Abstract
Selenium (Se) is an essential trace element for fish with more than 40 selenoproteins identified, many exhibiting antioxidant functions. This study investigated the effect of dietary Se supplementation on physiological parameters, selenoprotein and antioxidant enzyme gene expression in Atlantic bluefin tuna (ABT, Thunnus thynnus) larvae. First-feeding ABT larvae were divided into triplicate groups and fed rotifers Brachionus rotundiformis enriched with five different levels of Se (0, 3, 10, 30, and 100 µg Se·L−1) until 14 days after hatching. Both rotifers and ABT larvae effectively accumulated Se achieving maximum levels in the Se100 treatment (30.05 μg Se·g−1 and 194 ± 38 μg Se·g−1 dry mass, respectively). Larvae showed highest total length when fed Se3 rotifers, whereas flexion index was highest in larvae fed Se10. Selenium supplementation increased the gene expression of selenoproteins gpx1, msrb1, trxr2, selenom, selenop, and selenoe compared to the non-supplemented control (Se0), but only marginal differences were detected between supplementation levels. In contrast, expression of the antioxidant enzymes cat and sod1 were lowest in larvae fed Se100. To conclude, non-Se-enriched rotifers may be suboptimal for first feeding ABT larvae, which showed improved selenoprotein and antioxidant gene expression when fed a diet containing 4.42 μg Se·g−1 dry mass.
1. Introduction
Atlantic bluefin tuna (ABT, Thunnus thynnus, L.) is a high-value species on the market. In recent years, ABT juveniles have been captured from the wild and fattened using bait fish in so-called sea ranches [1]. Recently the production cycle of ABT has been closed, however low survival at larval stages dictates this is still far from commercial production [1,2]. As in other bluefin tuna species, ABT has a very sensitive larval stage characterized by several problematic issues including size variation, low swim bladder inflation rates and skeletal anomalies [3,4]. Improved knowledge of the nutritional requirements of larvae could be one solution to boost ABT production where no standard hatchery feeding protocol exists. Although some studies have been performed on different aspects of ABT nutrition [5,6,7,8] there is limited information regarding requirements and functionality for many micronutrients, especially at the larval stage.
Micromineral selenium (Se) is essential for health, normal growth and development in fish [9]. Although the Se requirement of ABT is currently unknown, wild tuna presents high levels of body Se that may be a response to counteract toxic effects of heavy metals prone to accumulate in this top predator species [10]. Fish obtain Se mainly from the diet with levels to satisfy requirements ranging between 0.15 and 1.85 µg·g−1, however, toxic effects can be observed at marginally higher levels [11]. Recent studies suggested that rotifers, which are also used as starter feeds for ABT, might not cover the nutritional requirements of marine finfish species [10,12]. As Se requirements are known to increase in response to oxidative stress, Se supplementation may prove especially beneficial in this fast-growing species at early life stages, which are especially sensitive to changing environmental conditions [13,14,15].
Oxidative stress derives from an imbalance between the production of reactive oxygen species (ROS) and the cellular antioxidant defence system [16,17]. While ROS are indispensable for cell signalling, excessive production leads to the oxidation of cellular compounds such as proteins and lipids. Cellular antioxidants can prevent, lower or reverse the damage caused by ROS. The antioxidant system is composed of nutritional compounds such as vitamins C and E that scavenge ROS, but also consists of several key enzymes including catalase (CAT), superoxide dismutase (SOD) and glutathione peroxidases (GPx) that catalyse the decomposition of hydrogen and lipid peroxides [18]. Seven GPx enzymes, that use glutathione as a substrate to reduce peroxides, have been described in fish out of which five were identified as selenoproteins [19]. Selenoproteins have selenocysteine incorporated at the active centre via a process of co-translational insertion in response to specific UGA codons and the presence of a cis-acting specific selenocysteine insertion sequence SECIS element [20]. Fish have one of the largest selenoproteoms identified among vertebrates [19]. Besides GPx, other selenoproteins are well characterized for their role in the antioxidant system including methionine sulfoxide reductase with its ability to recover oxidized methionine preventing the disruption of proteins or thioredoxin reductase with its ability to reduce oxidized thioredoxins [21,22,23]. Selenoprotein P was the first selenoprotein described to contain more than one selenocysteine. It acts as a seleno-transporter and its expression is often used to determine Se requirements [24]. Other selenoproteins are less well characterized to date, especially in fish, although many of them were shown to have antioxidant properties [25].
In the above context, the present study aimed to investigate the dose-dependent effect of dietary Se level on growth performance, the expression of selenoproteins and antioxidant enzymes, and oxidative status in ABT larvae.
2. Materials and Methods
2.1. Ethical Statement
This feeding trial was performed under the regulations on the protection of animals used for scientific purposes as defined by the European Directive 2010/63/EU (European Parliament and Council, 22 September 2010) and the Spanish legislation RD 53/2013 (BOE 8 February 2013). Additionally, ethical approval was granted by the Animal Welfare and Ethical Review Board (AWERB) of the University of Stirling, UK (ID TunaSe8456).
2.2. Atlantic Bluefin Tuna Larvae Rearing Conditions
The ABT eggs were obtained from naturally spawning ABT broodstock fish cultivated in floating net cages off the Cartagena coast, El Gorguel, SE Spain in July 2018. Collected eggs were transported under provision of oxygen to the Spanish Institute of Oceanography (IEO) Planta Experimental de Cultivos Marinos (Puerto de Mazarrón, Murcia, Spain) and placed in 100 L flow-through tanks equipped with oxygenized and sterilized seawater. To separate buoyant (viable) from non-buoyant (non-viable) eggs aeration and water flow were stopped after 1 h. All viable eggs were washed, disinfected (25 ppm iodine solution, Germiod, CENAVISA, SA, Reus, Spain) and counted. Fertilized eggs were kept in 1400 L cylindrical tanks under standardized conditions (24 °C water temperature, 37‰ salinity, 6.5 mg·L−1 dissolved oxygen and continuous photoperiod of 1000 lux) at a density of 8.5 eggs·L−1. Larvae hatched approximately 32 h after fertilization, with a hatching rate of almost 90%, and were fed the Se-enriched rotifers Brachionus rotundiformis from 2 days after hatching (dah). To create green-water, a mixture of the microalgae Isochrysis sp. (T-Iso) and Chlorella (V12 DHA-enriched, Pacific Trading Co., Fukuoka, Japan) were added to tanks at a density of 2–3 × 105 cells mL−1. During larvae feeding, photoperiod was maintained at 14 h light/10 h dark (light intensity about 1000 lux, from 7:30 h to 20:30 h), temperature ranged between 24–26 °C and daily water renewal was 100–200% tank volume·day−1. All incoming seawater was filtered at 10 µm size and UV sterilized. Larvae sinking (mainly at night) was prevented through the creation of an upwelling current which also helped to stabilize oxygen levels [5,6,26].
