Astaxanthin Induces Transcriptomic Responses Associated with Lifespan Extension in Caenorhabditis elegans
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Nematode Strains
2.3. Lifespan Assay
2.4. Sample Preparation
2.5. RNA-seq
2.6. Analyses of RNA-seq Datasets
2.7. PPI network Construction
2.8. miRNA Target Prediction
2.9. Oil Red O Staining
2.10. Quantitative RT-PCR Analyses
2.11. Statistical Analysis
3. Results
3.1. Astaxanthin Extends the Lifespan of C. elegans
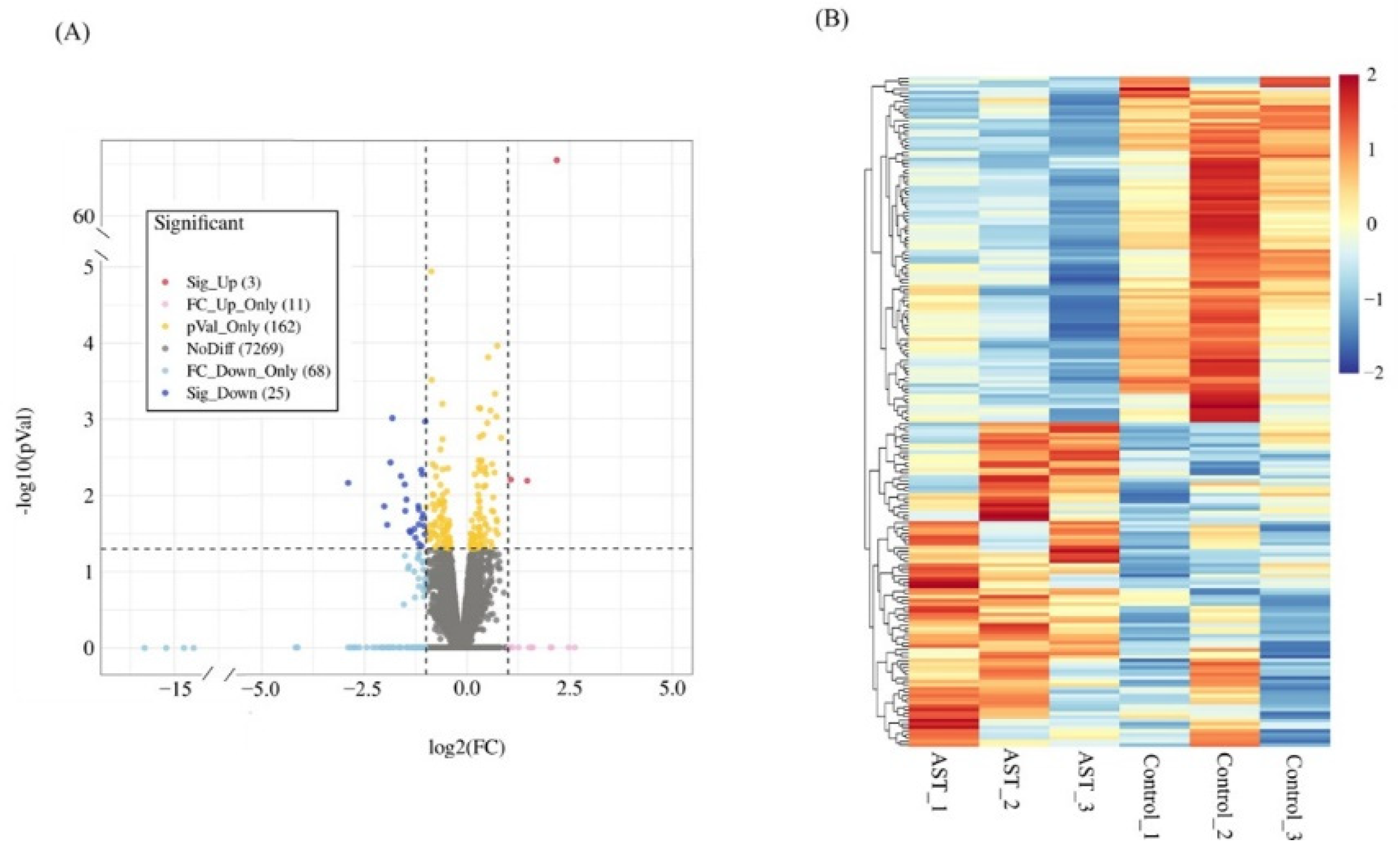
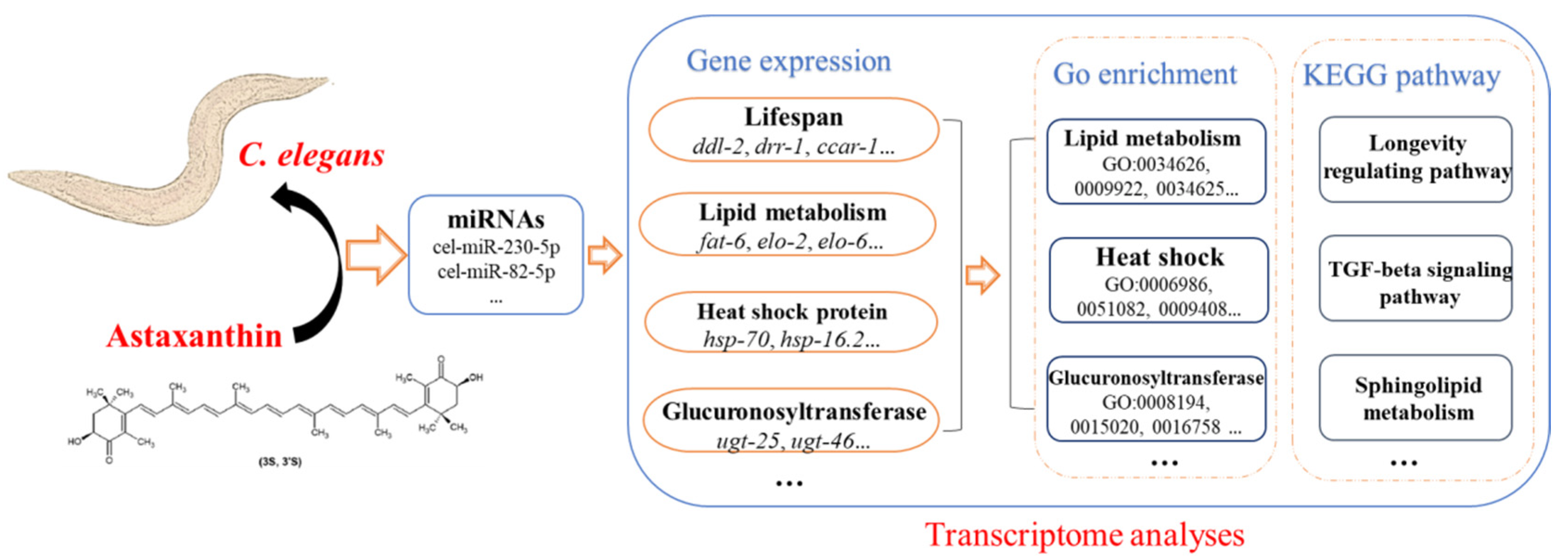
3.2. Differentially Expressed Genes Affected by Astaxanthin in C. elegans
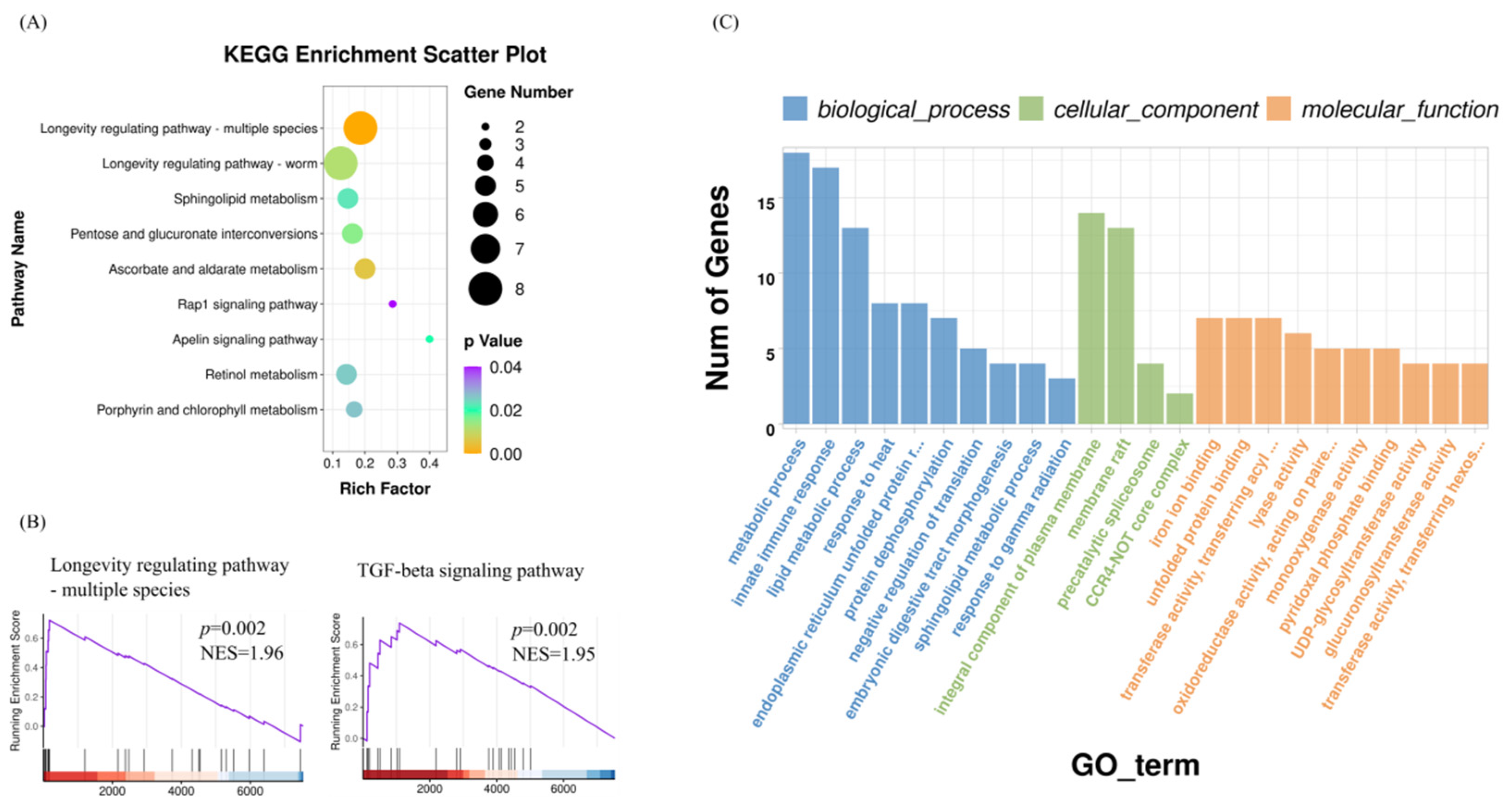
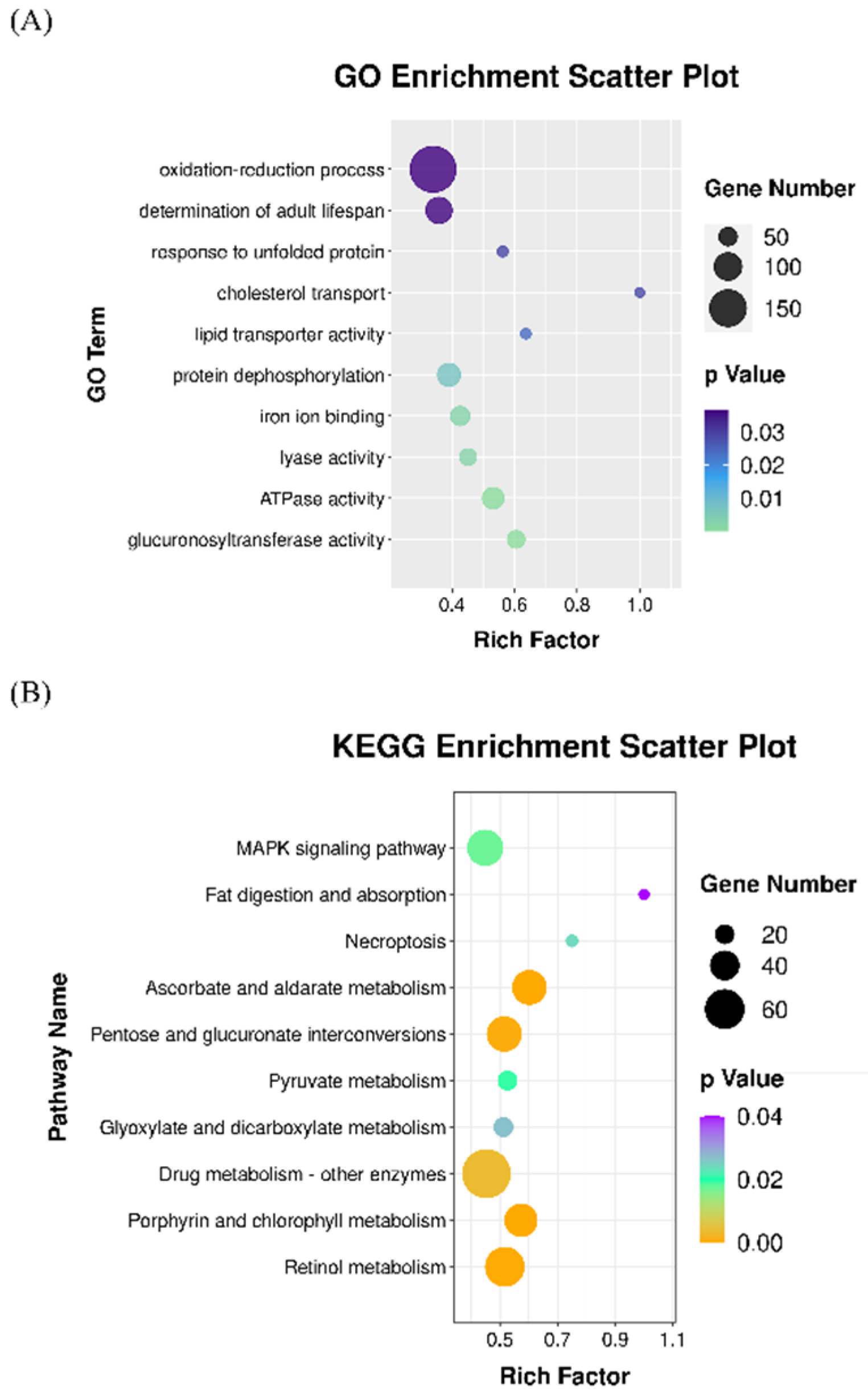
3.3. Enrichments of GO Terms and KEGG Pathways in the Transcriptomes of Astaxanthin-Treated C. elegans
3.4. PPI Network Analyses of DEGs
3.5. Differentially Expressed miRNA in C. elegans Treated with Astaxanthin
3.6. Effects of Astaxanthin on Lipid Metabolism in C. elegans
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sztretye, M.; Dienes, B.; Gönczi, M.; Czirják, T.; Csernoch, L.; Dux, L.; Szentesi, P.; Keller-Pintér, A. Astaxanthin: A potential mitochondrial-targeted antioxidant treatment in diseases and with aging. Oxid. Med. Cell. Longev. 2019, 2019, 3849692. [Google Scholar] [CrossRef] [Green Version]
- Fakhri, S.; Abbaszadeh, F.; Dargahi, L.; Jorjani, M. Astaxanthin: A mechanistic review on its biological activities and health benefits. Pharmacol. Res. 2018, 136, 1–20. [Google Scholar] [CrossRef]
- Guerin, M.; Huntley, M.E.; Olaizola, M. Haematococcus astaxanthin: Applications for human health and nutrition. Trends Biotechnol. 2003, 21, 210–216. [Google Scholar] [CrossRef]
