Assessment of Antioxidant, Immunomodulatory Activity of Oxidised Epigallocatechin-3-Gallate (Green Tea Polyphenol) and Its Action on the Main Protease of SARS-CoV-2—An In Vitro and In Silico Approach
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture and Cell Lines
2.2. Chemicals and Kits
2.3. Oxidation of EGCG
2.4. Cell Viability Assay
2.5. Antioxidant Activity
2.5.1. Total Antioxidant Assay
2.5.2. TBARS Assay
2.6. Cell Supernatant Collection
2.7. Inflammatory Markers
2.7.1. Human IL-6
2.7.2. Human IL-1β
2.7.3. TNF-α
2.8. In Silico Molecular Docking
2.9. Statistical Analysis
3. Results
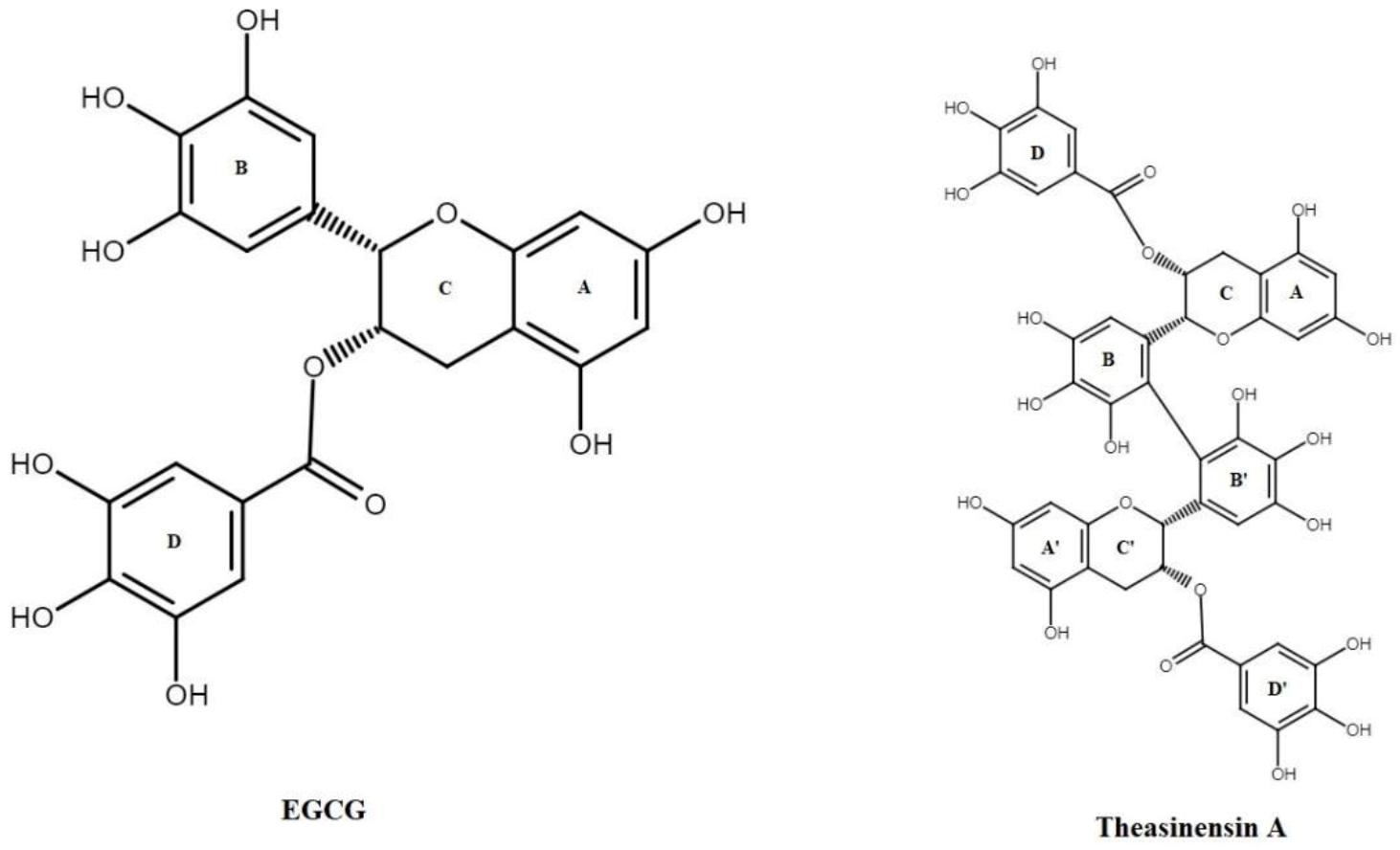
3.1. Oxidation of EGCG
3.2. Cell Viability Assay
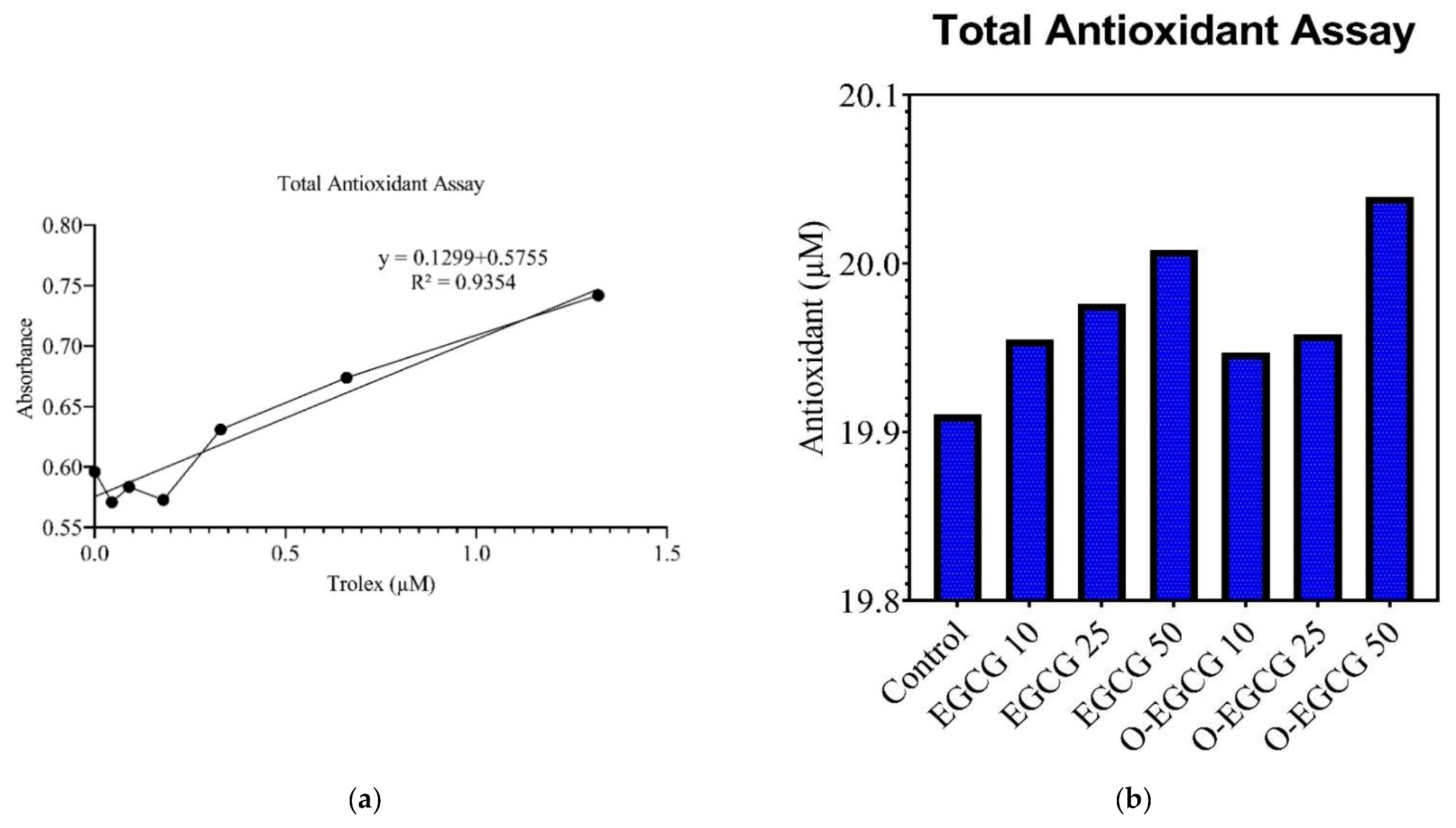
3.3. Antioxidant Assay
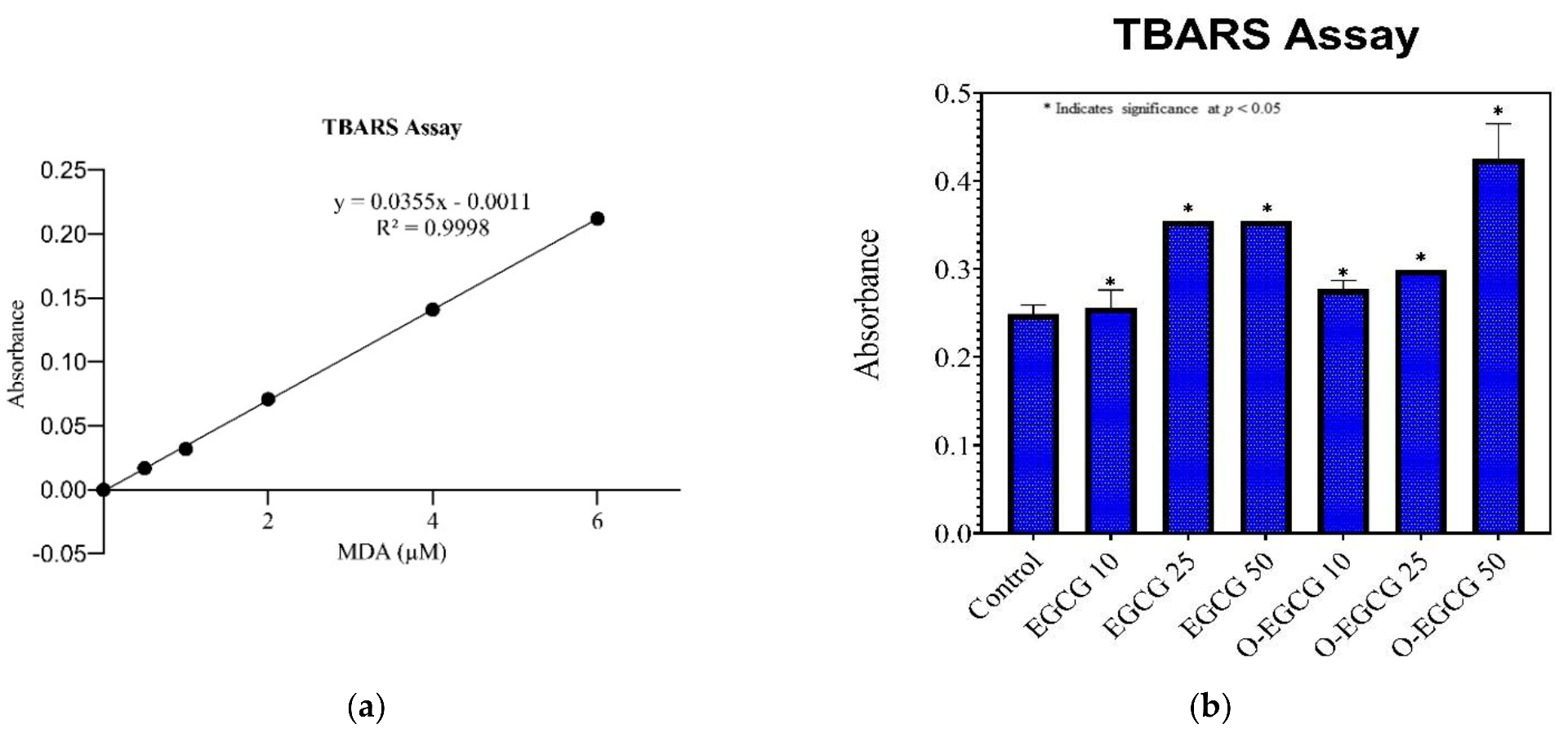
3.4. Thiobarbituric Acid Reactive Species Assay (TBARS)
3.5. Effect of EGCG and O-EGCG on Inflammatory Markers
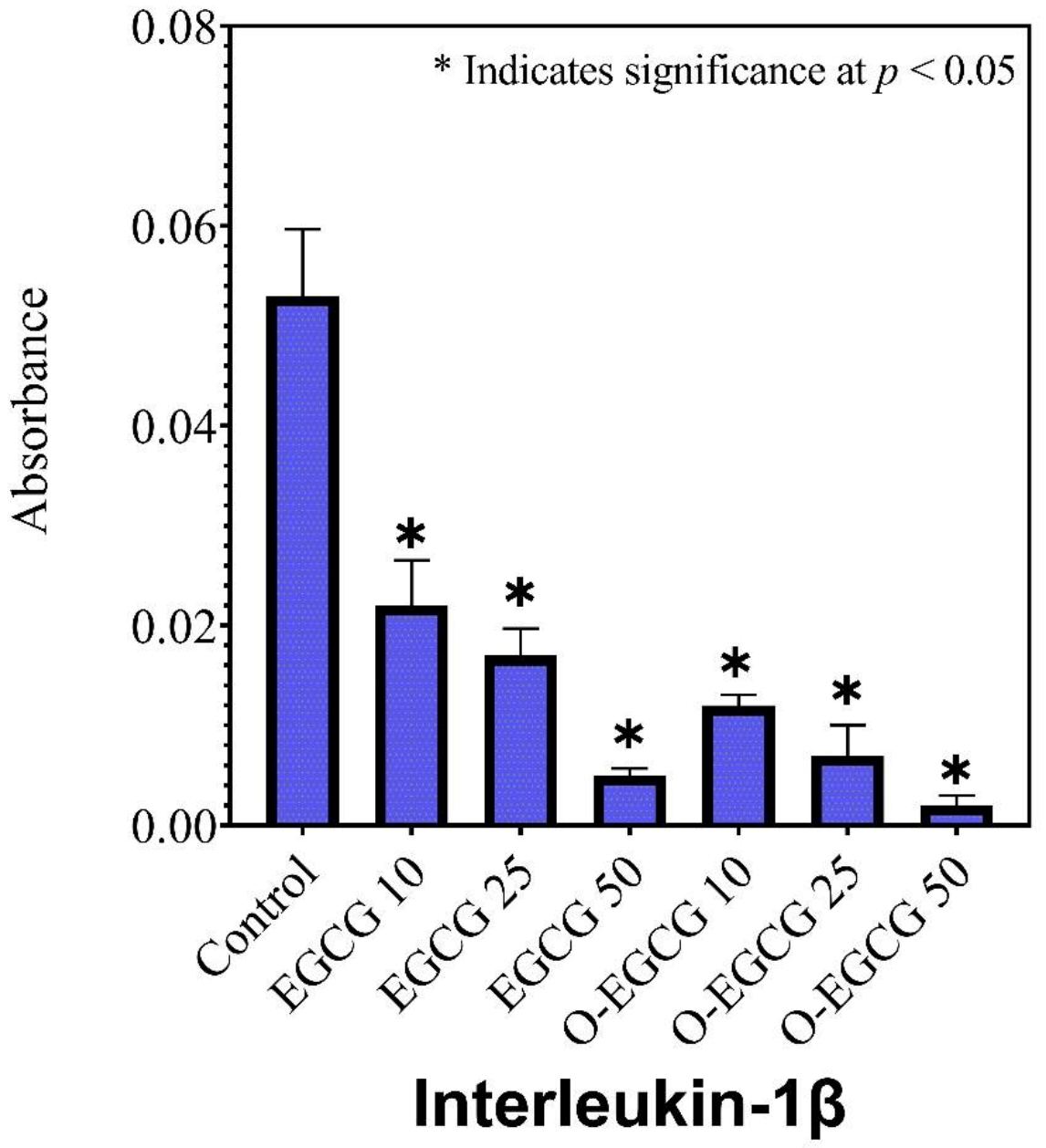
3.6. Human IL-1β
3.7. Human IL-6
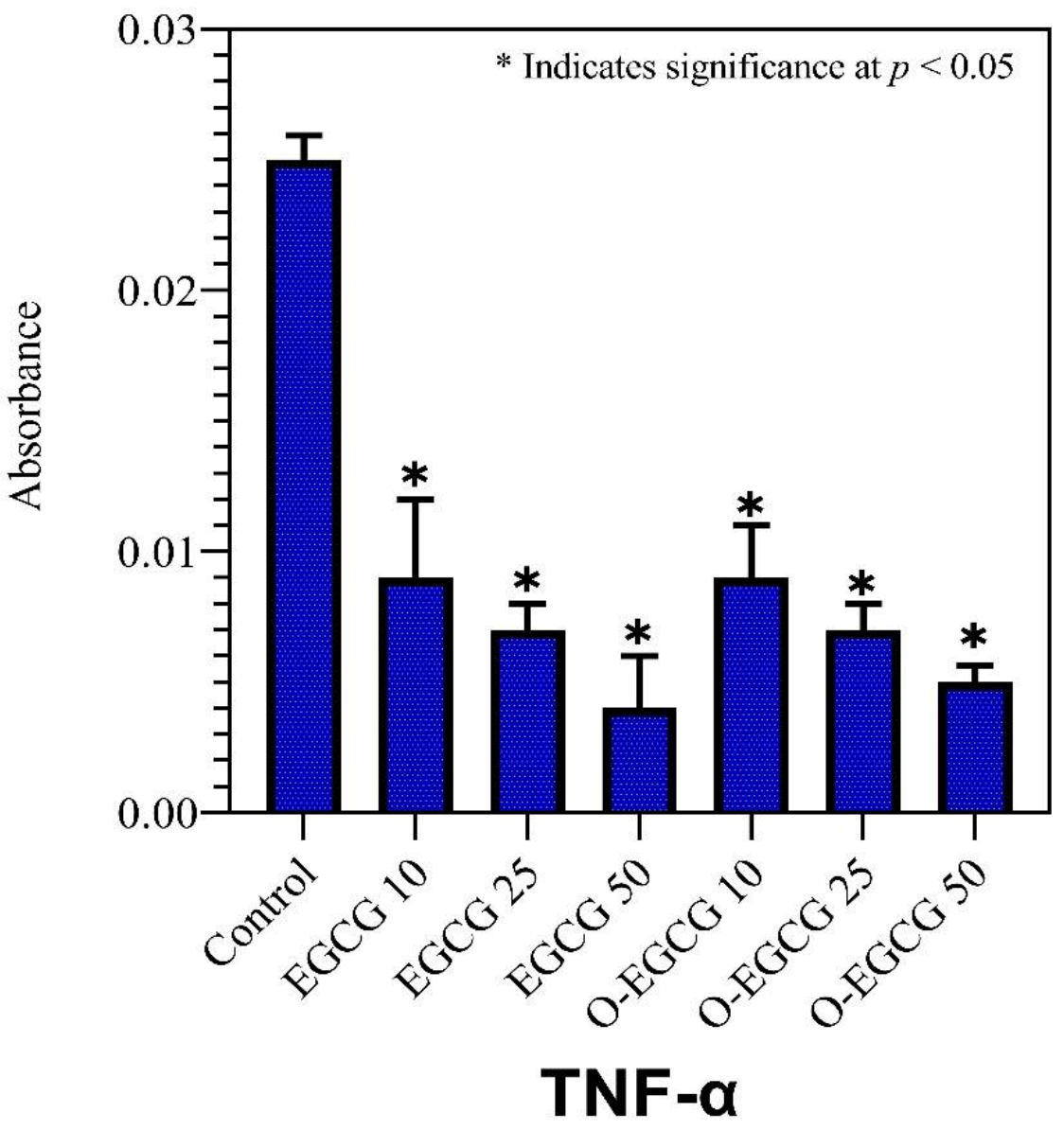
3.8. TNF-Alpha
3.9. In Silico Analysis
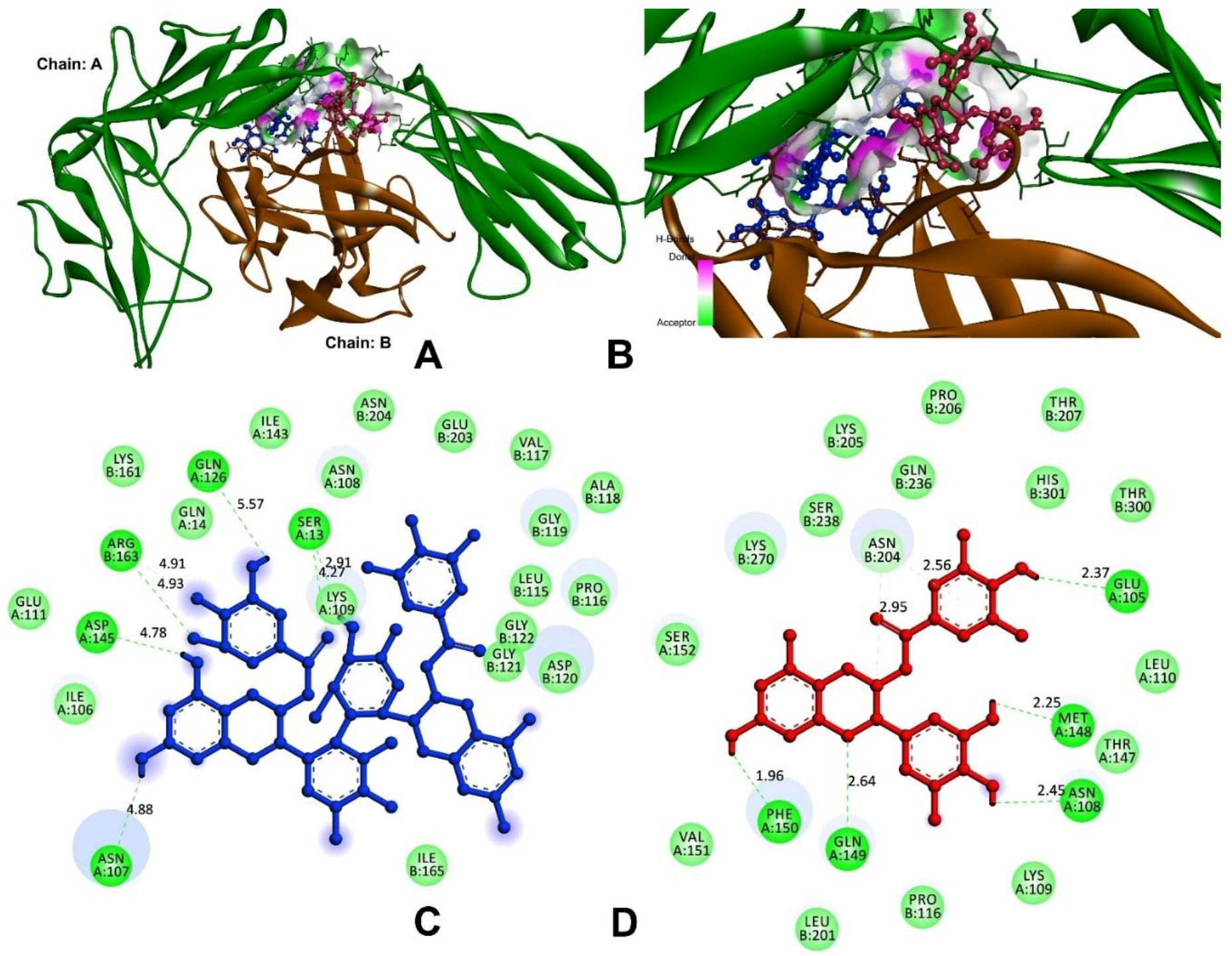
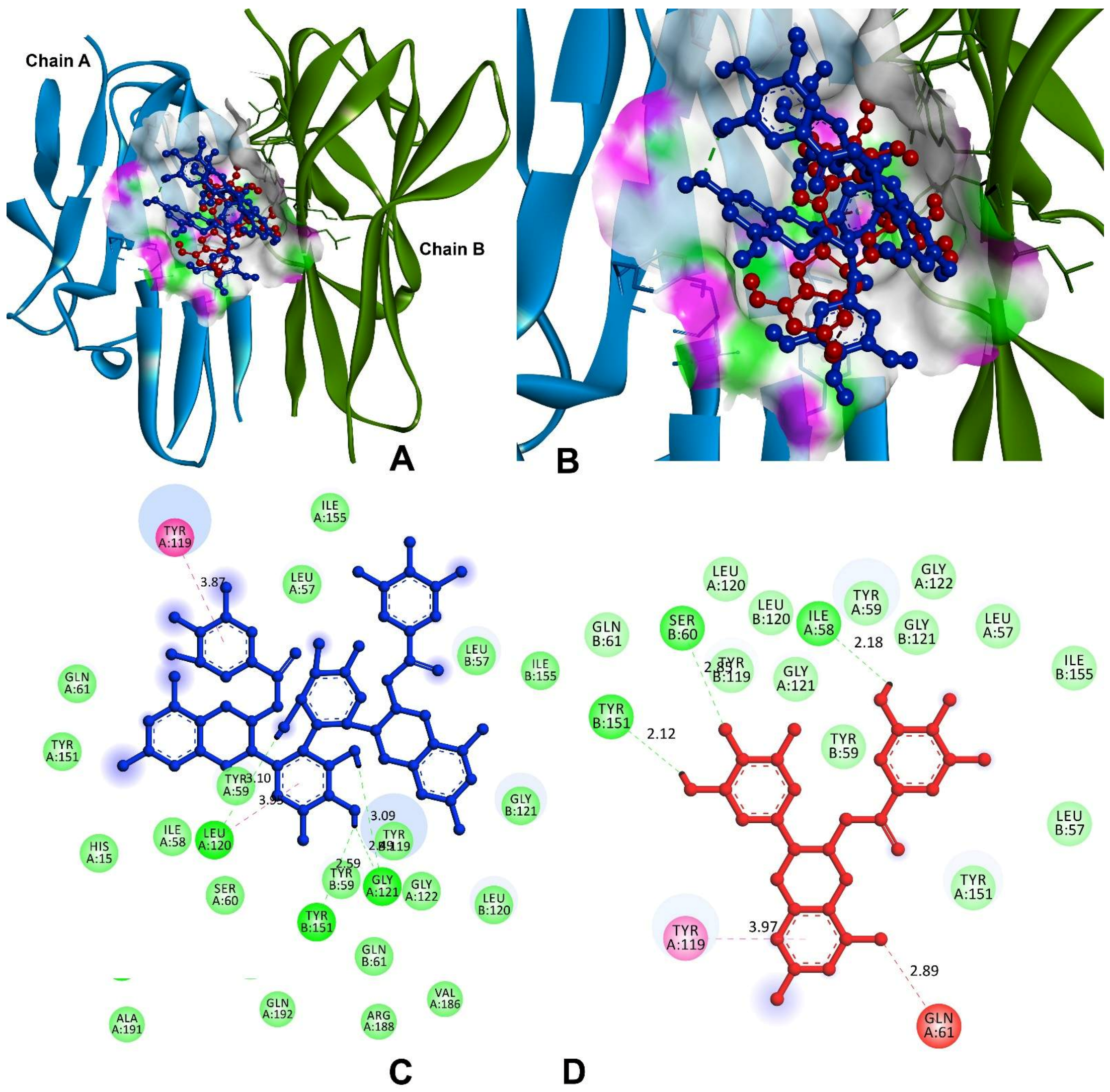
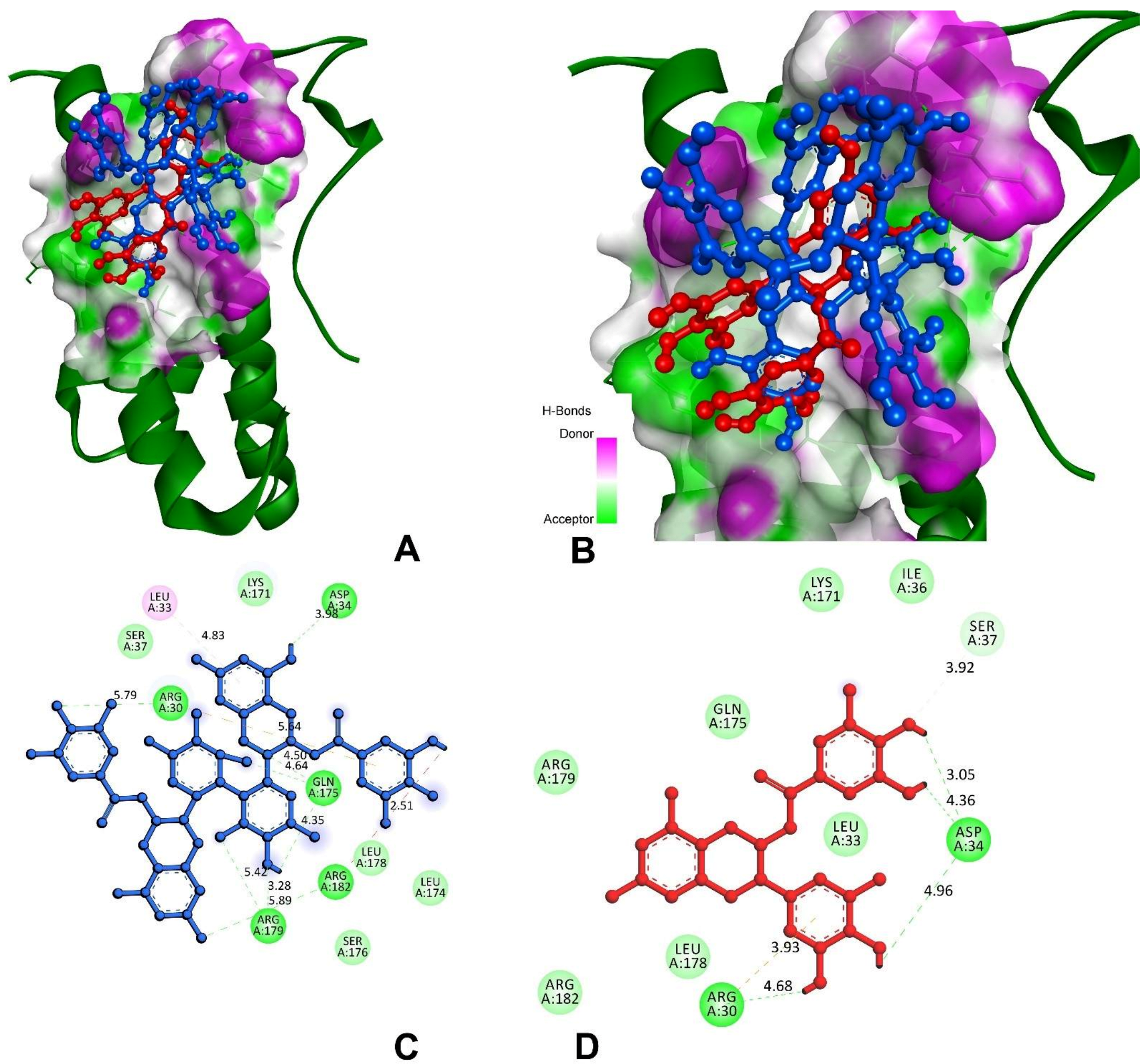
3.9.1. Anti-Inflammatory and Pro-Inflammatory Analysis
3.9.2. Inhibitor against Main Protease of SARS-CoV-2
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yahfoufi, N.; Alsadi, N.; Jambi, M.; Matar, C. The Immunomodulatory and Anti-Inflammatory Role of Polyphenols. Nutrients 2018, 10, 1618. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tekin, I.; Marotta, F. Polyphenols and Immune System. In Polyphenols: Prevention and Treatment of Human Disease, 2nd ed.; Academic Press: Cambridge, MA, USA, 2018; pp. 263–276. [Google Scholar]
- Zhang, X.; Li, J.; Li, Y.; Liu, Z.; Lin, Y.; Huang, J. Anti-melanogenic effects of epigallocatechin-3-gallate (EGCG), epicatechin-3-gallate (ECG) and gallocatechin-3-gallate (GCG) via down-regulation of cAMP/CREB/MITF signaling pathway in B16F10 melanoma cells. Fitoterapia 2020, 145, 104634. [Google Scholar] [CrossRef] [PubMed]