2.3. Dietary Treatments
Rotifer (Brachionus rotundiformis) grown on Algamac 3050 (Pacific Trading LTD, Kent, England) was supplemented with different levels of seleno-yeast, SelPlex®, Alltech (Meath, Ireland) and used as larvae feed. Rotifer enrichment took place for 18 h in culture medium supplemented with SelPlex® at the following doses ranging between non-supplemented to potentially excessive: 0.0 mg/106 rotifers (0 μg Se·L−1, Se0), 3 mg/106 rotifers (3 μg Se·L−1, Se3), 10 mg/106 rotifers (10 μg Se·L−1, Se10), 30 mg/106 rotifers (30 μg Se·L−1, Se30) and 100 mg/106 rotifers (100 μg Se·L−1, Se100). Selenium levels in rotifers were quantified as described in paragraph 2.6.1. The treatments and final analysed Se levels in freeze dried rotifers are summarized in Table 1.

Table 1.
Supplementation levels and analysed selenium concentrations in rotifer Brachionus rotundiformis enriched in culture medium for 18 h with different doses of seleno-yeast (SelPlex®) and used as starter feeds for ABT larvae in the feeding trial.
Each dietary treatment consisted of three triplicate 1500 L cylindro-conical tanks equipped with ABT larvae at a fish density of 10 larvae·L−1. Manual feeding ensured a constant prey density of 5 rotifers·mL−1 throughout the trial.
2.4. Sample Collection of Rotifers and ABT Larvae: Sampling for Growth Performance, Biochemical and Molecular Analysis
At sampling, individual fish were photographed and lengths, weight and developmental stage were determined on twenty-five anaesthetized fish (0.02% 2-phenoxyethanol, Sigma, Spain) per triplicate treatment. To determine the developmental stage larvae that achieved full flexion of the notochord by the end of the feeding trial (14 dah) were counted. Whole larvae were dried at 110 °C for 24 h and cooled in vacuo for 1 h before weighing in a precision balance to determine individual dry mass.
For each treatment three subsets of triplicate samples were collected to either determine dry mass (20 individuals), perform biochemical analysis (50 individuals) or molecular analysis (50 individuals). Larvae dedicated for molecular analysis were placed in 1.5 mL of RNAlater® (Sigma, Madrid, Spain) before all samples were snap-frozen in liquid nitrogen and stored at −80 °C until further analysis.
2.5. Larvae Biometry and Survival
At 1, 2, 3, 6, 8, 12 and 14 dah, 25 larvae per tank (replicate) were sampled for total length. Therefore, images were taken using a camera (Olympus SC20, OLYMPUS, Münster, Germany) connected to a microscope (Olympus SZ61-TR, Leica, Hamburg, Germany). Then, Image Pro 6.2 (Media cybernetics; Buckinghamshire, UK) software was used to determine total length. Survival (%) represents the difference in individual live larvae at the beginning and at the end of the trial.
2.6. Biochemical Analysis
2.6.1. Selenium Analysis
Selenium was measured in 40–80 mg of dried rotifers and ABT larvae. The samples were measured in triplicate by digestion in 5 mL of 65% nitric acid using a microwave digester (Mars6, CEM, Charlotte, NC, USA) with the following settings: 21 °C to 190 °C for 10 min at 800 W, then 190 °C for 20 min at 800 W and finally 30 min cooling. The digested samples were two times diluted in distilled water and 0.2 mL methanol, and total Se was measured by Inductively Coupled Plasma Mass Spectrometry (Thermo Scientific, XSeries2 ICP-MS, Waltham, MA, USA) using the carrier gases argon and hydrogen.
2.6.2. Total Lipid, Fatty Acid and TBARS Analysis
Total lipids of ABT larvae were extracted from pooled samples according to the method of Folch et al. (1957) [27]. Approximately 50 mg dried ABT larvae were homogenized and processed as described in detail previously [6] before the lipid content was determined gravimetrically.
Fatty acid methyl esters (FAME) were analysed from the extracted total lipids according to the method of Christie [28,29]. Gas-liquid chromatography (Agilent Technologies 7890B GC System) equipped with a 30 m × 0.32 mm i.d. fused silica capillary column (SUPELCOWAXTM-10, Supelco Inc., Bellefonte, PA, USA) was used to separate and quantify FAME as described previously in Betancor et al. (2017) [6].
Thiobarbituric acid reactive substances (TBARS), formed as a by-product of lipid peroxidation, were measured in total lipid extracts. Briefly, 0.1 mg of total lipids were evaporated to dryness under oxygen-free nitrogen. Next, 0.5 mL of 1% thiobarbituric acid, 0.5 mL of 10% trichloracetic acid and 50 µL of 0.02% BHT in ethanol were added and the samples placed in a 100 °C water bath for 20 min. Finally, absorbance was measured at 532 nm against a reagent blank.
2.7. Molecular Analysis
2.7.1. Tissue RNA Extraction and cDNA Synthesis
Total RNA extraction was performed on duplicate pools of 15 whole ABT larvae per tank (to provide 6 samples per dietary treatment) using 1 mL TriReagent® (Sigma-Aldrich) extraction buffer. Quantity and quality of RNA was measured using a Nanodrop ND-1000 (Labtech Int., East Sussex, UK) and additionally by electrophoresis running 200 ng of total RNA on a 1% agarose gel. Reverse transcription was performed with random primers on 2 µg total RNA using the High Capacity cDNA Reverse Transcription kit (Applied Biosystems, REF4368813, Warrington, UK) according to the manufacturers protocol.
2.7.2. Primer Preparation and qPCR
The primer sequences used in the quantitative real-time PCR (qPCR) are given in Table 2.

Table 2.
Primer sequences used to assay mRNA levels by real-time PCR.
Primers for gpx1, gpx4, cat, sod1, ef1a and bactin were already available [5]. The other primers were designed on gene sequences available in close related tuna species and on the sequence read archive (SRA) of ABT (SRX22557558, SRX2766917) by identifying and assembling the sequences. The efficiency of primers was evaluated on a serial dilution of ABT cDNA to verify it was >84% for all primer pairs.
