Identification and Characterization of Two Bibenzyl Glycosyltransferases from the Liverwort Marchantia polymorpha
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Chemicals
2.2. Sequence Alignment and Phylogenetic Analysis
2.3. RNA Extraction and cDNA Cloning
2.4. Heterologous Expression and Purification of Recombinant UGT Proteins
2.5. Enzyme Assay and Product Identification
2.6. In Vivo Functional Analysis of MpUGT737B1 in Escherichia coli
2.7. HPLC Analysis and LC-MS Analysis of the Product
2.8. Expression Patterns of the MpUGTs’ Response to UV Treatment
2.9. Subcellular Localization of MpUGTs
2.10. Homology Modeling and Molecular Docking of MpUGTs
3. Results
3.1. Selection and Phylogenetic Analysis of Candidate UGTs from M. polymorpha
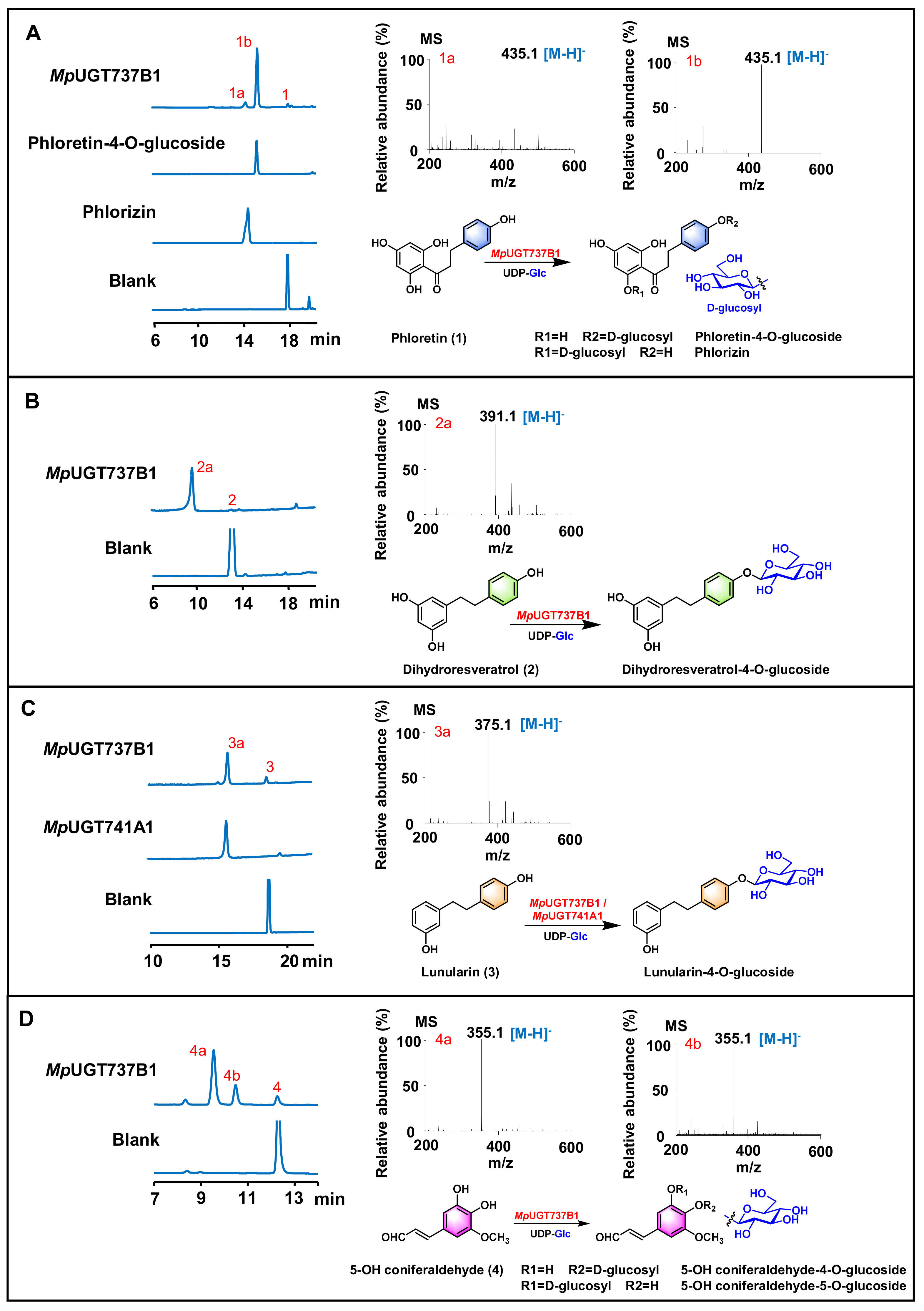
3.2. In Vitro Functional Characterization of Recombinant MpUGTs
3.3. Kinetic Analysis of MpUGT737B1 and MpUGT741A1
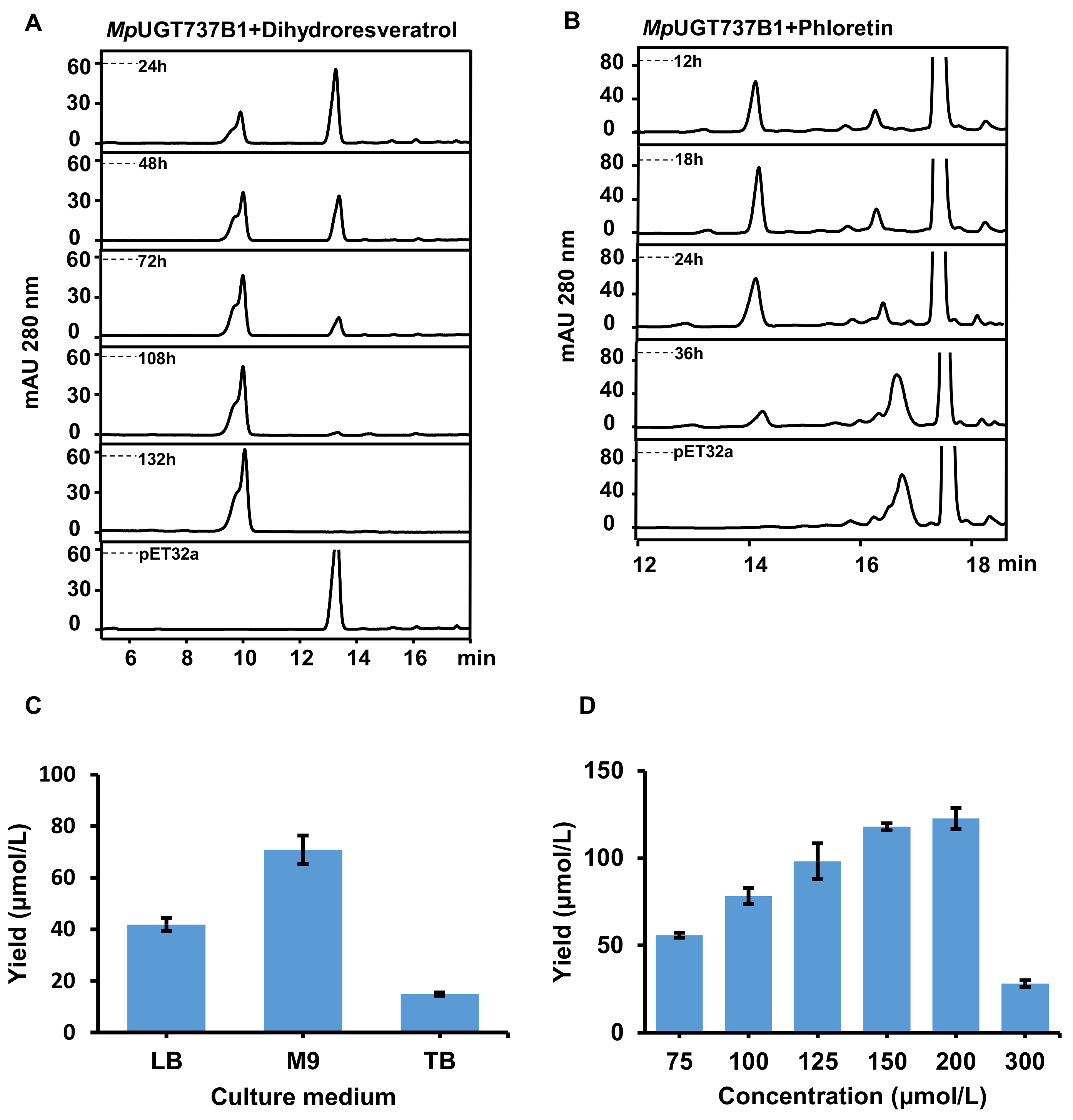
3.4. Bioconversion of Dihydroresveratrol and Phloretin into Their 4-O-Glucosides in E. coli
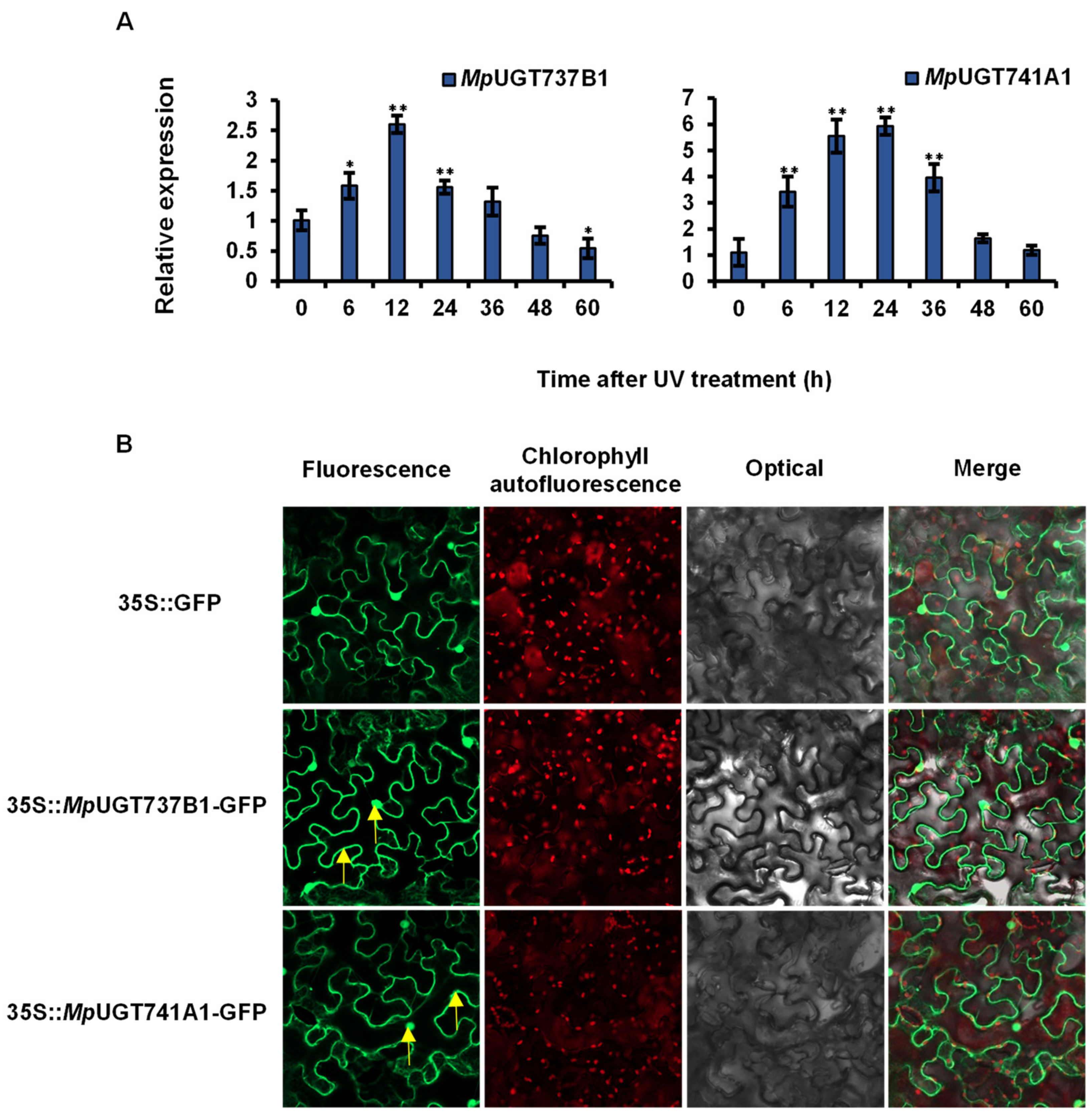
3.5. Analysis of Gene Expression Patterns after UV Treatment
3.6. Subcellular Localization of MpUGTs
3.7. Homology Modeling and Docking Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Agati, G.; Azzarello, E.; Pollastri, S.; Tattini, M. Flavonoids as antioxidants in plants: Location and functional significance. Plant Sci. 2012, 196, 67–76. [Google Scholar] [CrossRef] [PubMed]
- Asakawa, Y.; Ludwiczuk, A.; Novakovic, M.; Bukvicki, D.; Anchang, K.Y. Bis-bibenzyls, bibenzyls, and terpenoids in 33 genera of the Marchantiophyta (liverworts): Structures, synthesis, and bioactivity. J. Nat. Prod. 2022, 85, 729–762. [Google Scholar] [CrossRef] [PubMed]
- Asakawa, Y.; Ludwiczuk, A. Chemical constituents of bryophytes: Structures and biological activity. J. Nat. Prod. 2018, 81, 641–660. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.Q.; Li, W.; Chen, J.X.; Zhai, J.W.; Xu, H.Y.; Ni, L.; Wu, S.S. Chemical constituents and biological activity profiles on Pleione (Orchidaceae). Molecules 2019, 24, 3195. [Google Scholar] [CrossRef] [Green Version]