- Du, G.-J.; Zhang, Z.; Wen, X.-D.; Yu, C.; Calway, T.; Yuan, C.-S.; Wang, C.-Z. Epigallocatechin Gallate (EGCG) Is the Most Effective Cancer Chemopreventive Polyphenol in Green Tea. Nutrients 2012, 4, 1679–1691. [Google Scholar] [CrossRef] [PubMed]
- Srividhya, R.; Jyothilakshmi, V.; Arulmathi, K.; Senthilkumaran, V.; Kalaiselvi, P. Attenuation of senescence-induced oxidative exacerbations in aged rat brain by (−)-epigallocatechin-3-gallate. Int. J. Dev. Neurosci. 2008, 26, 217–223. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Han, Y.; Hu, Y.; Wu, X.; Wang, Y.; Zhang, X.; Fu, J.; Zou, X.; Zhang, J.; Chen, X.; et al. Targeting HO-1 by epigallocatechin-3-gallate reduces contrast-induced renal injury via anti-oxidative stress and anti-inflammation pathways. PLoS ONE 2016, 1, e0149032. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.-M.; An, J. Cytokines, inflammation and pain. Int. Anesthesiol. Clin. 2007, 45, 27–37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tanaka, T.; Narazaki, M.; Kishimoto, T. IL-6 en la inflamación, la inmunidad, y la enfermedad. Cold Spring Harb. Perspect. Biol. 2014, 10, a016295. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Wang, W.; Zhang, J.; Cui, H.; Ni, D.; Jiang, H. Dual effects of ascorbic acid on the stability of EGCG by the oxidation product dehydroascorbic acid promoting the oxidation and inhibiting the hydrolysis pathway. Food Chem. 2020, 337, 127639. [Google Scholar] [CrossRef]
- Wei, Y.; Chen, P.; Ling, T.; Wang, Y.; Dong, R.; Zhang, C.; Zhang, L.; Han, M.; Wang, D.; Wan, X.; et al. Certain (−)-epigallocatechin-3-gallate (EGCG) auto-oxidation products (EAOPs) retain the cytotoxic activities of EGCG. Food Chem. 2016, 204, 218–226. [Google Scholar] [CrossRef]
- Tanaka, T.; Watarumi, S.; Matsuo, Y.; Kamei, M.; Koun, I. Production of theasinensins A and D, epigallocatechin gallate dimers of black tea, by oxidation–reduction dismutation of dehydrotheasinensin A. Tetrahedron 2003, 59, 7939–7947. [Google Scholar] [CrossRef]
