Proteins Associated with Phagocytosis Alteration in Retinal Pigment Epithelial Cells Derived from Age-Related Macular Degeneration Patients
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients and hiPSC-RPE Cell Lines
2.2. Iron Treatment and Induction of Oxidative Stress
2.3. Isolation of Porcine POS
2.4. Analysis of Phagocytosis by Flow Cytometry
2.5. Total RNA Sequencing
2.6. Western Blot
3. Results
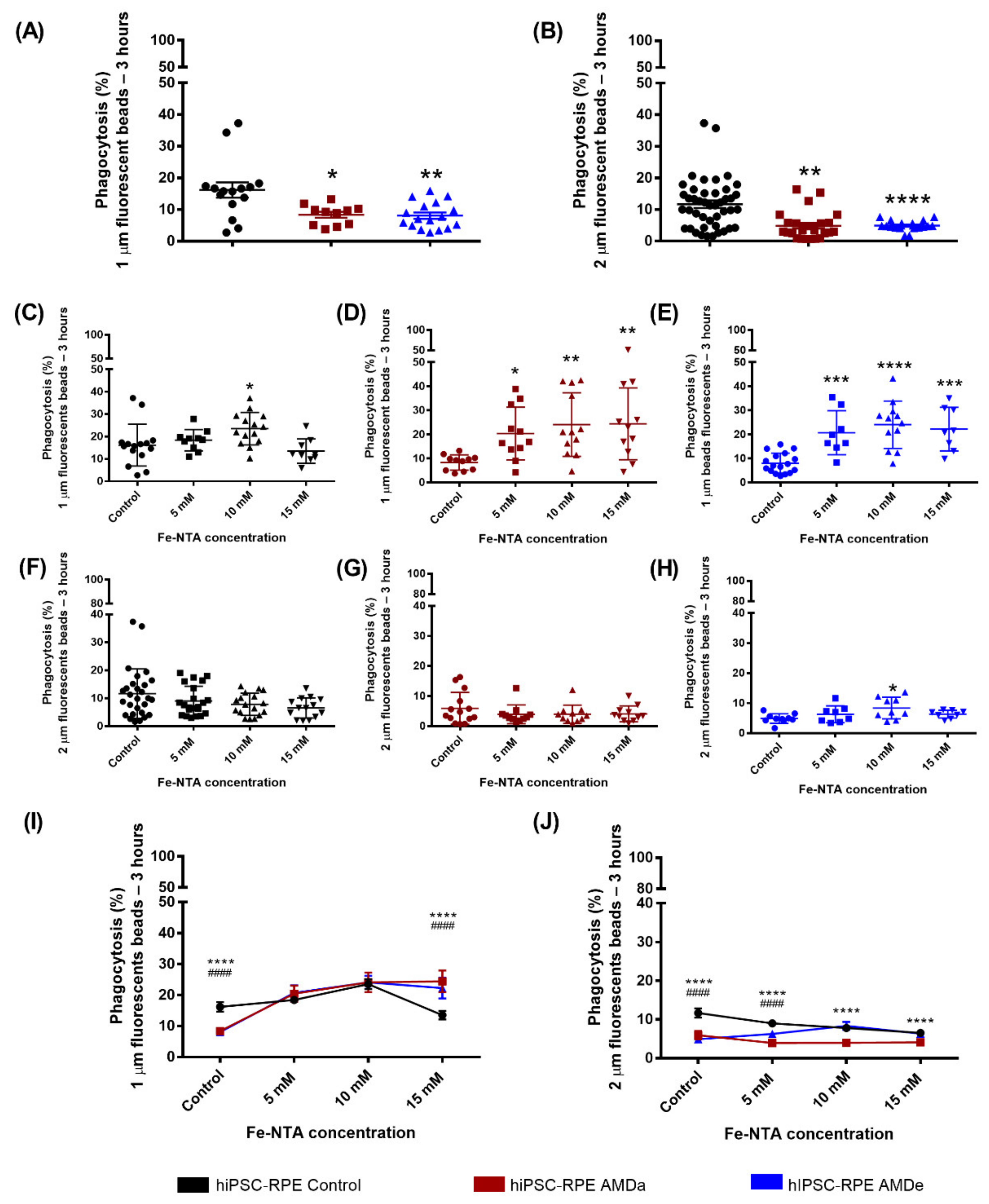
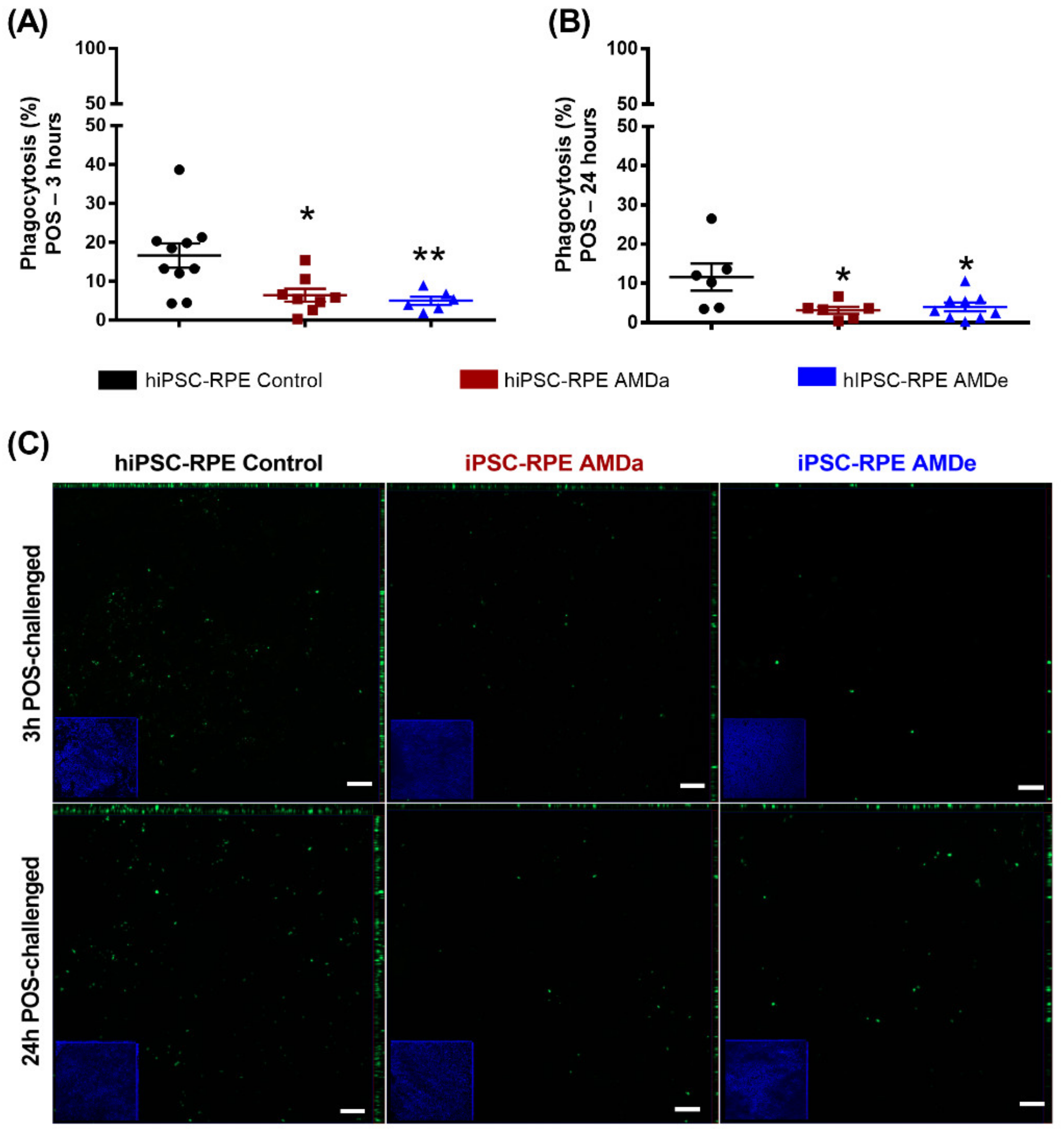
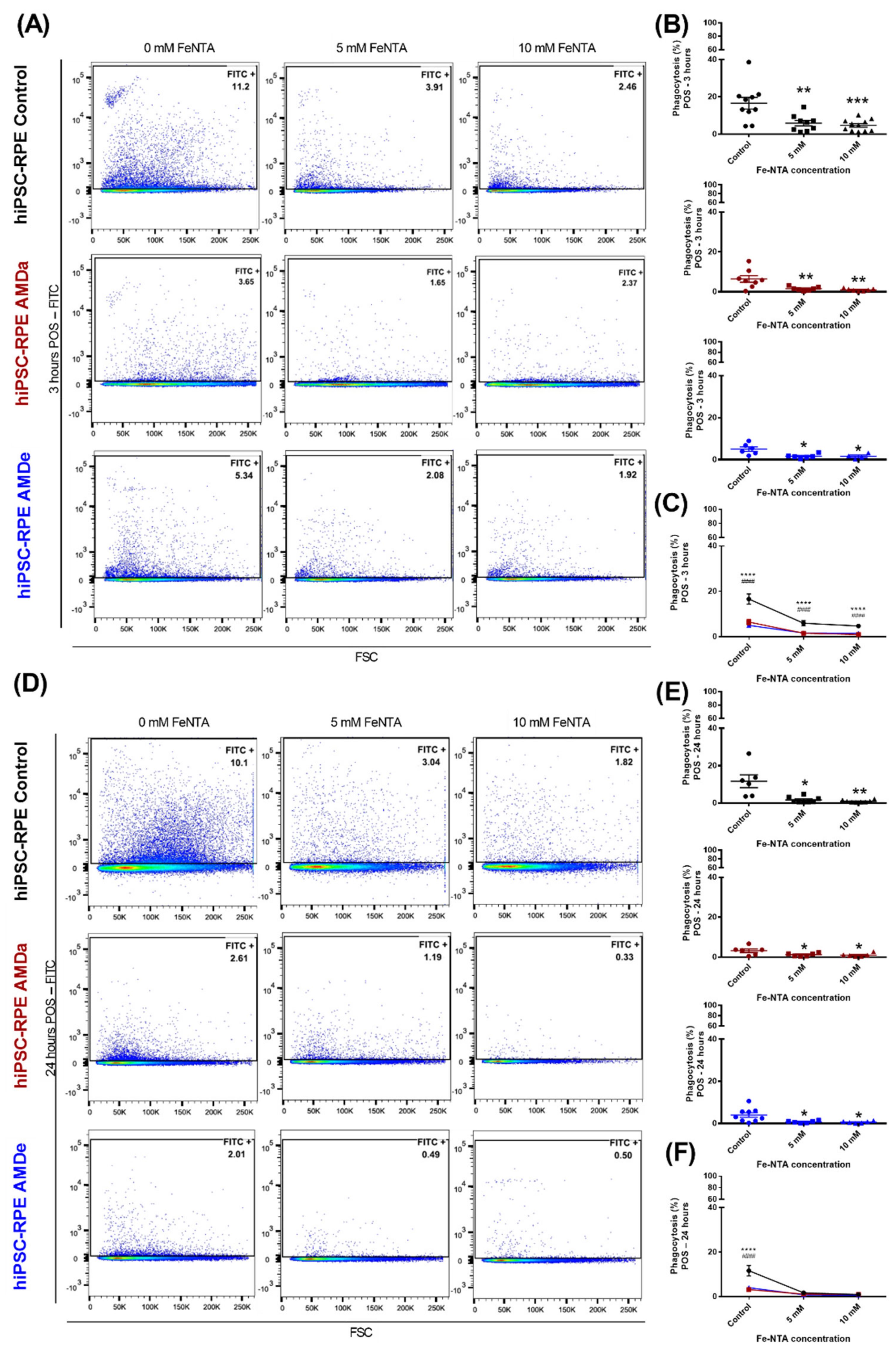
3.1. Non-Specific Phagocytosis Activity of hiPSC-RPE Control and AMD Cells in Basal Condition and under Oxidative Stress Condition
3.2. Specific Phagocytosis Activity of hiPSC-RPE Control and AMD Cells in Basal Condition and under Oxidative Stress Condition
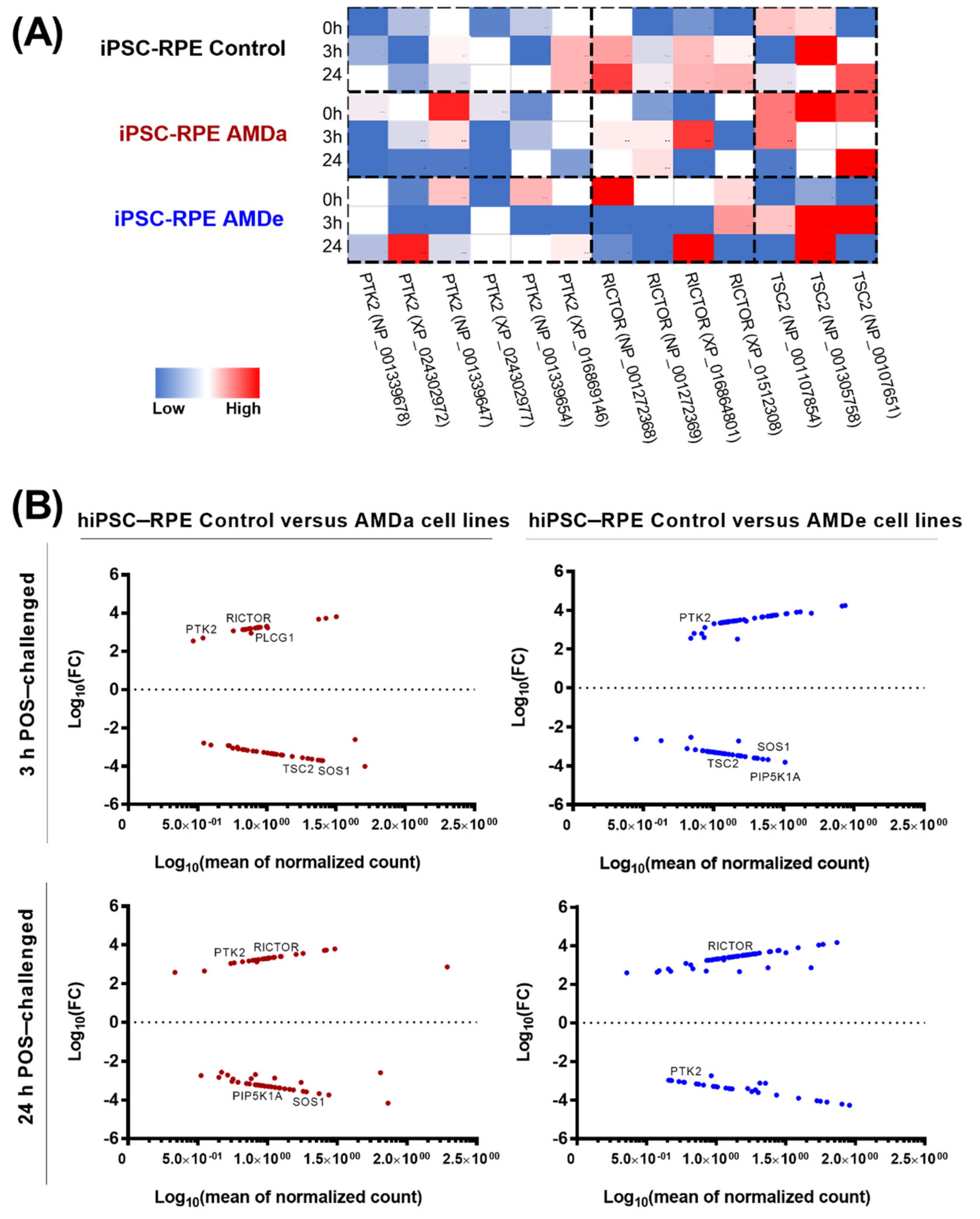
3.3. Pathways Activated during Specific Phagocytosis Activity in hiPSC-RPE Cells
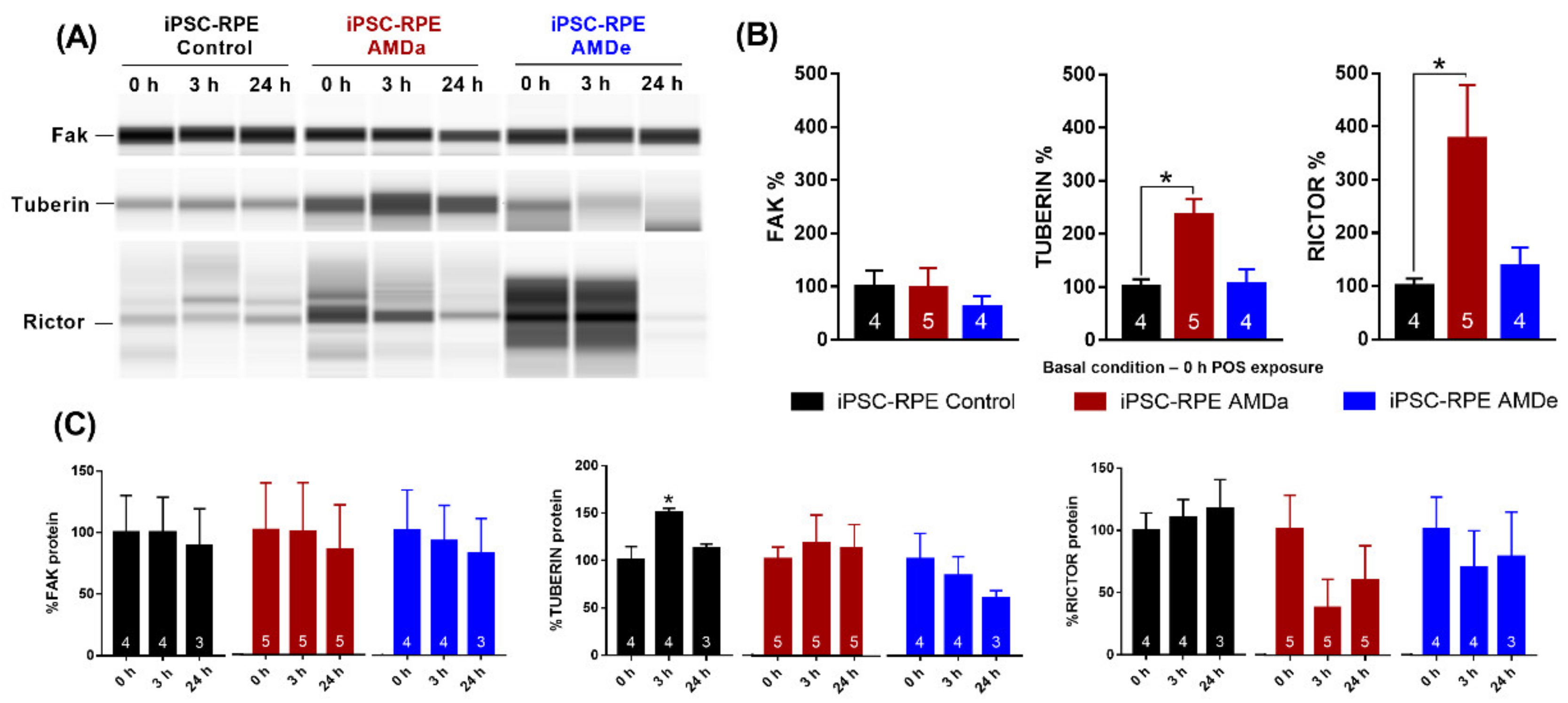
3.4. Difference between hiPSC Control Cells and AMD Cells in the PI3K/Akt, mTor and MEK/ERK Signaling Pathway Related to Specific Phagocytosis Activity
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Flaxman, S.R.; Bourne, R.R.A.; Resnikoff, S.; Ackland, P.; Braithwaite, T.; Cicinelli, M.V.; Das, A.; Jonas, J.B.; Keeffe, J.; Kempen, J.H.; et al. Global Causes of Blindness and Distance Vision Impairment 1990–2020: A Systematic Review and Meta-Analysis. Lancet Glob. Health 2017, 5, e1221–e1234. [Google Scholar] [CrossRef]
- Wong, W.L.; Su, X.; Li, X.; Cheung, C.M.G.; Klein, R.; Cheng, C.-Y.; Wong, T.Y. Global Prevalence of Age-Related Macular Degeneration and Disease Burden Projection for 2020 and 2040: A Systematic Review and Meta-Analysis. Lancet Glob. Health 2014, 2, E106–E116. [Google Scholar] [CrossRef]