The qPCR was performed in a qTower3 G real-time PCR Thermal Cycler (Analytic Jena GmbH, Jena, Germany) under the following conditions: 50 °C for 2 min, 95 °C for 10 min, then 34 cycles of 95 °C for 15 s, 60 °C for 30 s and 72 °C for 30 s, then 6 s at the annealing Tm. The total reaction volume of 10 µL compromised 5 µL Luminaris Color HiGreen qPCR Master Mix (Thermo Scientific, Hemel Hempsted, UK), 0.5 µL primer pairs at 10 pmol concentration, 1.5 µL deionised water and 2.5 µL cDNA (1/20 dilution). The housekeeping genes were measured in the same reaction volume but diluted at 1/200. A non-template control containing no cDNA was measured alongside each gene and the melting curves were systematically screened.
The relative expression levels of target genes were calculated as geometric means of the housekeeping genes ef1a and bactin using the ΔΔCT method on the average of the control treatment Se0 [30].
2.8. Statistical Analysis
Results are given as mean ± standard deviation. Data were analysed using statistical software R (R Development Core Team, 2021, Vienna, Austria). Normality and homogeneity were verified prior to model building resulting in rank transformation of gene expression data. Dietary treatments were analysed by one-way ANOVA followed by a Tukey’s HSD post hoc test and considered significantly different at p < 0.05.
3. Results
3.1. Performance of ABT Larvae during the Feeding Trial
ABT larvae fed Se3-rotifers showed the numerically highest survival, significantly better than those fed the Se10-rotifers (Table 3). Total length of larvae fed the non-supplemented rotifers was lower compared to larvae fed Se3- or Se30-rotifers with intermediate values for larvae fed the Se10 and Se100 treatments (Table 3). Similarly, ABT larvae fed Se0-rotifers had a lower dry mass than those fed Se30-rotifers. The flexion index was increased in larvae fed all Se-enriched treatments, but was highest in ABT larvae fed Se10- and Se30-rotifers.

Table 3.
Growth and survival of ABT larvae at 14 days after hatch fed rotifers Brachionus rotundiformis grown on Algamac 3050 Bio Marine® and enriched with graded levels of selenium (0, 3, 10, 30, and 100 mg of Se-yeast per 106 rotifers).
3.2. Whole Body Se Content of ABT Larvae
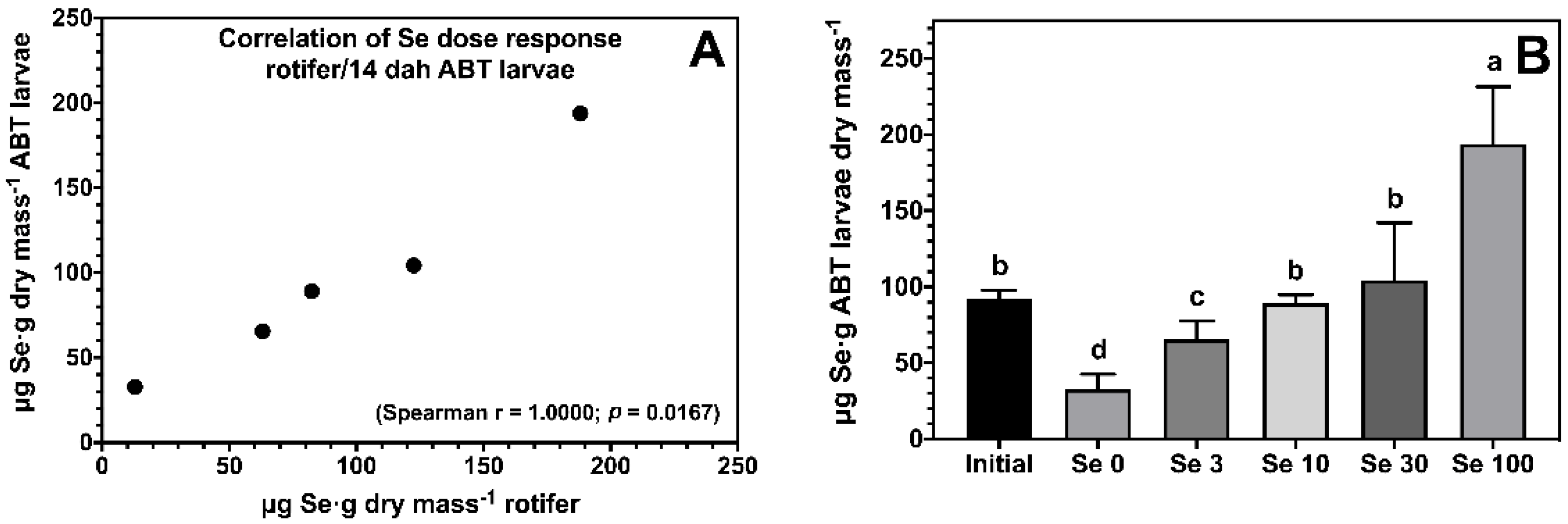
The enrichment of rotifers effectively increased body Se levels of ABT larvae, and showed a strong dose-dependent correlation (Figure 1A). All ABT larvae fed rotifers enriched with Se showed significantly higher body Se levels compared to the negative control treatment Se0 (Figure 1B). ABT larvae fed the Se100 treatment accumulated 194 ± 38 μg Se·g−1 dry mass followed by larvae fed Se10 and Se30, with larvae fed Se3 showing the lowest body Se levels within the supplemented treatments.
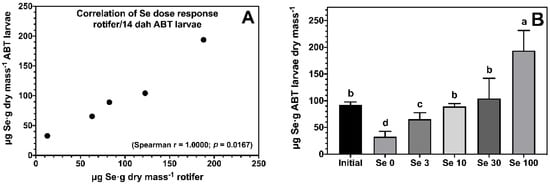
Figure 1.
Dose response correlation between selenium levels in rotifers Brachionus rotundiformis grown on Algamac 3050 Bio Marine® and enriched with 5 graded selenium levels (0, 3, 10, 30, and 100 mg of Se-yeast per 106 rotifers) and ABT larvae fed the rotifers from mouth-opening until 14 days post-hatching (A). Total body selenium levels (n = 3; mean ± SD) measured in ABT larvae at the end of the feeding trial (B). Bars not sharing a common superscript letter indicate significant differences (p < 0.05) between groups according to one-way ANOVA followed by Tukey’s HSD.
3.3. Total Lipid Fatty Acid Composition and TBARS Content of ABT Larvae
Feeding Se-enriched rotifers had no major impact on the fatty acid composition of ABT larvae other than some small, likely not biologically significant, variations in proportions of quantitively minor fatty acids, 18:3n − 6, 20:3n − 6 and 20:4n − 3 (Table 4). Similarly, the TBARS concentration as a measure of lipid peroxidation was not significant different between the groups (Table 4).

Table 4.
Total lipid (mg·g dw−1), fatty acid composition (% of total fatty acids) and TBARS content (nmol·g lipid−1) of ABT larvae fed rotifers Brachionus rotundiformis enriched with Algamac 3050 Bio Marine® and graded levels of selenium (0, 3, 10, 30, and 100 mg of Se-yeast per 106 rotifers) for 13 days.