- Weerawatanakorn, M.; Hung, W.-L.; Pan, M.-H.; Li, S.; Li, D.; Wan, X.; Ho, C.-T. Chemistry and health beneficial effects of oolong tea and theasinensins. Food Sci. Hum. Wellness 2015, 4, 133–146. [Google Scholar] [CrossRef] [Green Version]
- Hashimoto, M.; Ono, C.; Masuoka, C.; Ito, Y.; Sakata, Y.; Shimizu, K.; Nonaka, G.-I.; Nishioka, I.; Nohara, T. Evaluation of the antioxidative effect (in vitro) of tea polyphenols. Biosci. Biotechnol. Biochem. 2003, 67, 396–401. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Qin, S.; Xiao, J.; Tanigawa, S.; Uto, T.; Hashimoto, F.; Fujii, M.; Hou, D.X. A genome-wide microarray highlights the antiinflammatory genes targeted by oolong tea theasinensin A in macrophages. Nutr. Cancer 2011, 63, 1064–1073. [Google Scholar] [CrossRef]
- Hisanaga, A.; Ishida, H.; Sakao, K.; Sogo, T.; Kumamoto, T.; Hashimoto, F.; Hou, D.-X. Anti-inflammatory activity and molecular mechanism of Oolong tea theasinensin. Food Funct. 2014, 5, 1891–1897. [Google Scholar] [CrossRef]
- Jang, M.; Park, R.; Park, Y.-I.; Cha, Y.-E.; Yamamoto, A.; Lee, J.I.; Park, J. EGCG, a green tea polyphenol, inhibits human coronavirus replication in vitro. Biochem. Biophys. Res. Commun. 2021, 547, 23–28. [Google Scholar] [CrossRef]
- Tallei, T.E.; Tumilaar, S.G.; Niode, N.J.; Fatimawali; Kepel, B.J.; Idroes, R.; Effendi, Y.; Alam Sakib, S.; Bin Emran, T. Potential of Plant Bioactive Compounds as SARS-CoV-2 Main Protease (Mpro) and Spike (S) Glycoprotein Inhibitors: A Molecular Docking Study. Scientifica 2020, 2020, 6307457. [Google Scholar] [CrossRef]
- Norouzi, M.; Norouzi, S.; Ruggiero, A.; Khan, M.; Myers, S.; Kavanagh, K.; Vemuri, R. Type-2 Diabetes as a Risk Factor for Severe COVID-19 Infection. Microorganisms 2021, 9, 1211. [Google Scholar] [CrossRef]
- Zhu, N.; Huang, T.C.; Yu, Y.; LaVoie, E.J.; Yang, C.S.; Ho, C.T. Identification of oxidation products of (−)-epigallocatechin gallate and (−)-epigallocatechin with H2O2. J. Agric. Food Chem. 2000, 48, 979–981. [Google Scholar] [CrossRef]
- Vigers, G.P.A.; Anderson, L.J.; Caffes, P.; Brandhuber, B.J. Crystal structure of the type-I interleukin-1 receptor complexed with interleukin-1β. Nature 1997, 386, 190–194. [Google Scholar] [CrossRef]