- Nivison-Smith, L.; Milston, R.; Madigan, M.; Kalloniatis, M. Age-Related Macular Degeneration: Linking Clinical Presentation to Pathology. Optom. Vis. Sci. 2014, 91, 832–848. [Google Scholar] [CrossRef] [PubMed]
- Rakoczy, E.P. Gene Therapy for the Long Term Treatment of Wet AMD. Lancet 2017, 390, 6–7. [Google Scholar] [CrossRef]
- Kunchithapautham, K.; Atkinson, C.; Rohrer, B. Smoke Exposure Causes Endoplasmic Reticulum Stress and Lipid Accumulation in Retinal Pigment Epithelium through Oxidative Stress and Complement Activation. J. Biol. Chem. 2014, 289, 14534–14546. [Google Scholar] [CrossRef] [PubMed]
- Mullins, R.F.; Warwick, A.N.; Sohn, E.H.; Lotery, A.J. From Compliment to Insult: Genetics of the Complement System in Physiology and Disease in the Human Retina. Hum. Mol. Genet. 2017, 26, R51–R57. [Google Scholar] [CrossRef] [PubMed]
- Alberio, T.; Lopiano, L.; Fasano, M. Cellular Models to Investigate Biochemical Pathways in Parkinson’s Disease. FEBS J. 2012, 279, 1146–1155. [Google Scholar] [CrossRef]
- Malpass, K. Parkinson Disease: Induced Pluripotent Stem Cells-a New in Vitro Model to Investigate α-Synuclein Dysfunction in Parkinson Disease. Nat. Rev. Neurol. 2011, 7, 536. [Google Scholar] [CrossRef]
- Voisin, A.; Monville, C.; Plancheron, A.; Balbous, A.; Gaillard, A.; Leveziel, N. HRPE Cells Derived from Induced Pluripotent Stem Cells Are More Sensitive to Oxidative Stress than ARPE-19 Cells. Exp. Eye Res. 2018, 177, 76–86. [Google Scholar] [CrossRef]
- Voisin, A.; Monville, C.; Plancheron, A.; Béré, E.; Gaillard, A.; Leveziel, N. Cathepsin B PH-Dependent Activity Is Involved in Lysosomal Dysregulation in Atrophic Age-Related Macular Degeneration. Oxid. Med. Cell. Longev. 2019, 2019, 1–15. [Google Scholar] [CrossRef]