- Yazaki, K.; Yoshikoshi, C.; Oshiro, S.; Yanase, S. Supplemental cellular protection by a carotenoid extends lifespan via Ins/IGF-1 signaling in Caenorhabditis elegans. Oxid. Med. Cell. Longev. 2011, 2011, 596240. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Luo, Q.; Cao, Y.; Goulette, T.; Liu, X.; Xiao, H. Mechanism of different stereoisomeric astaxanthin in resistance to oxidative stress in Caenorhabditis elegans. J. Food Sci. 2016, 81, H2280–H2287. [Google Scholar] [CrossRef]
- Fu, M.; Zhang, X.; Zhang, X.; Yang, L.; Luo, S.; Liu, H. Autophagy plays a role in the prolongation of the life span of Caenorhabditis elegans by astaxanthin. Rejuvenation Res. 2020, 24, 185–205. [Google Scholar] [CrossRef]
- Liu, X.; Liu, H.; Chen, Z.; Xiao, J.; Cao, Y. DAF-16 acts as the “hub” of astaxanthin’s anti-aging mechanism to improve aging-related physiological functions in Caenorhabditis elegans. Food Funct. 2021, 12, 9098–9110. [Google Scholar] [CrossRef]
- Kim, S.S.; Lee, S.V. Non-Coding RNAs in Caenorhabditis elegans aging. Mol. Cells 2019, 42, 379–385. [Google Scholar] [CrossRef]
- Chatterjee, E.; Das, S. Non-coding RNAs in cardiac remodeling: Diversity in composition and function. Curr. Opin. Physiol. 2022, 26, 100534. [Google Scholar] [CrossRef]
- Liu, C.; Rennie, W.A.; Mallick, B.; Kanoria, S.; Long, D.; Wolenc, A.; Carmack, C.S.; Ding, Y. MicroRNA binding sites in C. elegans 3’ UTRs. RNA Biol. 2014, 11, 693–701. [Google Scholar] [CrossRef]
- Catalanotto, C.; Cogoni, C.; Zardo, G. MicroRNA in control of gene expression: An overview of nuclear functions. Int. J. Mol. Sci. 2016, 17, 1712. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith-Vikos, T.; Slack, F.J. MicroRNAs and their roles in aging. J. Cell Sci. 2012, 125, 7–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boehm, M.; Slack, F.A. A developmental timing microRNA and its target regulate life span in C. elegans. Science 2005, 310, 1954–1957. [Google Scholar] [CrossRef] [Green Version]