- Vigers, G.P.A.; Dripps, D.J.; Edwards, C.K.; Brandhuber, B.J. X-ray Crystal Structure of a Small Antagonist Peptide Bound to Interleukin-1 Receptor Type 1. J. Biol. Chem. 2000, 275, 36927–36933. [Google Scholar] [CrossRef] [Green Version]
- Somers, W.; Stahl, M.; Seehra, J.S. 1.9 Å crystal structure of interleukin 6: Implications for a novel mode of receptor dimerization and signaling. EMBO J. 1997, 16, 989–997. [Google Scholar] [CrossRef] [Green Version]
- Eck, M.J.; Sprang, S.R. The structure of tumor necrosis factor-α at 2.6 Å resolution: Implications for receptor binding. J. Biol. Chem. 1989, 264, 17595–17605. [Google Scholar] [CrossRef]
- Jin, Z.; Du, X.; Xu, Y.; Deng, Y.; Liu, M.; Zhao, Y.; Zhang, B.; Li, X.; Zhang, L.; Peng, C.; et al. Structure of M pro from SARS-CoV-2 and discovery of its inhibitors. Nature 2020, 582, 289–293. [Google Scholar] [CrossRef] [Green Version]
- Sliwoski, G.; Kothiwale, S.; Meiler, J.; Lowe, E.W. Computational methods in drug discovery. Pharmacol. Rev. 2014, 66, 334–395. [Google Scholar] [CrossRef] [Green Version]
- Khandare, A.L.; Validandi, V.; Manne, M.; Reddy, G.B.; Putcha, U.K.; Gourineni, S.R.; Nagalla, B. Tamarind fruit extract ameliorates fluoride toxicity by upregulating carbonic anhydrase II: A mechanistic study. Fluoride 2018, 51, 137–152. [Google Scholar]
- Munikumar, M.; Natarajan, P.; Amineni, U.; Krishna, K.R. Discovery of potential lumazine synthase antagonists for pathogens involved in bacterial meningitis: In silico study. Inf. Med. Unlocked 2019, 15, 100187. [Google Scholar] [CrossRef]
- Naik, V.R.; Munikumar, M.; Ramakrishna, U.; Srujana, M.; Goudar, G.; Naresh, P.; Kumar, B.N.; Hemalatha, R. Remdesivir (GS-5734) as a therapeutic option of 2019-nCOV main protease–in silico approach. J. Biomol. Struct. Dyn. 2021, 39, 4701–4714. [Google Scholar] [CrossRef]
