhao, Y.; Li, M. Impinging Flow Induces Expression of Monocyte Chemoattractant Protein-1 in Endothelial Cells Through Activation of the c-Jun N-Terminal Kinase/c-Jun/p38/c-Fos Pathway. World Neurosurg. 2022, 164, e681–e693. [Google Scholar] [CrossRef]
- Muthusamy, V.; Piva, T.J. The UV Response of the Skin: A Review of the MAPK, NFκB and TNFα Signal Transduction Pathways. Arch. Dermatol. Res. 2010, 302, 5–17. [Google Scholar] [CrossRef]
- Janson, N.D.; Jehanathan, N.; Jung, S.; Priyathilaka, T.T.; Nam, B.-H.; Kim, M.-J.; Lee, J. Insight into the Molecular Function and Transcriptional Regulation of Activator Protein 1 (AP-1) Components c-Jun/c-Fos Ortholog in Red Lip Mullet (Liza haematocheila). Fish Shellfish Immunol. 2019, 93, 597–611. [Google Scholar] [CrossRef]
- Briganti, S.; Picardo, M. Antioxidant Activity, Lipid Peroxidation and Skin Diseases. What’s New. J. Eur. Acad. Dermatol. Venereol. 2003, 17, 663–669. [Google Scholar] [CrossRef]
- Xiao, Z.; Yang, S.; Chen, J.; Li, C.; Zhou, C.; Hong, P.; Sun, S.; Qian, Z.-J. Trehalose against UVB-Induced Skin Photoaging by Suppressing MMP Expression and Enhancing Procollagen I Synthesis in HaCaT Cells. J. Funct. Foods 2020, 74, 104198. [Google Scholar] [CrossRef]
- Kim, H.M.; Lee, D.E.; Park, S.D.; Kim, Y.-T.; Kim, Y.J.; Jeong, J.W.; Jang, S.S.; Ahn, Y.-T.; Sim, J.-H.; Huh, C.-S.; et al. Oral Administration of Lactobacillus plantarum HY7714 Protects Hairless Mouse Against Ultraviolet B-Induced Photoaging. J. Microbiol. Biotechnol. 2014, 24, 1583–1591. [Google Scholar] [CrossRef] [PubMed]
- Wlaschek, M.; Tantcheva-Poór, I.; Naderi, L.; Ma, W.; Schneider, L.A.; Razi-Wolf, Z.; Schüller, J.; Scharffetter-Kochanek, K. Solar UV Irradiation and Dermal Photoaging. J. Photochem. Photobiol. B 2001, 63, 41–51. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).