- de Lencastre, A.; Pincus, Z.; Zhou, K.; Kato, M.; Lee, S.S.; Slack, F.J. MicroRNAs both promote and antagonize longevity in C. elegans. Curr. Biol. 2010, 20, 2159–2168. [Google Scholar] [CrossRef] [Green Version]
- Zhao, S.; Fung-Leung, W.P.; Bittner, A.; Ngo, K.; Liu, X. Comparison of RNA-Seq and microarray in transcriptome profiling of activated T cells. PLoS ONE 2014, 9, e78644. [Google Scholar] [CrossRef]
- Peng, L.; Wang, L.; Yang, Y.F.; Zou, M.M.; He, W.Y.; Wang, Y.; Wang, Q.; Vasseur, L.; You, M.S. Transcriptome profiling of the Plutella xylostella (Lepidoptera: Plutellidae) ovary reveals genes involved in oogenesis. Gene 2017, 637, 90–99. [Google Scholar] [CrossRef]
- Gong, P.; Donohue, K.B.; Mayo, A.M.; Wang, Y.; Hong, H.; Wilbanks, M.S.; Barker, N.D.; Guan, X.; Gust, K.A. Comparative toxicogenomics of three insensitive munitions constituents 2,4-dinitroanisole, nitroguanidine and nitrotriazolone in the soil nematode Caenorhabditis elegans. BMC Syst. Biol. 2018, 12, 92. [Google Scholar] [CrossRef] [Green Version]
- Yen, K.; Le, T.T.; Bansal, A.; Narasimhan, S.D.; Cheng, J.X.; Tissenbaum, H.A. A comparative study of fat storage quantitation in nematode Caenorhabditis elegans using label and label-free methods. PLoS ONE 2010, 5, e12810. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [Green Version]
- Savage-Dunn, C.; Padgett, R.W. The TGF-beta family in Caenorhabditis elegans. Cold Spring Harb. Perspect. Biol. 2017, 9, a022178. [Google Scholar] [CrossRef]