- Manne, M.; Goudar, G.; Varikasuvu, S.R.; Khetagoudar, M.C.; Kanipakam, H.; Natarajan, P.; Ummiti, D.M.; Yenagi, V.A.; Chinthakindi, S.; Dharani, P.; et al. Cordifolioside: Potent inhibitor against Mpro of SARS-CoV-2 and immunomodulatory through human TGF-β and TNF-α. 3 Biotech 2021, 11, 136. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [Green Version]
- Azam, S.S.; Abbasi, S.W. Molecular docking studies for the identification of novel melatoninergic inhibitors for acetylserotonin-O-methyltransferase using different docking routines. Theor. Biol. Med. Model. 2013, 10, 63. [Google Scholar] [CrossRef] [Green Version]
- Ram, T.S.; Munikumar, M.; Raju, V.N.; Devaraj, P.; Boiroju, N.K.; Hemalatha, R.; Prasad, P.V.V.; Gundeti, M.; Sisodia, B.S.; Pawar, S.; et al. In silico evaluation of the compounds of the ayurvedic drug, AYUSH-64, for the action against the SARS-CoV-2 main protease. J. Ayurveda Integr. Med. 2022, 13, 100413. [Google Scholar] [CrossRef] [PubMed]
- Pradeepkiran, J.A.; Munikumar, M.; Hema, K.; Natarajan, P.; Sainath, S. Molecular docking and dynamics simulations of novel drug targets. In Brucella Melitensis; Academic Press: Cambridge, MA, USA, 2021; pp. 79–131. [Google Scholar]
- Munikumar, M.; Krishna, V.S.; Reddy, V.S.; Rajeswari, B.; Sriram, D.; Rao, M.V. In silico design of small peptides antagonist against leptin receptor for the treatment of obesity and its associated immune-mediated diseases. J. Mol. Graph. Model. 2018, 82, 20–36. [Google Scholar] [CrossRef] [PubMed]
- Halim, S.A.; Jawad, M.; Ilyas, M.; Mir, Z.; Mirza, A.A.; Husnain, T. In silico identification of novel IL-1β inhibitors to target protein–protein interfaces. Comput. Biol. Chem. 2015, 58, 158–166. [Google Scholar] [CrossRef] [PubMed]
- He, M.M.; Smith, A.S.; Oslob, J.D.; Flanagan, W.M.; Braisted, A.C.; Whitty, A.; Cancilla, M.T.; Wang, J.; Lugovskoy, A.A.; Yoburn, J.C.; et al. Small-Molecule Inhibition of TNF-α. Science 2005, 310, 1022–1025. [Google Scholar] [CrossRef]
- Wang, F.; Chen, C.; Liu, X.; Yang, K.; Xu, X.; Yang, H. Crystal structure of feline infectious peritonitis virus main protease in complex with synergetic dual inhibitors. J. Virol. 2016, 90, 1910–1917. [Google Scholar] [CrossRef] [Green Version]