- Winkel-Shirley, B. Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 2001, 126, 485–493. [Google Scholar] [CrossRef] [Green Version]
- Eckermann, C.; Schröder, G.; Eckermann, S.; Strack, D.; Schmidt, J.; Schneider, B.; Schröder, J. Stilbenecarboxylate biosynthesis: A new function in the family of chalcone synthase-related proteins. Phytochemistry 2003, 62, 271–286. [Google Scholar] [CrossRef]
- Dias, M.C.; Pinto, D.; Silva, A.M.S. Plant flavonoids: Chemical characteristics and biological activity. Molecules 2021, 26, 5377. [Google Scholar] [CrossRef]
- Allegrone, G.; Pollastro, F.; Magagnini, G.; Taglialatela-Scafati, O.; Seegers, J.; Koeberle, A.; Werz, O.; Appendino, G. The bibenzyl canniprene inhibits the production of pro-inflammatory eicosanoids and selectively accumulates in some Cannabis sativa strains. J. Nat. Prod. 2017, 80, 731–734. [Google Scholar] [CrossRef]
- Vitalini, S.; Cicek, S.S.; Granica, S.; Zidorn, C. Dihydroresveratrol type dihydrostilbenoids: Chemical diversity, chemosystematics, and bioactivity. Curr. Med. Chem. 2017, 25, 1194–1240. [Google Scholar] [CrossRef]
- Vogt, T. Glycosyltransferases in plant natural product synthesis: Characterization of a supergene family. Trends Plant 2000, 5, 380–386. [Google Scholar] [CrossRef]
- Jones, P.; Vogt, T. Glycosyltransferases in secondary plant metabolism: Tranquilizers and stimulant controllers. Planta 2001, 213, 164–174. [Google Scholar] [CrossRef]
- Mackenzie, P.I.; Owens, I.S.; Burchell, B.; Bock, K.W.; Bairoch, A.; Bélanger, A.; Fournel-Gigleux, S.; Green, M.; Hum, D.W.; Iyanagi, T.; et al. The UDP glycosyltransferase gene superfamily: Recommended nomenclature update based on evolutionary divergence. Pharmacogenetics 1997, 7, 255–269. [Google Scholar] [CrossRef]
- Jadhav, S.; Patel, K.A.; Dholakia, B.B.; Khan, B.M. Structural characterization of a flavonoid glycosyltransferase from Withania somnifera. Bioinformation 2012, 8, 943–949. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Hou, B. Glycosyltransferases: Key players involved in the modification of plant secondary metabolites. Front. Biol. China 2009, 4, 39–46. [Google Scholar] [CrossRef]