- Gems, D.; McElwee, J.J. Broad spectrum detoxification: The major longevity assurance process regulated by insulin/IGF-1 signaling? Mech. Ageing Dev. 2005, 126, 381–387. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, X.; Wang, H.; Hu, T. Identification of key genes and pathways in abdominal aortic aneurysm by integrated bioinformatics analysis. J. Int. Med. Res. 2020, 48, 300060519894437. [Google Scholar] [CrossRef] [Green Version]
- Castro, C.; Sar, F.; Shaw, W.R.; Mishima, M.; Miska, E.A.; Griffin, J.L. A metabolomic strategy defines the regulation of lipid content and global metabolism by Δ9 desaturases in Caenorhabditis elegans. BMC Genom. 2012, 13, 36. [Google Scholar] [CrossRef] [Green Version]
- Syntichaki, P.; Samara, C.; Tavernarakis, N. The vacuolar H+ -ATPase mediates intracellular acidification required for neurodegeneration in C. elegans. Curr. Biol. 2005, 15, 1249–1254. [Google Scholar] [CrossRef] [Green Version]
- Imanikia, S.; Ozbey, N.P.; Krueger, C.; Casanueva, M.O.; Taylor, R.C. Neuronal XBP-1 activates intestinal lysosomes to improve proteostasis in C. elegans. Curr. Biol. 2019, 29, 2322–2338. [Google Scholar] [CrossRef] [Green Version]
- Ren, H.; Zhang, H. Wnt signaling controls temporal identities of seam cells in Caenorhabditis elegans. Dev. Biol. 2010, 345, 144–155. [Google Scholar] [CrossRef] [Green Version]
- Hoier, E.F.; Mohler, W.A.; Kim, S.K.; Hajnal, A. The Caenorhabditis elegans APC-related gene apr-1 is required for epithelial cell migration and Hox gene expression. Genes Dev. 2000, 14, 874–886. [Google Scholar] [CrossRef]
- Huang, Y.; Gao, X.C.; Ren, H.T.; Xiong, J.L.; Sun, X.H. Characterization of conserved and novel miRNAs using deep sequencing and prediction of miRNA targets in Crucian carp (Carassius auratus). Gene 2017, 635, 61–68. [Google Scholar] [CrossRef]