- Yang, H.; Bartlam, M.; Rao, Z. Drug design targeting the main protease, the Achilles’ heel of coronaviruses. Curr. Pharm. Des. 2006, 12, 4573–4590. [Google Scholar] [CrossRef]
- Silva, A.M.; Martins-Gomes, C.; Fangueiro, J.F.; Andreani, T.; Souto, E.B. Comparison of antiproliferative effect of epigallocatechin gallate when loaded into cationic solid lipid nanoparticles against different cell lines. Pharm. Dev. Technol. 2019, 24, 1243–1249. [Google Scholar] [CrossRef] [Green Version]
- Kang, H.W. Inhibitory Effect of Cancer Cells Proliferation from Epigallocatechin-3-O-gallate. J. Food Nutr. Res. 2015, 3, 281–284. [Google Scholar] [CrossRef] [Green Version]
- Shen, X.; Zhang, Y.; Feng, Y.; Zhang, L.; Li, J.; Xie, Y.-A.; Luo, X. Epigallocatechin-3-gallate inhibits cell growth, induces apoptosis and causes S phase arrest in hepatocellular carcinoma by suppressing the AKT pathway. Int. J. Oncol. 2014, 44, 791–796. [Google Scholar] [CrossRef] [Green Version]
- Valcic, S.; Burr, J.A.; Timmermann, B.N.; Liebler, D.C. Antioxidant Chemistry of Green Tea Catechins. New Oxidation Products of (−)-Epigallocatechin Gallate and (−)-Epigallocatechin from Their Reactions with Peroxyl Radicals. Chem. Res. Toxicol. 2000, 13, 801–810. [Google Scholar] [CrossRef]
- Kim, H.-S.; Quon, M.; Kim, J.-A. New insights into the mechanisms of polyphenols beyond antioxidant properties; lessons from the green tea polyphenol, epigallocatechin 3-gallate. Redox Biol. 2014, 2, 187–195. [Google Scholar] [CrossRef] [Green Version]
- Diaz, P.; Jeong, S.C.; Lee, S.; Khoo, C.; Koyyalamudi, S.R. Antioxidant and anti-inflammatory activities of selected medicinal plants and fungi containing phenolic and flavonoid compounds. Chin. Med. 2012, 7, 26. [Google Scholar] [CrossRef] [Green Version]
- Yashin, A.; Yashin, Y.; Nemzer, B. Determination of Antioxidant Activity in Tea Extracts, and Their Total Antioxidant Content. Am. J. Biomed. Sci. 2011. Available online: http://www.nwpii.com/ajbms/papers/AJBMS_2011_4_09.pdf (accessed on 18 October 2021).