- Li, P.; Li, Y.J.; Zhang, F.J.; Zhang, G.Z.; Jiang, X.Y.; Yu, H.M.; Hou, B.K. The Arabidopsis UDP-glycosyltransferases UGT79B2 and UGT79B3, contribute to cold, salt and drought stress tolerance via modulating anthocyanin accumulation. Plant J. Cell Mol. Biol. 2017, 89, 85–103. [Google Scholar] [CrossRef] [Green Version]
- Li, P.; Li, Y.J.; Wang, B.; Yu, H.M.; Li, Q.; Hou, B.K. The Arabidopsis UGT87A2, a stress-inducible family 1 glycosyltransferase, is involved in the plant adaptation to abiotic stresses. Physiol. Plant. 2017, 159, 416–432. [Google Scholar] [CrossRef]
- König, S.; Feussner, K.; Kaever, A.; Landesfeind, M.; Thurow, C.; Karlovsky, P.; Gatz, C.; Polle, A.; Feussner, I. Soluble phenylpropanoids are involved in the defense response of Arabidopsis against Verticillium longisporum. New Phytol. 2014, 202, 823–837. [Google Scholar] [CrossRef]
- Lim, E.K.; Jackson, R.G.; Bowles, D.J. Identification and characterisation of Arabidopsis glycosyltransferases capable of glucosylating coniferyl aldehyde and sinapyl aldehyde. FEBS Lett. 2005, 579, 2802–2806. [Google Scholar] [CrossRef] [Green Version]
- Chen, K.; Hu, Z.M.; Song, W.; Wang, Z.; Ye, M. Diversity of O-glycosyltransferases contributes to the biosynthesis of flavonoid and triterpenoid glycosides in glycyrrhiza uralensis. ACS Synth. Biol. 2019, 8, 1858–1866. [Google Scholar] [CrossRef]
- Ono, E.; Yu, H.; Horikawa, M.; Kunikane-Doi, S.; Imai, H.; Takahashi, S.; Kawai, Y.; Ishiguro, M.; Fukui, Y.; Nakayama, T. Functional differentiation of the glycosyltransferases that contribute to the chemical diversity of bioactive flavonol glycosides in grapevines (Vitis vinifera). Plant Cell 2010, 22, 2856–2871. [Google Scholar] [CrossRef] [Green Version]
- Xie, L.; Cao, Y.; Zhao, Z.; Ren, C.; Xing, M.; Wu, B.; Zhang, B.; Xu, C.; Chen, K.; Li, X. Involvement of MdUGT75B1 and MdUGT71B1 in flavonol galactoside/glucoside biosynthesis in apple fruit. Food Chem. 2020, 312, 126124. [Google Scholar] [CrossRef]