- Saini, J.S.; Corneo, B.; Miller, J.D.; Kiehl, T.R.; Wang, Q.; Boles, N.C.; Blenkinsop, T.A.; Stern, J.H.; Temple, S. Nicotinamide Ameliorates Disease Phenotypes in a Human IPSC Model of Age-Related Macular Degeneration. Cell Stem Cell 2017, 20, 635–647. [Google Scholar] [CrossRef] [PubMed]
- Ach, T.; Tolstik, E.; Messinger, J.D.; Zarubina, A.V.; Heintzmann, R.; Curcio, C.A. Lipofuscin Redistribution and Loss Accompanied by Cytoskeletal Stress in Retinal Pigment Epithelium of Eyes With Age-Related Macular DegenerationLipofuscin Redistribution in AMD. Investig. Ophthalmol. Vis. Sci. 2015, 56, 3242–3252. [Google Scholar] [CrossRef] [PubMed]
- Terluk, M.R.; Kapphahn, R.J.; Soukup, L.M.; Gong, H.; Gallardo, C.; Montezuma, S.R.; Ferrington, D.A. Investigating Mitochondria as a Target for Treating Age-Related Macular Degeneration. J. Neurosci. 2015, 35, 7304–7311. [Google Scholar] [CrossRef] [PubMed]
- Olchawa, M.M.; Pilat, A.K.; Szewczyk, G.M.; Sarna, T.J.; Sarna, T. Inhibition of Phagocytic Activity of ARPE-19 Cells by Free Radical Mediated Oxidative Stress. Free Radic. Res. 2016, 50, 887–897. [Google Scholar] [CrossRef] [PubMed]
- Bosch, E.; Horwitz, J.; Bok, D. Phagocytosis of Outer Segments by Retinal Pigment Epithelium: Phagosome-Lysosome Interaction. J. Histochem. Cytochem. 1993, 41, 253–263. [Google Scholar] [CrossRef] [PubMed]
- Dalvi, S.; Galloway, C.A.; Winschel, L.; Hashim, A.; Soto, C.; Tang, C.; MacDonald, L.A.; Singh, R. Environmental Stress Impairs Photoreceptor Outer Segment (POS) Phagocytosis and Degradation and Induces Autofluorescent Material Accumulation in HiPSC-RPE Cells. Cell Death Discov. 2019, 5, 96. [Google Scholar] [CrossRef]