- Johnson, A.A.; Stolzing, A. The role of lipid metabolism in aging, lifespan regulation, and age-related disease. Aging Cell 2019, 18, e13048. [Google Scholar] [CrossRef] [Green Version]
- Zheng, J.; Greenway, F.L. Caenorhabditis elegans as a model for obesity research. Int. J. Obes. 2012, 36, 186–194. [Google Scholar] [CrossRef]
- Srinivasan, S. Regulation of body fat in Caenorhabditis elegans. Annu. Rev. Physiol. 2015, 77, 161–178. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Zou, X.; Ding, Y.; Wang, H.; Wu, X.; Liang, B. Comparative genomics and functional study of lipid metabolic genes in Caenorhabditis elegans. BMC Genom. 2013, 14, 164. [Google Scholar] [CrossRef] [Green Version]
- Watts, J.L.; Ristow, M. Lipid and carbohydrate metabolism in Caenorhabditis elegans. Genetics 2017, 207, 413–446. [Google Scholar] [CrossRef]
- Goncalves, J.; Wan, Y.; Garcia, L.R. Stearoyl-CoA desaturases sustain cholinergic excitation and copulatory robustness in metabolically aging C. elegans males. iScience 2022, 25, 104082. [Google Scholar] [CrossRef]
- Dixit, A.; Sandhu, A.; Modi, S.; Shashikanth, M.; Koushika, S.P.; Watts, J.L.; Singh, V. Neuronal control of lipid metabolism by STR-2 G protein-coupled receptor promotes longevity in Caenorhabditis elegans. Aging Cell 2020, 19, e13160. [Google Scholar] [CrossRef]
- Mutlu, A.S.; Duffy, J.; Wang, M.C. Lipid metabolism and lipid signals in aging and longevity. Dev. Cell 2021, 56, 1394–1407. [Google Scholar] [CrossRef]
- Benedetto, A.; Gems, D. Autophagy promotes visceral aging in wild-type C. elegans. Autophagy 2019, 15, 731–732. [Google Scholar] [CrossRef] [Green Version]