- Dai, X.; Zhuang, J.; Wu, Y.; Wang, P.; Zhao, G.; Liu, Y.; Jiang, X.; Gao, L.; Xia, T. Identification of a flavonoid glucosyltransferase involved in 7-OH site glycosylation in tea plants (Camellia sinensis). Sci. Rep. 2017, 7, 5926. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Sun, Z.; Liu, D.; Li, X.; Rehman, R.U.; Wang, H.; Wu, Z. Apple phlorizin attenuates oxidative stress in Drosophila melanogaster. J. Food Biochem. 2019, 43, e12744. [Google Scholar]
- Yahyaa, M.; Davidovich-Rikanati, R.; Eyal, Y.; Sheachter, A.; Marzouk, S.; Lewinsohn, E.; Ibdah, M. Identification and characterization of UDP-glucose: Phloretin 4′-O-glycosyltransferase from Malus x domestica Borkh. Phytochemistry 2016, 130, 47–55. [Google Scholar] [CrossRef]
- Jugd, H.; Nguy, D.; Moller, I.; Cooney, J.M.; Atkinson, R.G. Isolation and characterization of a novel glycosyltransferase that converts phloretin to phlorizin, a potent antioxidant in apple. FEBS J. 2010, 275, 3804–3814. [Google Scholar] [CrossRef]
- Wu, S.Y.; Fu, Y.H.; Zhou, Q.; Bai, M.; Chen, G.Y.; Han, C.R.; Song, X.P. A new dihydrochalcone glycoside from the stems of Homalium stenophyllum. Nat. Prod. Res. 2017, 32, 953–958. [Google Scholar] [CrossRef]
- Shimakage, R.; Nihei, K.I. Synthesis, structural revision, and tyrosinase inhibitory activity of proposed phloretin-4-O-β-D-glucopyranoside from Homalium stenophyllum. Nat. Prod. Res. 2020, 14, 1803–1811. [Google Scholar] [CrossRef] [PubMed]
- Oode, C.; Shimada, W.; Izutsu, Y.; Yokota, M.; Iwadate, T.; Nihei, K. Synthesis of dihydroresveratrol glycosides and evaluation of their activity against melanogenesis in B16F0 melanoma cells. Eur. J. Med. Chem. 2014, 87, 862–867. [Google Scholar] [CrossRef] [PubMed]
- Zhu, T.T.; Liu, H.; Wang, P.Y.; Ni, R.; Cheng, A.X. Functional characterization of UDP-glucosyltransferases from the liverwort Plagiochasma appendiculatum and their potential for biosynthesizing flavonoid 7-O-glucosides. Plant Sci. 2020, 299, 110577. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.C.; Xiong, R.L.; Zhu, T.T.; Ni, R.; Cheng, A.X. Cloning and functional characterization of three flavonoid O-glucosyltransferase genes from the liverworts Marchantia emarginata and Marchantia paleacea. Plant Physiol. Biochem. 2021, 166, 495–504. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Wu, Y.; Zhao, Y.; Han, X.; Lou, H.; Cheng, A. Molecular cloning and biochemical characterization of two cinnamyl alcohol dehydrogenases from a liverwort Plagiochasma appendiculatum. Plant Physiol. Biochem. 2013, 70, 133–141. [Google Scholar] [CrossRef]
- Koichiro, T.; Joel, D.; Masatoshi, N.; Sudhir, K. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007, 24, 1596–1599. [Google Scholar]
- Gambino, G.; Perrone, I.; Gribaudo, I. A rapid and effective method for RNA extraction from different tissues of grapevine and other woody plants. Phytochem. Anal. PCA 2010, 19, 520–525. [Google Scholar] [CrossRef]
- Jung, W.; Yu, O.; Lau, S.M.C.; O’Keefe, D.P.; Odell, J.; Fader, G.; Mcgonigle, B. Identification and expression of isoflavone synthase, the key enzyme for biosynthesis of isoflavones in legumes. Nat. Biotechnol. 2000, 18, 208–212. [Google Scholar] [CrossRef]
- Hood, E.E.; Gelvin, S.B.; Melchers, L.S.; Hoekema, A. New Agrobacterium helper plasmids for gene transfer to plants. Transgenic Res. 1993, 2, 208–218. [Google Scholar] [CrossRef]
- Maharjan, R.; Fukuda, Y.; Shimomura, N.; Nakayama, T.; Ozaki, S.I. An ambidextrous polyphenol glycosyltransferase PaGT2 from Phytolacca americana. Biochemistry 2020, 59, 2551–2561. [Google Scholar] [CrossRef]
- Xie, K.; Chen, R.; Li, J.; Wang, R.; Chen, D.; Dou, X.; Dai, J. Exploring the catalytic promiscuity of a new glycosyltransferase from Carthamus tinctorius. Org. Lett. 2014, 16, 4874–4877. [Google Scholar] [CrossRef]
- Hectors, K.; Prinsen, E.; Coen, W.D.; Jansen, M.; Guisez, Y. Arabidopsis thaliana plants acclimated to low dose rates of ultraviolet B radiation show specific changes in morphology and gene expression in the absence of stress symptoms. New Phytol. 2007, 175, 255–270. [Google Scholar] [CrossRef]