- He, F.; Agosto, M.A.; Nichols, R.M.; Wensel, T.G. Multiple Phosphatidylinositol(3)Phosphate Roles in Retinal Pigment Epithelium Membrane Recycling. BioRxiv 2020. [Google Scholar] [CrossRef]
- Mukherjee, P.K.; Marcheselli, V.L.; de Rivero Vaccari, J.C.; Gordon, W.C.; Jackson, F.E.; Bazan, N.G. Photoreceptor Outer Segment Phagocytosis Attenuates Oxidative Stress-Induced Apoptosis with Concomitant Neuroprotectin D1 Synthesis. Proc. Natl. Acad. Sci. USA 2007, 104, 13158–13163. [Google Scholar] [CrossRef]
- Mao, Y.; Finnemann, S.C. Essential Diurnal Rac1 Activation during Retinal Phagocytosis Requires Avβ5 Integrin but Not Tyrosine Kinases Focal Adhesion Kinase or Mer Tyrosine Kinase. Mol. Biol. Cell 2012, 23, 1104–1114. [Google Scholar] [CrossRef]
- D’Cruz, P.M.; Yasumura, D.; Weir, J.; Matthes, M.T.; Abderrahim, H.; LaVail, M.M.; Vollrath, D. Mutation of the Receptor Tyrosine Kinase Gene Mertk in the Retinal Dystrophic RCS Rat. Hum. Mol. Genet. 2000, 9, 645–651. [Google Scholar] [CrossRef]
- Olchawa, M.; Szewczyk, G.; Zaręba, M.; Piłat, A.; Bzowska, M.; Mikołajczyk, T.; Sarna, T. Sub-Lethal Photodynamic Damage to ARPE-19 Cells Transiently Inhibits Their Phagocytic Activity†. Photochem. Photobiol. 2010, 86, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Inana, G.; Murat, C.; An, W.; Yao, X.; Harris, I.R.; Cao, J. RPE Phagocytic Function Declines in Age-Related Macular Degeneration and Is Rescued by Human Umbilical Tissue Derived Cells. J. Transl. Med. 2018, 16, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Parinot, C.; Rieu, Q.; Chatagnon, J.; Finnemann, S.C.; Nandrot, E.F. Large-Scale Purification of Porcine or Bovine Photoreceptor Outer Segments for Phagocytosis Assays on Retinal Pigment Epithelial Cells. J. Vis. Exp. JoVE 2014, 94, e52100. [Google Scholar] [CrossRef] [PubMed]