- Ezcurra, M.; Benedetto, A.; Sornda, T.; Gilliat, A.F.; Au, C.; Zhang, Q.; van Schelt, S.; Petrache, A.L.; Wang, H.; de la Guardia, Y.; et al. C. elegans eats its own intestine to make yolk leading to multiple senescent pathologies. Curr. Biol. 2018, 28, 2544–2556. [Google Scholar] [CrossRef] [Green Version]
- Alper, S. Model systems to the rescue: The relationship between aging and innate immunity. Commun. Integr. Biol. 2010, 3, 409–414. [Google Scholar] [CrossRef]
- DeVeale, B.; Brummel, T.; Seroude, L. Immunity and aging: The enemy within? Aging Cell 2004, 3, 195–208. [Google Scholar] [CrossRef]
- Prithika, U.; Deepa, V.; Balamurugan, K. External induction of heat shock stimulates the immune response and longevity of Caenorhabditis elegans towards pathogen exposure. Innate Immun. 2016, 22, 466–478. [Google Scholar] [CrossRef] [Green Version]
- Hsu, A.-L.; Murphy, C.T.; Kenyon, C. Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science 2003, 300, 1142–1145. [Google Scholar] [CrossRef] [Green Version]
- Imanikia, S.; Hylands, P.; Sturzenbaum, S.R. The double mutation of cytochrome P450’s and fatty acid desaturases affect lipid regulation and longevity in C. elegans. Biochem. Biophys. Rep. 2015, 2, 172–178. [Google Scholar] [CrossRef] [Green Version]
- Cutler, R.G.; Thompson, K.W.; Camandola, S.; Mack, K.T.; Mattson, M.P. Sphingolipid metabolism regulates development and lifespan in Caenorhabditis elegans. Mech. Ageing Dev. 2014, 143–144, 9–18. [Google Scholar] [CrossRef] [Green Version]
- Horikawa, M.; Nomura, T.; Hashimoto, T.; Sakamoto, K. Elongation and desaturation of fatty acids are critical in growth, lipid metabolism and ontogeny of Caenorhabditis elegans. J. Biochem. 2008, 144, 149–158. [Google Scholar] [CrossRef]
- Brock, T.J.; Browse, J.; Watts, J.L. Genetic regulation of unsaturated fatty acid composition in C. elegans. PLoS Genet. 2006, 2, e108. [Google Scholar] [CrossRef] [Green Version]