3.4. Gene Expression
3.4.1. Expression of Selenoproteins in ABT Larvae
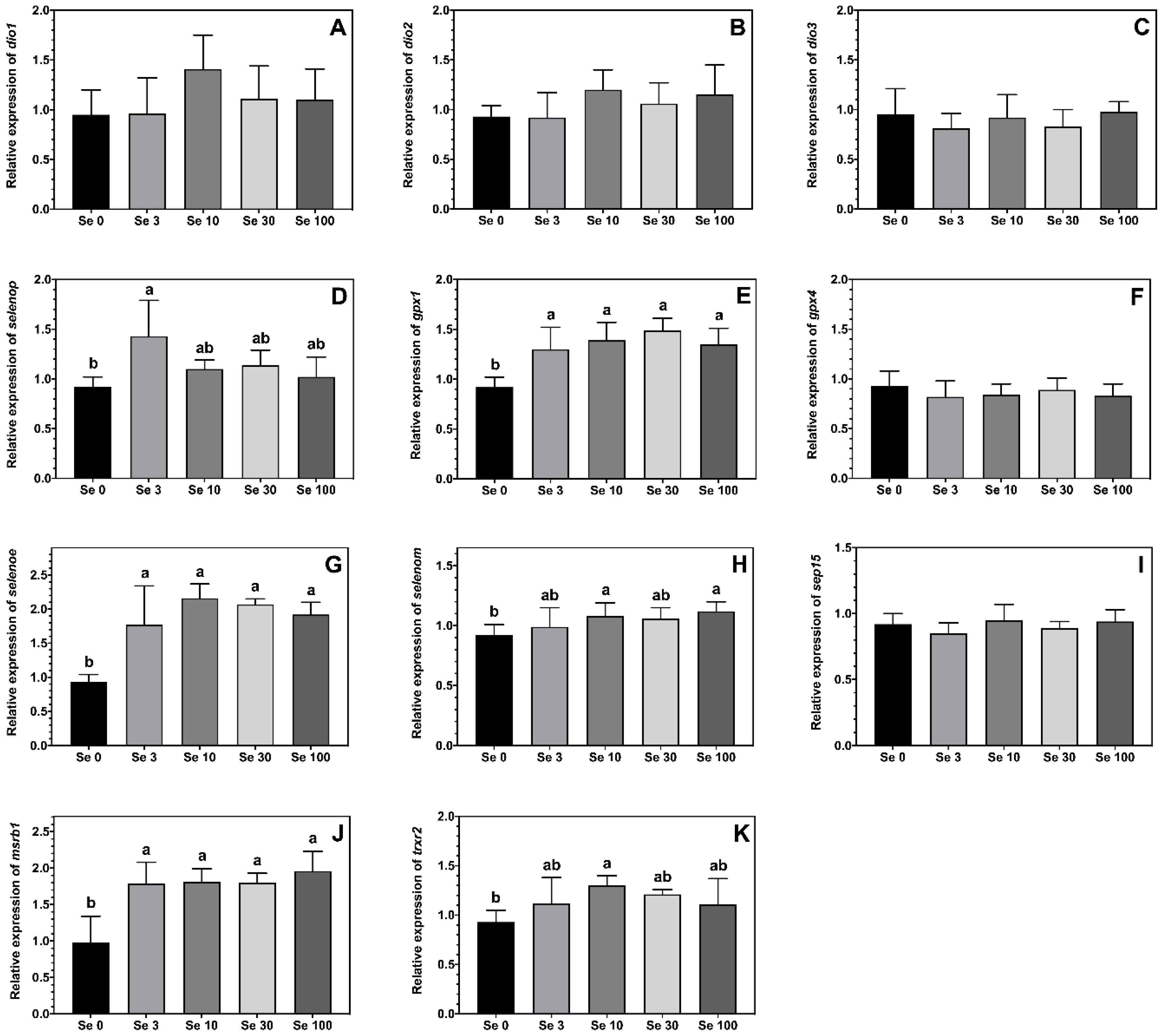
The expression levels of the selenoproteins gpx1, msrb1 and selenoe were higher in ABT larvae fed all the Se-enriched rotifers compared to the non-enriched Se0 treatment (Figure 2E,G,J). A similar pattern was observed for the expression levels of selenop, trxr2 and selenom, although the differences were only statistically significant between larvae fed Se0 vs. Se3 for selenop, Se0 vs. Se10 for trxr2, and Se0 vs. both Se10 and Se100 for selenom, with fish from other treatments showing intermediate values (Figure 2D,H,K). The feeding of Se-enriched rotifers had no significant effect on the expression levels of gpx4, sep15, dio1, dio2 and dio3 in the ABT larvae (Figure 2A–C,F,I).
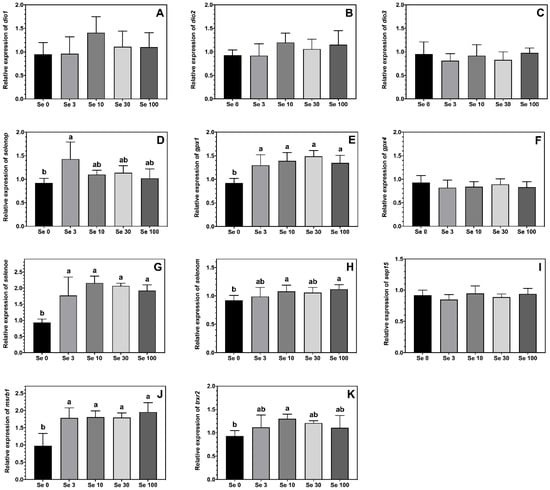
Figure 2.
Expression levels (means ± SD, n = 6) of selenoproteins iodothyronine deiodinase 1, dio1 (A); iodothyronine deiodinase 2, dio2 (B); iodothyronine deiodinase 3, dio3 (C); selenoprotein p, selenop (D); glutathione peroxidase 1, gpx1 (E); glutathione peroxidase 4, gpx4 (F); selenoprotein E, selenoe (G); selenoprotein M, selenom (H); 15-kda selenoprotein, sep15 (I); methionine sulfoxide reductase 1, msrb1 (J) and thioredoxin reductase 2, trxr2 (K) measured by real-time PCR. Data were normalized to a geometric mean of ef1a and bactin and are presented as fold-changes of mRNA abundance compared with Se0. One-way ANOVA on ranks followed by Tukey’s HSD postdoc test was used to detect significant differences as displayed through uncommon superscript letters.