- Leng, N.; Li, Y.; McIntosh, B.E.; Nguyen, B.K.; Duffin, B.; Tian, S.; Thomson, J.A.; Dewey, C.N.; Stewart, R.; Kendziorski, C. EBSeq-HMM: A Bayesian Approach for Identifying Gene-Expression Changes in Ordered RNA-Seq Experiments. Bioinformatics 2015, 31, 2614–2622. [Google Scholar] [CrossRef] [PubMed]
- Samuel, W.; Jaworski, C.; Postnikova, O.A.; Kutty, R.K.; Duncan, T.; Tan, L.X.; Poliakov, E.; Lakkaraju, A.; Redmond, T.M. Appropriately Differentiated ARPE-19 Cells Regain Phenotype and Gene Expression Profiles Similar to Those of Native RPE Cells. Mol. Vis. 2017, 23, 60. [Google Scholar]
- Bulloj, A.; Duan, W.; Finnemann, S.C. PI 3-Kinase Independent Role for AKT in F-Actin Regulation during Outer Segment Phagocytosis by RPE Cells. Exp. Eye Res. 2013, 113, 9–18. [Google Scholar] [CrossRef]
- Chen, H.; Lukas, T.J.; Du, N.; Suyeoka, G.; Neufeld, A.H. Dysfunction of the Retinal Pigment Epithelium with Age: Increased Iron Decreases Phagocytosis and Lysosomal Activity. Investig. Opthalmol. Vis. Sci. 2009, 50, 1895. [Google Scholar] [CrossRef]
- Wong, R.W.; Richa, D.C.; Hahn, P.; Green, W.R.; Dunaief, J.L. Iron toxicity as a potential factor in AMD. Retina 2007, 27, 997–1003. [Google Scholar] [CrossRef]
- Chiang, C.-K.; Tworak, A.; Kevany, B.M.; Xu, B.; Mayne, J.; Ning, Z.; Figeys, D.; Palczewski, K. Quantitative Phosphoproteomics Reveals Involvement of Multiple Signaling Pathways in Early Phagocytosis by the Retinal Pigmented Epithelium. J. Biol. Chem. 2017, 292, 19826–19839. [Google Scholar] [CrossRef]