- Zhou, K.; Hu, L.; Li, P.; Gong, X.; Ma, F. Genome-wide identification of glycosyltransferases converting phloretin to phloridzin in Malus species. Plant Sci. 2017, 265, 131–145. [Google Scholar] [CrossRef]
- Le, R.J.; Brigitte, H.; Anne, C.; Simon, H.; Godfrey, N. Glycosylation is a major regulator of phenylpropanoid availability and biological activity in plants. Front. Plant Sci. 2016, 7, 735. [Google Scholar]
- Huang, X.X.; Wang, Y.; Lin, J.S.; Chen, L.; Li, Y.J.; Liu, Q.; Wang, G.F.; Xu, F.; Liu, L.; Hou, B.K. The novel pathogen-responsive glycosyltransferase UGT73C7 mediates the redirection of phenylpropanoid metabolism and promotes SNC1-dependent Arabidopsis immunity. Plant J. 2021, 107, 149–165. [Google Scholar] [CrossRef] [PubMed]
- Lanot, A.; Hodge, D.; Jackson, R.G.; George, G.L.; Bowles, D.J. The glucosyltransferase UGT72E2 is responsible for monolignol 4-O-glucoside production in Arabidopsis thaliana. Plant J. 2010, 48, 286–295. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, S.; Badger, M.R. Photoprotection in plants: A new light on photosystem II damage. Trends Plant Sci. 2011, 16, 53–60. [Google Scholar] [CrossRef] [PubMed]
- Peng, M.; Shahzad, R.; Gul, A.; Subthain, H.; Shen, S.; Lei, L.; Zheng, Z.; Zhou, J.; Lu, D.; Wang, S. Differentially evolved glucosyltransferases determine natural variation of rice flavone accumulation and UV-tolerance. Nat. Commun. 2017, 8, 1975. [Google Scholar] [CrossRef]

| Substrate | MpUGT737B1 | MpUGT741A1 | |
|---|---|---|---|
| Flavones | apigenin | trace a | ND b |
| luteolin | ND | trace | |
| chrysoeriol | trace | ND | |
| Flavonols | quercetin | ND | ND |
| kaempferol | ND | ND | |
| isorhamnetin | ND | ND | |
| Flavanones | naringenin | trace | 10.98 ± 0.56 |
| hesperetin | ND | trace | |
| liquiritigenin | ND | trace | |
| pinocembrin | trace | trace | |
| Chalcones | isoliquiritigenin | ND | ND |
| Isoflavones | genistein | ND | ND |
| Dihydrochalcone | phloretin | 95.29 ± 1.49 c | ND |
| Bibenzyls | lunularin | 90.38 ± 2.32 | 99.66 ± 0.48 |
| lunularic acid | ND | ND | |
| dihydroresveratrol | 98.84 ± 1.65 | ND | |
| Stilbenes | resveratrol | ND | ND |
| Coumarins | esculetin | ND | ND |