3.4.2. Expression of Other Antioxidant Defence Genes in ABT Larvae
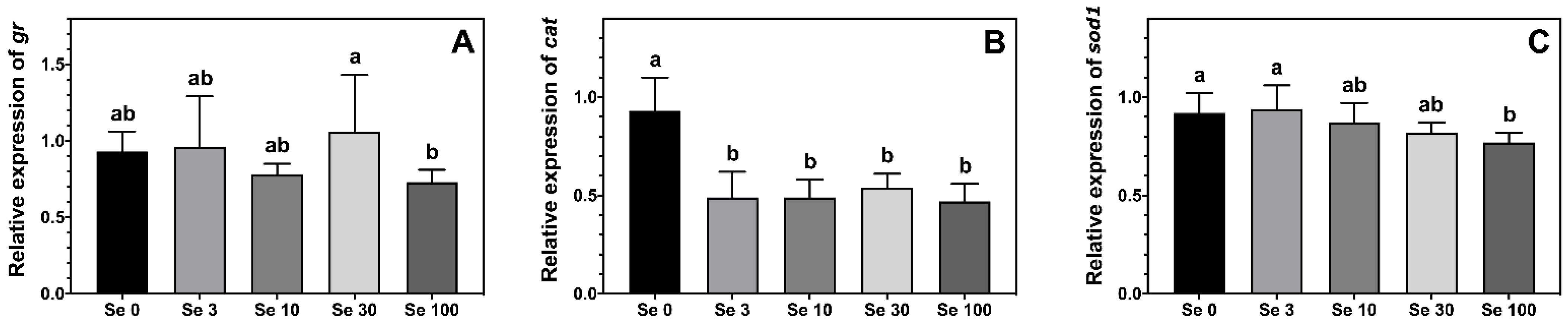
The expression level of gr was highest in ABT larvae fed Se30-rotifers compared to those fed the Se100 treatment, while larvae fed rotifers enriched with 0–10 mg doses of Se-yeast displayed an intermediate level of expression (Figure 3A). The expression of the antioxidant enzyme cat was lower in ABT larvae fed all the Se-enriched rotifers compared to larvae fed the control supplemented rotifers (Figure 3B). Additionally, the expression of sod1 in ABT larvae reflected the Se-enrichment in the rotifers (Figure 3C). In this sense, the highest sod1 expression was measured in the two lowest Se treatments Se0 and Se3, while the lowest expression was observed in larvae fed the Se100 rotifers, the highest Se-enrichment. On the other hand, ABT larvae fed Se10- and Se30-rotifers, the medium enrichment treatments, displayed intermediate values for sod1 expression.
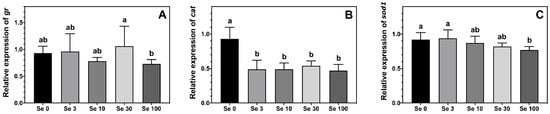
Figure 3.
Expression levels (means ± SD, n = 6) of antioxidant enzymes gluthatione reductase, gr (A); catalase, cat (B) and superoxide dismutase 1, sod1 (C) measured by real-time PCR. Data were normalized to a geometric mean of ef1a and bactin and are presented as fold-changes of mRNA abundance compared with Se0. One-way ANOVA on ranks followed by Tukey’s HSD postdoc test was used to detect significant differences as displayed through uncommon superscript letters.
4. Discussion
The Se requirement for ABT is currently unknown. In a dose-dependent feeding trial this study could show for the first time that optimized Se feeding has the potential to improve ABT larval performance under aquaculture conditions. The further molecular analysis revealed that these changes were accompanied with modifications in the antioxidant metabolism of the fish.
Selenium enrichment had no significant effect on the survival of ABT larvae in the present trial. This was similar to a previous study in Senegalese sole (Solea Senegalensis) larvae where no differences in survival were detected by enriching rotifers with 3 mg Se-yeast per million [31]. The higher survival previously described in cod (Gadus morhua) larvae fed Se-supplemented rotifers might be a result of simultaneous co-enrichment with iodone, as enrichment with Se alone had no significant effect on survival of red sea bream (Pagrus major) larvae [32,33]. In the present study, the lower survival observed in ABT larvae fed rotifers enriched with 10 mg compared to 3 mg Se-yeast per million rotifers does not appear to be a result of excessive Se supplementation as decreased survival was not observed at the highest Se enrichment level, in line with previous observations in red drum (Sciaenops Ocellatus) larvae where adverse effects on growth were only reported at Se supplementation levels above 50 mg/L [34].
In the present trial, growth, in terms of total body length and weight, was lowest in ABT larvae fed unenriched rotifers, however, a significant difference was only detected in larvae fed the rotifers enriched with 3 and 30 mg Se-yeast. The improved growth with Se supplementation observed in this study might be related to accelerated development as the flexion index was significantly higher in larvae fed all Se-enriched treatments compared to the non-supplemented control. Indeed, an earlier study in red sea bream larvae showed similar effects in response to dietary Se supplementation [33]. Interestingly, a study on Senegalese sole reported that Se enrichment modified the activity of the thyroid hormone T4 during critical phases of larval development [31,35]. The iodothyronine deiodinases (DIOs) are selenoproteins, therefore, their expression can be enhanced by higher levels of tissue Se. In the present study, whole-body Se content in ABT larvae increased in a dose-dependent manner with Se supplementation levels. Nonetheless, the expression of selenoproteins follows a principle of hierarchy and, in comparison to other selenoproteins, the expression of DIOs, where absence can be lethal, is known to be generally stable and will rather decrease with pronounced Se deficiency [36]. This could explain why no significant differences were observed between treatments for the expression of DIOs, but also with some of the other selenoproteins in the present study. Indeed, the Se content of the control non-supplemented rotifers, with 0.10 µg Se g−1 rotifers, in this study, is lower than the Se requirements reported for other fish species, which range between 0.15 and 1.85 µg Se g−1 diet in comparison to the lowest supplementation level which provided 4.42 µg Se g−1 rotifers which is similar to the optimal dietary Se level reported for red drum larvae [11,34]. Consistent with this, the expression of selenop, which is often used as an indicator for Se requirements, was higher in ABT larvae fed the lowest supplementation treatment (Se3) compared to the non-supplemented control, but was not affected by higher enrichment levels in this study. Similarly, the expression of seleno-dependent GPx is known to increase according to Se level making it an indicator for optimal Se status of fish larvae [37]. In the present study, the expression of gpx1, but not gpx4, was elevated by Se supplementation but, indeed, gpx4 ranks higher in selenoprotein hierarchy [36]. Glutathione peroxidases are key enzymes for the reduction of lipid peroxides using glutathione as a substrate [38]. The current data show that other selenoproteins with an active role in the antioxidant system were also upregulated in ABT larvae fed the Se-enriched rotifers. These included msrb1 with its ability to recover oxidized methionine thus preventing the disruption of proteins, and trxr2 with its ability to reduce oxidized thioredoxins [21,22,23]. In contrast, the thioredoxin-like protein sep15, which is involved in antioxidant protection of the thyroid gland was not affected by Se supplementation in the present trial [39]. Nevertheless, in this study, the total Se requirements of ABT larvae might be masked by the mobilization of parentally transferred Se as indicated by the higher total body Se levels detected in the initial control samples compared to larvae at final sampling. The Se requirements in ABT broodstock are currently unknown, but as previously demonstrated in rainbow trout (Oncorhynchus mykiss) parentally transferred Se can induce modifications in the antioxidant metabolism of first feeding fry [40].