- Ryeom, S.W.; Silverstein, R.L.; Scotto, A.; Sparrow, J.R. Binding of Anionic Phospholipids to Retinal Pigment Epithelium May Be Mediated by the Scavenger Receptor CD36. J. Biol. Chem. 1996, 271, 20536–20539. [Google Scholar] [CrossRef][Green Version]
- Nandrot, E.F.; Anand, M.; Almeida, D.; Atabai, K.; Sheppard, D.; Finnemann, S.C. Essential Role for MFG-E8 as Ligand for αvβ5 Integrin in Diurnal Retinal Phagocytosis. Proc. Natl. Acad. Sci. USA 2007, 104, 12005–12010. [Google Scholar] [CrossRef] [PubMed]
- Goruppi, S.; Ruaro, E.; Varnum, B.; Schneider, C. Requirement of Phosphatidylinositol 3-Kinase-Dependent Pathway and Src for Gas6-Axl Mitogenic and Survival Activities in NIH 3T3 Fibroblasts. Mol. Cell. Biol. 1997, 17, 4442–4453. [Google Scholar] [CrossRef] [PubMed]
- Beatty, S.; Koh, H.; Phil, M.; Henson, D.; Boulton, M. The Role of Oxidative Stress in the Pathogenesis of Age-Related Macular Degeneration. Surv. Ophthalmol. 2000, 45, 115–134. [Google Scholar] [CrossRef]
- Yuodelis, C.; Hendrickson, A. A Qualitative and Quantitative Analysis of the Human Fovea during Development. Vision Res. 1986, 26, 847–855. [Google Scholar] [CrossRef]
- Mustafi, D.; Engel, A.H.; Palczewski, K. Structure of Cone Photoreceptors. Prog. Retin. Eye Res. 2009, 28, 289–302. [Google Scholar] [CrossRef]
- Mazzoni, F.; Safa, H.; Finnemann, S.C. Understanding Photoreceptor Outer Segment Phagocytosis: Use and Utility of RPE Cells in Culture. Exp. Eye Res. 2014, 126, 51–60. [Google Scholar] [CrossRef]