| Phenylpropyl | caffeic acid | ND | ND |
| caffeoyl aldehyde | 62.40 ± 1.19 | trace | |
| coniferaldehyde | 70.37 ± 1.32 | ND | |
| coniferyl alcohol | 74.45 ± 2.25 | ND | |
| 5-OH coniferyl alcohol | ND | ND | |
| 5-OH coniferaldehyde | 90.39 ± 0.36 | ND | |
| sinapaldehyde | 14.78 ± 0.21 | ND | |
| sinapyl alcohol | ND | ND |
| Enzyme | Substrate | Km (μM) | Vmax | kcat | kcat/Km |
|---|---|---|---|---|---|
| (nmol mg−1 min−1) | (s−1) | (M−1 s−1) | |||
| Phloretin | 50.2 ± 10.6 | 70.0 ± 5.5 | 0.062 ± 0.005 | 1244.2 | |
| MpUGT737B1 | Dihydroresveratrol | 45.6 ± 9.7 | 100.3 ± 6.9 | 0.089 ± 0.006 | 1960.4 |
| Lunularin | 39.3 ± 7.5 | 37.3 ± 2.2 | 0.033 ± 0.002 | 846.2 | |
| MpUGT741A1 | Lunularin | 89.2 ± 17.6 | 150.4 ± 12.3 | 0.124 ± 0.012 | 1504.6 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xiong, R.-L.; Zhang, J.-Z.; Liu, X.-Y.; Deng, J.-Q.; Zhu, T.-T.; Ni, R.; Tan, H.; Sheng, J.-Z.; Lou, H.-X.; Cheng, A.-X. Identification and Characterization of Two Bibenzyl Glycosyltransferases from the Liverwort Marchantia polymorpha. Antioxidants 2022, 11, 735. https://doi.org/10.3390/antiox11040735
Xiong R-L, Zhang J-Z, Liu X-Y, Deng J-Q, Zhu T-T, Ni R, Tan H, Sheng J-Z, Lou H-X, Cheng A-X. Identification and Characterization of Two Bibenzyl Glycosyltransferases from the Liverwort Marchantia polymorpha. Antioxidants. 2022; 11(4):735. https://doi.org/10.3390/antiox11040735
Chicago/Turabian StyleXiong, Rui-Lin, Jiao-Zhen Zhang, Xin-Yan Liu, Jian-Qun Deng, Ting-Ting Zhu, Rong Ni, Hui Tan, Ju-Zheng Sheng, Hong-Xiang Lou, and Ai-Xia Cheng. 2022. "Identification and Characterization of Two Bibenzyl Glycosyltransferases from the Liverwort Marchantia polymorpha" Antioxidants 11, no. 4: 735. https://doi.org/10.3390/antiox11040735
APA StyleXiong, R.-L., Zhang, J.-Z., Liu, X.-Y., Deng, J.-Q., Zhu, T.-T., Ni, R., Tan, H., Sheng, J.-Z., Lou, H.-X., & Cheng, A.-X. (2022). Identification and Characterization of Two Bibenzyl Glycosyltransferases from the Liverwort Marchantia polymorpha. Antioxidants, 11(4), 735. https://doi.org/10.3390/antiox11040735