- Severino, J.F.; Goodman, B.A.; Kay, C.W.; Stolze, K.; Tunega, D.; Reichenauer, T.G.; Pirker, K.F. Free radicals generated during oxidation of green tea polyphenols: Electron paramagnetic resonance spectroscopy combined with density functional theory calculations. Free Radic. Biol. Med. 2009, 46, 1076–1088. [Google Scholar] [CrossRef]
- Sang, S.; Lambert, J.D.; Ho, C.-T.; Yang, C.S. The chemistry and biotransformation of tea constituents. Pharmacol. Res. 2011, 64, 87–99. [Google Scholar] [CrossRef]
- Saffari, Y.; Sadrzadeh, S. Green tea metabolite EGCG protects membranes against oxidative damage in vitro. Life Sci. 2004, 74, 1513–1518. [Google Scholar] [CrossRef]
- Huang, C.-C.; Fang, J.-Y.; Wu, W.-B.; Chiang, H.-S.; Wei, Y.-J.; Hung, C.-F. Protective effects of (−)-epicatechin-3-gallate on UVA-induced damage in HaCaT keratinocytes. Arch. Dermatol. Res. 2005, 296, 473–481. [Google Scholar] [CrossRef]
- Glei, M.; Pool-Zobel, B. The main catechin of green tea, (−)-epigallocatechin-3-gallate (EGCG), reduces bleomycin-induced DNA damage in human leucocytes. Toxicol. In Vitro 2006, 20, 295–300. [Google Scholar] [CrossRef]
- Cao, T.; Zhang, X.; Yang, D.; Wang, Y.-Q.; Qiao, Z.-D.; Huang, J.-M.; Zhang, P. Antioxidant effects of epigallocatechin-3-gallate on the aTC1-6 pancreatic alpha cell line. Biochem. Biophys. Res. Commun. 2017, 495, 693–699. [Google Scholar] [CrossRef]
- Luo, K.-W.; Xia, J.; Cheng, B.-H.; Gao, H.-C.; Fu, L.-W.; Luo, X.-L. Tea polyphenol EGCG inhibited colorectal-cancer-cell proliferation and migration via downregulation of STAT3. Gastroenterol. Rep. 2020, 9, 59–70. [Google Scholar] [CrossRef]
- Huang, S.-C.; Kao, Y.-H.; Shih, S.-F.; Tsai, M.-C.; Lin, C.-S.; Chen, L.W.; Chuang, Y.-P.; Tsui, P.-F.; Ho, L.-J.; Lai, J.-H.; et al. Epigallocatechin-3-gallate exhibits immunomodulatory effects in human primary T cells. Biochem. Biophys. Res. Commun. 2021, 550, 70–76. [Google Scholar] [CrossRef]
- Lambert, J.D.; Elias, R.J. The antioxidant and pro-oxidant activities of green tea polyphenols: A role in cancer prevention. Arch. Biochem. Biophys. 2010, 501, 65–72. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shii, T.; Miyamoto, M.; Matsuo, Y.; Tanaka, T.; Kouno, I. Biomimetic One-Pot Preparation of a Black Tea Polyphenol Theasinensin A from Epigallocatechin Gallate by Treatment with Copper(II) Chloride and Ascorbic Acid. Chem. Pharm. Bull. 2011, 59, 1183–1185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hong, J.; Lu, H.; Meng, X.; Ryu, J.-H.; Hara, Y.; Yang, C.S. Stability, cellular uptake, biotransformation, and efflux of tea polyphenol (−)-epigallocatechin-3-gallate in HT-29 human colon adenocarcinoma cells. Cancer Res. 2002, 62, 7241–7246. [Google Scholar] [PubMed]
- Hou, D.X.; Masuzaki, S.; Tanigawa, S.; Hashimoto, F.; Chen, J.; Sogo, T.; Fujii, M. Oolong tea theasinensins attenuate cyclooxygenase-2 expression in lipopolysaccharide (LPS)-activated mouse macrophages: Structure–activity relationship and molecular mechanisms. J. Agric. Food Chem. 2010, 58, 12735–12743. [Google Scholar] [CrossRef]
- Battista, E.; Scognamiglio, P.L.; Di Luise, N.; Raucci, U.; Donati, G.; Rega, N.; Netti, P.A.; Causa, F. Turn-on fluorescence detection of protein by molecularly imprinted hydrogels based on supramolecular assembly of peptide multi-functional blocks. J. Mater. Chem. B 2018, 6, 1207–1215. [Google Scholar] [CrossRef] [Green Version]
- Platella, C.; Raucci, U.; Rega, N.; D’Atri, S.; Levati, L.; Roviello, G.N.; Fuggetta, M.P.; Musumeci, D.; Montesarchio, D. Shedding light on the interaction of polydatin and resveratrol with G-quadruplex and duplex DNA: A biophysical, computational and biological approach. Int. J. Biol. Macromol. 2020, 151, 1163–1172. [Google Scholar] [CrossRef]
- Wahedi, H.M.; Ahmad, S.; Abbasi, S.W. Stilbene-based natural compounds as promising drug candidates against COVID-19. J. Biomol. Struct. Dyn. 2021, 39, 3225–3234. [Google Scholar] [CrossRef]
- Perrella, F.; Coppola, F.; Petrone, A.; Platella, C.; Montesarchio, D.; Stringaro, A.; Ravagnan, G.; Fuggetta, M.; Rega, N.; Musumeci, D. Interference of Polydatin/Resveratrol in the ACE2:Spike Recognition during COVID-19 Infection. A Focus on Their Potential Mechanism of Action through Computational and Biochemical Assays. Biomolecules 2021, 11, 1048. [Google Scholar] [CrossRef]



| Sample (µM) | EGCG | O-EGCG |
|---|---|---|
| % Viability | ||
| 10 | 85.55 ± 0.01 | 77.37 ± 0.10 |
| 20 | 67.97 ± 0.05 | 74.03 ± 0.17 |
| 30 | 65.36 ± 0.08 | 71.82 ± 0.15 |
| 40 | 51.52 ± 0.08 | 48.80 ± 0.11 |
| 50 | 46.74 ± 0.05 | 44.23 ± 0.22 |
| 60 | 34.51 ± 0.12 | 38.73 ± 0.13 |
| 70 | 33.25 ± 0.10 | 30.68 ± 0.09 |
| 80 | 10.42 ± 0.04 | 23.07 ± 0.11 |
| 90 | 4.99 ± 0.04 | 6.09 ± 0.03 |
| 100 | 0.91 ± 0.04 | −1.15 ± 0.05 |
| Trolox (µM) | Absorbance | Sample | Absorbance | Sample | Antioxidant (µM) |