- Finnemann, S.C. Focal Adhesion Kinase Signaling Promotes Phagocytosis of Integrin-Bound Photoreceptors. EMBO J. 2003, 22, 4143–4154. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, J.; Cai, J.; Sternberg, P. Altered MTOR Signaling in Senescent Retinal Pigment Epithelium. Investig. Ophthalmol. Vis. Sci. 2010, 51, 5314–5319. [Google Scholar] [CrossRef]
- Oh, W.J.; Jacinto, E. MTOR Complex 2 Signaling and Functions. Cell Cycle 2011, 10, 2305–2316. [Google Scholar] [CrossRef]
- Morrison Joly, M.; Williams, M.M.; Hicks, D.J.; Jones, B.; Sanchez, V.; Young, C.D.; Sarbassov, D.D.; Muller, W.J.; Brantley-Sieders, D.; Cook, R.S. Two Distinct MTORC2-Dependent Pathways Converge on Rac1 to Drive Breast Cancer Metastasis. Breast Cancer Res. 2017, 19, 74. [Google Scholar] [CrossRef]
- Sit, S.-T.; Manser, E. Rho GTPases and Their Role in Organizing the Actin Cytoskeleton. J. Cell Sci. 2011, 124, 679–683. [Google Scholar] [CrossRef] [PubMed]
- Goncharova, E.A.; Goncharov, D.A.; Li, H.; Pimtong, W.; Lu, S.; Khavin, I.; Krymskaya, V.P. MTORC2 Is Required for Proliferation and Survival of TSC2-Null Cells. Mol. Cell. Biol. 2011, 31, 2484–2498. [Google Scholar] [CrossRef] [PubMed]
- Sethna, S.; Chamakkala, T.; Gu, X.; Thompson, T.C.; Cao, G.; Elliott, M.H.; Finnemann, S.C. Regulation of Phagolysosomal Digestion by Caveolin-1 of the Retinal Pigment Epithelium Is Essential for Vision. J. Biol. Chem. 2016, 291, 6494–6506. [Google Scholar] [CrossRef] [PubMed]
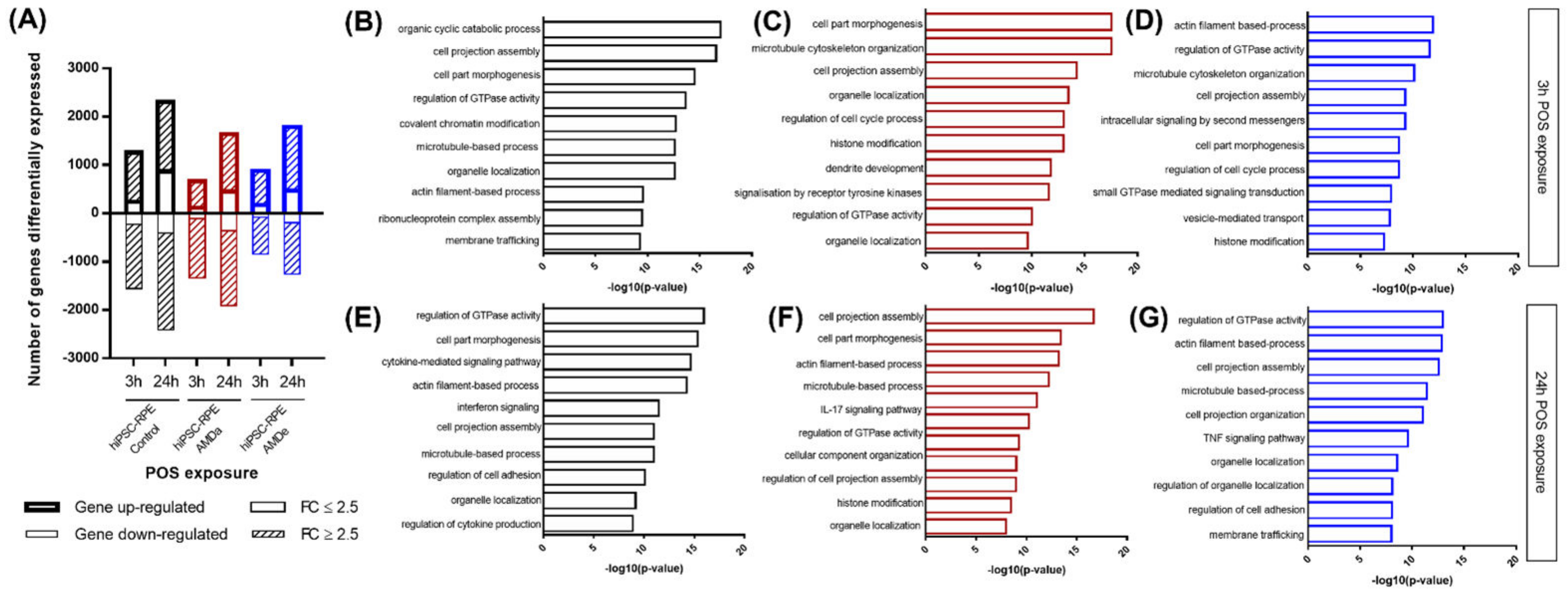
| Comparison between hiPSC-RPE Control, AMDa and AMDe Cells of Selected GO Biological Process Involved in 3 h and 24 h—POS Challenged | ||||
|---|---|---|---|---|
| POS-Challenged Time | 3 h | 24 h | ||
| hiPSC-RPE Cell Lines | Control vs. AMDa | Control vs. AMDe | Control vs. AMDa | Control vs. AMDe |
| GO Biological Process | Log10(p-value) | |||