- Yue, Y.; Shen, P.; Chang, A.L.; Qi, W.; Kim, K.H.; Kim, D.; Park, Y. trans-Trismethoxy resveratrol decreased fat accumulation dependent on fat-6 and fat-7 in Caenorhabditis elegans. Food Funct. 2019, 10, 4966–4974. [Google Scholar] [CrossRef]
- Ackerman, D.; Gems, D. The mystery of C. elegans aging: An emerging role for fat. Distant parallels between C. elegans aging and metabolic syndrome? Bioessays 2012, 34, 466–471. [Google Scholar] [CrossRef]
- Brock, T.J.; Browse, J.; Watts, J.L. Fatty acid desaturation and the regulation of adiposity in Caenorhabditis elegans. Genetics 2007, 176, 865–875. [Google Scholar] [CrossRef] [Green Version]
- Shi, X.; Li, J.; Zou, X.; Greggain, J.; Rodkaer, S.V.; Faergeman, N.J.; Liang, B.; Watts, J.L. Regulation of lipid droplet size and phospholipid composition by stearoyl-CoA desaturase. J. Lipid Res. 2013, 54, 2504–2514. [Google Scholar] [CrossRef]
- Jaeger, S.; Barends, S.; Giege, R.; Eriani, G.; Martin, F. Expression of metazoan replication-dependent histone genes. Biochimie 2005, 87, 827–834. [Google Scholar] [CrossRef]
- Prigent, C.; Dimitrov, S. Phosphorylation of serine 10 in histone H3, what for? J. Cell Sci. 2003, 116, 3677–3685. [Google Scholar] [CrossRef] [Green Version]
- Yi, S.J.; Kim, K. New Insights into the Role of Histone Changes in Aging. Int. J. Mol. Sci. 2020, 21, 8241. [Google Scholar] [CrossRef]
- Bannister, A.J.; Kouzarides, T. Regulation of chromatin by histone modifications. Cell Res. 2011, 21, 381–395. [Google Scholar] [CrossRef]
- Molina-Serrano, D.; Kyriakou, D.; Kirmizis, A. Histone modifications as an intersection between diet and longevity. Front. Genet. 2019, 10, 192. [Google Scholar] [CrossRef]