Contrary to our expectations, the expression levels of selenoproteins in ABT larvae showed few differences among the Se-enriched groups, which indicated that the lowest supplementation level might have already provided sufficient Se to support adequate selenoprotein production in this study. Surplus Se may not have been translated to selenoproteins, but rather stored as other forms. In the blood of wild tuna, more than 98% of total Se was identified as selenoneine, an organic Se analogue of ergothionine with strong antioxidant properties [41]. However, the question is if similar findings could be expected in tuna originating from closed-cycled aquaculture. It is currently unknown whether selenoneine originates from the diet and accumulates in the tissue of tuna as a top predator species or if it is synthesised. Selenoneine was detected in mackerels, a common prey for tuna, but high levels of selenoneine were also detected in beluga whales, but not their prey [42]. Selenoneine is synthesised by similar enzymes as ergothionine, a process so far only described in a limited number of bacterial and fungal species but never in vertebrates [43]. Nevertheless, selenoneine could be a product of specific microorganisms present in the gut microbiome of tuna species and, indeed, could represent a potent antioxidant besides selenocysteine, which is the active Se form in selenoproteins, and should therefore be analysed in future studies.
It has been suggested that the vital role of Se in the antioxidant system can also help to prevent oxidative stress resulting from the provision of live feeds which are often specifically enriched with high levels of n − 3 long-chain polyunsaturated fatty acids that can promote pro-oxidant conditions. Supplementation with Se (and iodine) was reported in a previous study to lead to modifications in the fatty acid profile of cod larvae [32]. However, in the present study the fatty acid profiles of ABT larvae displayed no differences between larvae fed rotifers supplemented with Se, with similar results previously observed in red sea bream [33]. Additionally, the levels of TBARS, which are by-products of lipid oxidation, were not significantly different between ABT larvae fed the different treatments giving no indication of elevated oxidative stress in any of the present treatments. However, the fatty acid profile of fish larvae predominantly reflects the fatty acid profile of the diet, i.e., the rotifers [44]. Live feeds that do not bring a standardised nutritional profile add a factor of uncertainty when comparing studies [12,32]. This is especially true for investigations on microminerals, which can substantially deviate between different batches of live feeds. In the present study, another indicator that indeed supports changes in the cellular redox state of ABT larvae by Se supplementation, especially at higher levels, is the reduced gene expression of the two antioxidant enzymes sod1 and cat. While these enzymes are not selenoproteins and therefore not directly regulated by Se supply, they could be regulated indirectly by the impact of Se on redox status through redox sensitive transcription factors [45,46]. This shift in the transcription of antioxidant genes should be further investigated in future studies focusing on the effects of post-transcriptional enzyme regulation and the glutathione metabolism.
5. Conclusions
The present study showed that rotifers without Se enrichment might provide levels of Se that are suboptimal for ABT larvae at first feeding. Se supplementation improved growth performance as well as selenoprotein gene expression. Improved oxidative status in ABT fed Se-enriched rotifers was indicated by lower antioxidant enzyme gene expression, albeit not supported by measures of lipid oxidation. The lack of significant differences in selenoprotein gene expression between larvae fed higher levels of Se supplementation suggested that a dietary Se level of 4.42 μg·g−1 DM may be sufficient to satisfy Se requirements in ABT larvae.
Author Contributions
Conceptualization, M.B.B., D.R.T. and G.M.; methodology, M.S.; validation, M.S., F.d.l.G. and A.O.; formal analysis, P.W. and G.M.; investigation, P.W., M.B.B., A.O., F.d.l.G. and G.M.; resources, M.B.B. and G.M.; data curation, P.W. and G.M.; writing—original draft preparation, P.W. and G.M.; writing—review and editing, M.B.B. and D.R.T.; visualization, G.M.; supervision, M.B.B. and G.M.; project administration, M.B.B.; funding acquisition, M.B.B. and G.M. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Consejería de Innovación, Ciencia y Empresa de la Junta de Andalucía, Proyecto de Excelencia de Promoción General del Conocimiento [Ref. RNM 733, 2012), and Programa Estatal de Investigación del Ministerio de Economía y Competitividad [Ref. RTC2016-5835-2-P01].
Institutional Review Board Statement
The animal study protocol was approved by the Institutional Review Board (or Ethics Committee) of the University of Stirling (TunaSe 8456, 04/08/2022).
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are contained within the article.