|---|---|---|---|---|---|
| 1.32 | 0.7418 | Control | 0.6526 | Control | 19.91033 |
| 0.66 | 0.6737 | EGCG 10 | 0.6971 | S 10 | 19.95483 |
| 0.33 | 0.6309 | EGCG 25 | 0.7184 | S 25 | 19.97613 |
| 0.18 | 0.5727 | EGCG 50 | 0.7505 | S 50 | 20.00823 |
| 0.09 | 0.5835 | O-EGCG 10 | 0.6895 | P 10 | 19.94723 |
| 0.045 | 0.5709 | O-EGCG25 | 0.7003 | P 25 | 19.95803 |
| Blank | 0.596 | O-EGCG 50 | 0.782 | P 50 | 20.03973 |
| Standard Concentration | Absorbance | Sample | Average Absorbance | Standard Deviation |
|---|---|---|---|---|
| 0 | 0 | Control | 0.249 | 0.018 |
| 0.5 | 0.017 | EGCG 10 | 0.256 | 0.023 |
| 1 | 0.032 | EGCG 25 | 0.355 | 0.016 |
| 2 | 0.071 | EGCG 50 | 0.355 | 0.015 |
| 4 | 0.141 | O-EGCG 10 | 0.277 | 0.019 |
| 6 | 0.212 | O-EGCG25 | 0.299 | 0.024 |
| O-EGCG 50 | 0.425 | 0.040 | ||
| Sample | IL-1β | IL-6 | TNF-α |
|---|---|---|---|
| Absorbance ± SD | |||
| Control | 0.053 ± 0.007 | 0.135 ± 0.010 | 0.025 ± 0.001 |
| EGCG 10 | 0.022 ± 0.005 | 0.109 ± 0.003 | 0.009 ± 0.003 |
| EGCG 25 | 0.017 ± 0.003 | 0.104 ± 0.004 | 0.007 ± 0.001 |
| EGCG 50 | 0.005 ± 0.001 | 0.091 ± 0.006 | 0.004 ± 0.002 |
| O-EGCG 10 | 0.012 ± 0.001 | 0.095 ± 0.006 | 0.009 ± 0.004 |
| O-EGCG25 | 0.007 ± 0.003 | 0.087 ± 0.002 | 0.007 ± 0.001 |
| O-EGCG 50 | 0.002 ± 0.001 | 0.079 ± 0.009 | 0.005 ± 0.001 |
| Compound (PubChem ID) | Docking Score (kcal/mol) | H-Bonds | Bond Length (Å) |
|---|---|---|---|
| Interleukin-1 (IL-1) | |||
| Theasinensin A | −8.9 | H80-A:SER13:O | 4.27 |
| A: SER13:HG-N:O25 | 2.91 | ||
| H41-A:ASN107:OD1 | 4.88 | ||
| H39-A:GLN126:OE1 | 5.57 | ||
| H39-A:ASP145:OD1 | 4.78 | ||
| B:ARG163:HH11-N:O36 | 4.93 | ||
| B:ARG163:CD-N:O34 | 4.91 | ||
| EGCG | −8.3 | H34-A:GLU105:OE2 | 2.37 |
| H34-A:ASN108:O | 2.45 | ||
| H37-A:MET148:O | 2.25 | ||
| A:GLN149:HE21-N:O1 | 2.64 | ||
| H-A:PHE150:O41 | 1.96 | ||
| C2-B:ASN204:OD1 | 2.95 | ||
| B:ASN204:HD21-π | 2.56 | ||
| Tumour necrosis factor-alpha (TNF-α) | |||
| Theasinensin A | −8.4 | A:TYR119-π | 3.87 |
| H77-A:LEU120:O | 3.10 | ||
| A:LEU120:CO-π | 3.95 | ||
| H45-A:GLY121:O | 2.49 | ||
| H46-A:GLY121:O | 3.09 | ||
| H45-B:TYR151:OH | 2.59 | ||
| EGCG | −7.2 | H20-A:ILE58:O | 2.18 |
| A:GLN61:OE1-NO38 | 2.89 | ||
| A:TYR119-π | 3.97 | ||
| B:SER60:HN-O34 | 2.85 | ||
| H37-B:TYR151:OH | 2.12 | ||
| Interleukin-6 (IL-6) | |||
| Theasinensin A | −7.2 | A:ARG30:HH11-O69 | 5.79 |
| H39-A:ASP34:OD1 | 3.98 | ||
| A:GLN175:HE21-O77 | 4.64 | ||
| A:GLN175:HE22-O14 | 4.50 | ||
| H45-A:GLN175:O | 4.35 | ||
| A:ARG179:HE-O46 | 5.42 | ||
| A:ARG179:HH21-O44 | 3.28 | ||
| A:ARG182:HH22-O75 | 5.89 | ||
| A:ARG182:HH22-H33 | 2.51 | ||
| EGCG | −6.4 | A:ARG30:HH11-π | 3.92 |
| H33-A:ARG30:O | 4.68 | ||
| H35-A:ASP34:OD1 | 4.96 | ||
| H25-A:ASP34:OD1 | 4.36 | ||
| H23-A:ASP34:OD1 | 3.05 | ||
| A:SER37:CB-O22 | 3.14 | ||
| Main protease (Mpro) of SARS-CoV-2 | |||
| Theasinensin A | −8.70 | A:HIS41:HE2-O60 | 4.64 |
| A:MET49:SD-π | 7.51 | ||
| H80-A:PHE140:O | 4.90 | ||
| H82-A:LEU141:O | 6.12 | ||
| A:ASN142:HD21-O36 | 5.25 | ||
| A:CYS145:SG-O58 | 3.65 | ||
| A:CYS145:SG-π | 5.37 | ||
| A:HIS163:HE2-O81 | 2.15 | ||
| H76-A:MET165:SD | 2.44 | ||
| EGCG | −7.77 | H41-A:THR190:O | 2.64 |
| H34-A:LEU141:O | 2.25 | ||
| A:ASN142:HN-O22 | 2.84 | ||
| H37-A:ASN142:OD1 | 2.50 | ||
| A:MET165:SD-π | 5.13 | ||
| A:GLU166:OE1-π | 3.99 | ||
| H41-A:ARG188:O | 2.18 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ungarala, R.; Munikumar, M.; Sinha, S.N.; Kumar, D.; Sunder, R.S.; Challa, S. Assessment of Antioxidant, Immunomodulatory Activity of Oxidised Epigallocatechin-3-Gallate (Green Tea Polyphenol) and Its Action on the Main Protease of SARS-CoV-2—An In Vitro and In Silico Approach. Antioxidants 2022, 11, 294. https://doi.org/10.3390/antiox11020294
Ungarala R, Munikumar M, Sinha SN, Kumar D, Sunder RS, Challa S. Assessment of Antioxidant, Immunomodulatory Activity of Oxidised Epigallocatechin-3-Gallate (Green Tea Polyphenol) and Its Action on the Main Protease of SARS-CoV-2—An In Vitro and In Silico Approach. Antioxidants. 2022; 11(2):294. https://doi.org/10.3390/antiox11020294
Chicago/Turabian StyleUngarala, Ramakrishna, Manne Munikumar, Sukesh Narayan Sinha, Dileshwar Kumar, R. Shyam Sunder, and Suresh Challa. 2022. "Assessment of Antioxidant, Immunomodulatory Activity of Oxidised Epigallocatechin-3-Gallate (Green Tea Polyphenol) and Its Action on the Main Protease of SARS-CoV-2—An In Vitro and In Silico Approach" Antioxidants 11, no. 2: 294. https://doi.org/10.3390/antiox11020294
APA StyleUngarala, R., Munikumar, M., Sinha, S. N., Kumar, D., Sunder, R. S., & Challa, S. (2022). Assessment of Antioxidant, Immunomodulatory Activity of Oxidised Epigallocatechin-3-Gallate (Green Tea Polyphenol) and Its Action on the Main Protease of SARS-CoV-2—An In Vitro and In Silico Approach. Antioxidants, 11(2), 294. https://doi.org/10.3390/antiox11020294