| Regulation of GTPase activity | −9.474354469 | −17.7943 | −12.0125 | −13.1224 |
| Cell junction organization | −4.06929 | −7.56126 | −3.61127 | −7.53024 |
| Cell part morphogenesis | −7.300745101 | −16.7688 | −4.77497 | −9.28681 |
| Microtubule-based process | −6.04778 | −10.1841 | −6.04778 | −9.19358 |
| Plasma membrane bounded cell projection assembly | −7.26267 | −11.7729 | −7.26267 | −8.91932 |
| Organelle localization | −4.66769 | −14.0398 | −4.7191 | −10.326 |
| Membrane trafficking | −4.7262 | −12.4136 | −3.62451 | −5.06347 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Voisin, A.; Gaillard, A.; Balbous, A.; Leveziel, N. Proteins Associated with Phagocytosis Alteration in Retinal Pigment Epithelial Cells Derived from Age-Related Macular Degeneration Patients. Antioxidants 2022, 11, 713. https://doi.org/10.3390/antiox11040713
Voisin A, Gaillard A, Balbous A, Leveziel N. Proteins Associated with Phagocytosis Alteration in Retinal Pigment Epithelial Cells Derived from Age-Related Macular Degeneration Patients. Antioxidants. 2022; 11(4):713. https://doi.org/10.3390/antiox11040713
Chicago/Turabian StyleVoisin, Audrey, Afsaneh Gaillard, Anaïs Balbous, and Nicolas Leveziel. 2022. "Proteins Associated with Phagocytosis Alteration in Retinal Pigment Epithelial Cells Derived from Age-Related Macular Degeneration Patients" Antioxidants 11, no. 4: 713. https://doi.org/10.3390/antiox11040713
APA StyleVoisin, A., Gaillard, A., Balbous, A., & Leveziel, N. (2022). Proteins Associated with Phagocytosis Alteration in Retinal Pigment Epithelial Cells Derived from Age-Related Macular Degeneration Patients. Antioxidants, 11(4), 713. https://doi.org/10.3390/antiox11040713