Acknowledgments
We wish to express our gratitude to the technical staff at Laboratory of Marine Aquaculture (IEO), Puerto de Mazarrón (Murcia), Spain and Nutritional Analytical Services (NAS), Institute of Aquaculture, University of Stirling, UK who supported the experimental trail and sample analysis performed in this study.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Van Beijnen, J. The Closed Cycle Aquaculture of Atlantic Bluefin Tuna in Europe: Current Status, Market Perceptions and Future Perspectives. Academia 2017. [Google Scholar] [CrossRef]
- de la Gándara, F.; Ortega, A.; Buentello, A. Chapter 6—Tuna Aquaculture in Europe. In Advances in Tuna Aquaculture; Benetti, D.D., Partridge, G.J., Buentello, A., Eds.; Academic Press: San Diego, CA, USA, 2016; pp. 115–157. ISBN 978-0-12-411459-3. [Google Scholar]
- Sawada, Y.; Okada, T.; Miyashita, S.; Murata, O.; Kumai, H. Completion of the Pacific Bluefin Tuna Thunnus Orientalis (Temminck et Schlegel) Life Cycle. Aquac. Res. 2005, 36, 413–421. [Google Scholar] [CrossRef]
- Woolley, L.D.; Qin, J.G. Swimbladder Inflation and Its Implication to the Culture of Marine Finfish Larvae. Rev. Aquac. 2010, 2, 181–190. [Google Scholar] [CrossRef]
- Betancor, M.B.; Ortega, A.; de la Gándara, F.; Tocher, D.R.; Mourente, G. Molecular Aspects of Lipid Metabolism, Digestibility and Antioxidant Status of Atlantic Bluefin Tuna (T. Thynnus L.) Larvae during First Feeding. Aquaculture 2017, 479, 357–369. [Google Scholar] [CrossRef]
- Betancor, M.B.; Ortega, A.; de la Gándara, F.; Tocher, D.R.; Mourente, G. Lipid Metabolism-Related Gene Expression Pattern of Atlantic Bluefin Tuna (Thunnus Thynnus L.) Larvae Fed on Live Prey. Fish Physiol. Biochem. 2017, 43, 493–516. [Google Scholar] [CrossRef]
- Koven, W.; Nixon, O.; Allon, G.; Gaon, A.; El Sadin, S.; Falcon, J.; Besseau, L.; Escande, M.; Vassallo Agius, R.; Gordin, H.; et al. The Effect of Dietary DHA and Taurine on Rotifer Capture Success, Growth, Survival and Vision in the Larvae of Atlantic Bluefin Tuna (Thunnus Thynnus). Aquaculture 2018, 482, 137–145. [Google Scholar] [CrossRef]
- Morais, S.; Mourente, G.; Ortega, A.; Tocher, J.A.; Tocher, D.R. Expression of Fatty Acyl Desaturase and Elongase Genes, and Evolution of DHA:EPA Ratio during Development of Unfed Larvae of Atlantic Bluefin Tuna (Thunnus Thynnus L.). Aquaculture 2011, 313, 129–139. [Google Scholar] [CrossRef]
- Khan, K.U.; Zuberi, A.; Fernandes, J.B.K.; Ullah, I.; Sarwar, H. An Overview of the Ongoing Insights in Selenium Research and Its Role in Fish Nutrition and Fish Health. Fish Physiol. Biochem. 2017, 43, 1689–1705. [Google Scholar] [CrossRef]
- Kljaković-Gašpić, Z.; Tičina, V. Mercury and Selenium Levels in Archive Samples of Wild Atlantic Bluefin Tuna from the Mediterranean Sea. Chemosphere 2021, 284, 131402. [Google Scholar] [CrossRef]
- Prabhu, P.A.J.; Schrama, J.W.; Kaushik, S.J. Mineral Requirements of Fish: A Systematic Review. Rev. Aquac. 2016, 8, 172–219. [Google Scholar] [CrossRef]
- Hamre, K.; Srivastava, A.; RØnnestad, I.; Mangor-Jensen, A.; Stoss, J. Several Micronutrients in the Rotifer Brachionus Sp. May Not Fulfil the Nutritional Requirements of Marine Fish Larvae. Aquac. Nutr. 2008, 14, 51–60. [Google Scholar] [CrossRef]
- Dawood, M.A.O.; Koshio, S.; Zaineldin, A.I.; Van Doan, H.; Moustafa, E.M.; Abdel-Daim, M.M.; Angeles Esteban, M.; Hassaan, M.S. Dietary Supplementation of Selenium Nanoparticles Modulated Systemic and Mucosal Immune Status and Stress Resistance of Red Sea Bream (Pagrus Major). Fish Physiol. Biochem. 2019, 45, 219–230. [Google Scholar] [CrossRef] [PubMed]
- Küçükbay, F.Z.; Yazlak, H.; Karaca, I.; Sahin, N.; Tuzcu, M.; Cakmak, M.N.; Sahin, K. The Effects of Dietary Organic or Inorganic Selenium in Rainbow Trout (Oncorhynchus Mykiss) under Crowding Conditions. Aquac. Nutr. 2009, 15, 569–576. [Google Scholar] [CrossRef]
- Rider, S.A.; Davies, S.J.; Jha, A.N.; Fisher, A.A.; Knight, J.; Sweetman, J.W. Supra-Nutritional Dietary Intake of Selenite and Selenium Yeast in Normal and Stressed Rainbow Trout (Oncorhynchus Mykiss): Implications on Selenium Status and Health Responses. Aquaculture 2009, 295, 282–291. [Google Scholar] [CrossRef]
- Filho, D.W. Reactive Oxygen Species, Antioxidants and Fish Mitochondria. Front. Biosci. 2007, 12, 1229. [Google Scholar] [CrossRef]
- Sies, H. Oxidative Stress: Concept and Some Practical Aspects. Antioxidants 2020, 9, 852. [Google Scholar] [CrossRef]
- Chowdhury, S.; Saikia, S.K. Oxidative Stress in Fish: A Review. J. Sci. Res. 2020, 12, 145–160. [Google Scholar] [CrossRef]
- Mariotti, M.; Ridge, P.G.; Zhang, Y.; Lobanov, A.V.; Pringle, T.H.; Guigo, R.; Hatfield, D.L.; Gladyshev, V.N. Composition and Evolution of the Vertebrate and Mammalian Selenoproteomes. PLoS ONE 2012, 7, e33066. [Google Scholar] [CrossRef]
- Labunskyy, V.M.; Hatfield, D.L.; Gladyshev, V.N. Selenoproteins: Molecular Pathways and Physiological Roles. Physiol. Rev. 2014, 94, 739–777. [Google Scholar] [CrossRef]
- Ingold, I.; Conrad, M. Oxidative Stress, Selenium Redox Systems Including GPX/TXNRD Families. In Selenium; Michalke, B., Ed.; Molecular and Integrative Toxicology; Springer International Publishing: Cham, Switzerland, 2018; pp. 111–135. ISBN 978-3-319-95390-8. [Google Scholar]
- dos Santos, S.L.; Petropoulos, I.; Friguet, B. The Oxidized Protein Repair Enzymes Methionine Sulfoxide Reductases and Their Roles in Protecting against Oxidative Stress, in Ageing and in Regulating Protein Function. Antioxidants 2018, 7, 191. [Google Scholar] [CrossRef]
- Lu, J.; Holmgren, A. The Thioredoxin Antioxidant System. Free Radic. Biol. Med. 2014, 66, 75–87. [Google Scholar] [CrossRef] [PubMed]
- Saito, Y. Selenium Transport Mechanism via Selenoprotein P—Its Physiological Role and Related Diseases. Font. Nutr. 2021, 8, 685517. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Roh, Y.J.; Han, S.-J.; Park, I.; Lee, H.M.; Ok, Y.S.; Lee, B.C.; Lee, S.-R. Role of Selenoproteins in Redox Regulation of Signaling and the Antioxidant System: A Review. Antioxidants 2020, 9, 383. [Google Scholar] [CrossRef]
- García, A.O. Cultivo Integral de dos Especies de Escómbridos: Atún Rojo del Atlántico (Thunnus thynnus, L. 1758) y Bonito Atlántico (Sarda sarda, Bloch 1793). Ph.D. Thesis, Murcia University, Murcia, Spain, 2015; 224p. [Google Scholar]
- Folch, J.; Lees, M.; Sloane Stanley, G.H. A Simple Method for the Isolation and Purification of Total Lipids from Animal Tissues. J. Biol. Chem. 1957, 226, 497–509. [Google Scholar] [CrossRef]
- Christie, W.W.; Han, X. Chapter 9—Isolation of Fatty Acids and Identification by Spectroscopic and Related Techniques. In Lipid Analysis, 4th ed.; Christie, W.W., Han, X., Eds.; Oily Press Lipid Library Series; Woodhead Publishing: Sawston, UK, 2012; pp. 181–211. ISBN 978-0-9552512-4-5. [Google Scholar]
- Christie, W.W. Lipid Analysis. Trends Food Sci. Technol. 1996, 11, 145. [Google Scholar] [CrossRef]
- Pfaffl, M.W.; Horgan, G.W.; Dempfle, L. Relative Expression Software Tool (REST©) for Group-Wise Comparison and Statistical Analysis of Relative Expression Results in Real-Time PCR. Nucleic. Acids Res. 2002, 30, e36. [Google Scholar] [CrossRef]
- Ribeiro, A.R.A.; Ribeiro, L.; Sæle, Ø.; Hamre, K.; Dinis, M.T.; Moren, M. Selenium Supplementation Changes Glutathione Peroxidase Activity and Thyroid Hormone Production in Senegalese Sole (Solea Senegalensis) Larvae. Aquac. Nutr. 2012, 18, 559–567. [Google Scholar] [CrossRef]
- Hamre, K.; Mollan, T.A.; Øystein, S.; Erstad, B. Rotifers Enriched with Iodine and Selenium Increase Survival in Atlantic Cod (Gadus Morhua) Larvae. Aquaculture 2008, 284, 190–195. [Google Scholar] [CrossRef]
- Kim, H.-J.; Sakakura, Y.; Maruyama, I.; Nakamura, T.; Takiyama, K.; Fujiki, H.; Hagiwara, A. Feeding Effect of Selenium Enriched Rotifers on Larval Growth and Development in Red Sea Bream Pagrus Major. Aquaculture 2014, 432, 273–277. [Google Scholar] [CrossRef]
- Juhász, P.; Lengyel, S.; Udvari, Z.; Sándor, A.N.; Stündl, L. Optimised Selenium Enrichment of Artemia Sp. Feed to Improve Red Drum (Sciaenops Ocellatus) Larvae Rearing. Acta Biol. Hung. 2017, 68, 255–266. [Google Scholar] [CrossRef]
- Ribeiro, A.R.A.; Ribeiro, L.; Sæle, Ø.; Dinis, M.T.; Moren, M. Iodine and Selenium Supplementation Increased Survival and Changed Thyroid Hormone Status in Senegalese Sole (Solea Senegalensis) Larvae Reared in a Recirculation System. Fish Physiol. Biochem. 2012, 38, 725–734. [Google Scholar] [CrossRef]
- Sunde, R.A. Selenoproteins: Hierarchy, Requirements, and Biomarkers. In Selenium: Its Molecular Biology and Role in Human Health; Hatfield, D.L., Berry, M.J., Gladyshev, V.N., Eds.; Springer: New York, NY, USA, 2012; pp. 137–152. ISBN 978-1-4614-1025-6. [Google Scholar]
- Penglase, S.; Nordgreen, A.; van der Meeren, T.; Olsvik, P.A.; Sæle, Ø.; Sweetman, J.W.; Baeverfjord, G.; Helland, S.; Hamre, K. Increasing the Level of Selenium in Rotifers (Brachionus Plicatilis ‘Cayman’) Enhances the MRNA Expression and Activity of Glutathione Peroxidase in Cod (Gadus Morhua L.) Larvae. Aquaculture 2010, 306, 259–269. [Google Scholar] [CrossRef]
- Brigelius-Flohé, R.; Flohé, L. Regulatory Phenomena in the Glutathione Peroxidase Superfamily. Antioxid. Redox Signal. 2020, 33, 498–516. [Google Scholar] [CrossRef] [PubMed]
- Schomburg, L.; Köhrle, J. On the Importance of Selenium and Iodine Metabolism for Thyroid Hormone Biosynthesis and Human Health. Mol. Nutr. Food Res. 2008, 52, 1235–1246. [Google Scholar] [CrossRef] [PubMed]
- Wischhusen, P.; Parailloux, M.; Geraert, P.A.; Briens, M.; Bueno, M.; Mounicou, S.; Bouyssiere, B.; Prabhu, P.A.J.; Kaushik, S.J.; Fauconneau, B.; et al. Effect of parental selenium in rainbow trout (Oncorhynchus mykiss) broodstock on antioxidant status, its parental transfer and oxidative status in the progeny. Aquaculture 2019, 507, 126–138. [Google Scholar] [CrossRef]
- Yamashita, Y.; Yamashita, M. Identification of a Novel Selenium-Containing Compound, Selenoneine, as the Predominant Chemical Form of Organic Selenium in the Blood of Bluefin Tuna. J. Biol. Chem. 2010, 285, 18134–18138. [Google Scholar] [CrossRef]
- Achouba, A.; Dumas, P.; Ouellet, N.; Little, M.; Lemire, M.; Ayotte, P. Selenoneine Is a Major Selenium Species in Beluga Skin and Red Blood Cells of Inuit from Nunavik. Chemosphere 2019, 229, 549–558. [Google Scholar] [CrossRef]
- Pluskal, T.; Ueno, M.; Yanagida, M. Genetic and Metabolomic Dissection of the Ergothioneine and Selenoneine Biosynthetic Pathway in the Fission Yeast, S. Pombe, and Construction of an Overproduction System. PLoS ONE 2014, 9, e97774. [Google Scholar] [CrossRef]
- Cavrois-Rogacki, T.; Rolland, A.; Migaud, H.; Davie, A.; Monroig, O. Enriching Artemia Nauplii with Selenium from Different Sources and Interactions with Essential Fatty Acid Incorporation. Aquaculture 2020, 520, 734677. [Google Scholar] [CrossRef]
- Glorieux, C.; Zamocky, M.; Sandoval, J.M.; Verrax, J.; Calderon, P.B. Regulation of Catalase Expression in Healthy and Cancerous Cells. Free Radic. Biol. Med. 2015, 87, 84–97. [Google Scholar] [CrossRef]
- Miao, L.; St. Clair, D.K. Regulation of Superoxide Dismutase Genes: Implications in Disease. Free Radic. Biol. Med. 2009, 47, 344–356. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).