- Huang, B.; Zhong, D.; Zhu, J.; An, Y.; Gao, M.; Zhu, S.; Dang, W.; Wang, X.; Yang, B.; Xie, Z. Inhibition of histone acetyltransferase GCN5 extends lifespan in both yeast and human cell lines. Aging Cell 2020, 19, e13129. [Google Scholar] [CrossRef] [Green Version]
- Subasic, D.; Brümmer, A.; Wu, Y.; Pinto, S.M.; Imig, J.; Keller, M.; Jovanovic, M.; Lightfoot, H.L.; Nasso, S.; Götze, S.; et al. Cooperative target mRNA destabilization and translation inhibition by miR-58 microRNA family in C. elegans. Genome Res. 2015, 25, 1680–1691. [Google Scholar] [CrossRef] [Green Version]
- Lucanic, M.; Graham, J.; Scott, G.; Bhaumik, D.; Benz, C.C.; Hubbard, A.; Lithgow, G.J.; Melov, S. Age-related micro-RNA abundance in individual C. elegans. Aging 2013, 5, 394–411. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Zhang, W.; Dong, M. The miR-58 microRNA family is regulated by insulin signaling and contributes to lifespan regulation in Caenorhabditis elegans. Sci. China Life Sci. 2018, 61, 1060–1070. [Google Scholar] [CrossRef]
- Kato, M.; Chen, X.; Inukai, S.; Zhao, H.; Slack, F.J. Age-associated changes in expression of small, noncoding RNAs, including microRNAs, in C. elegans. RNA 2011, 17, 1804–1820. [Google Scholar] [CrossRef]
- Ke, J.P.; Yu, J.Y.; Gao, B.; Hu, F.L.; Xu, F.Q.; Yao, G.; Bao, G.H. Two new catechins from Zijuan green tea enhance the fitness and lifespan of Caenorhabditis elegans via insulin-like signaling pathways. Food Funct. 2022, 13, 9299–9310. [Google Scholar] [CrossRef]
- Chen, H.; Li, R.; Zhao, F.; Luan, L.; Han, T.; Li, Z. Betulinic acid increases lifespan and stress resistance via insulin/IGF-1 signaling pathway in Caenorhabditis elegans. Front. Nutr. 2022, 9, 960239. [Google Scholar] [CrossRef]
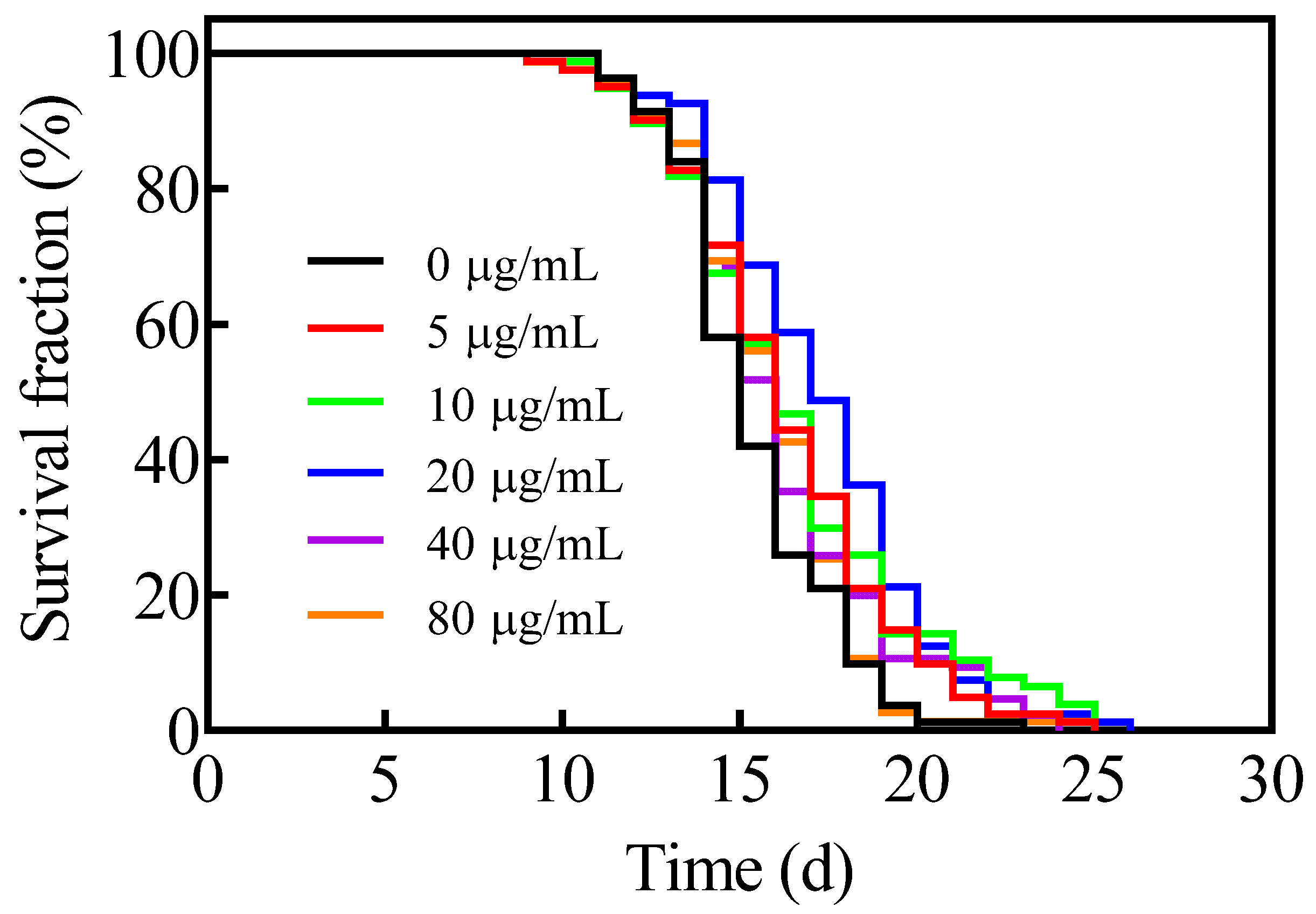

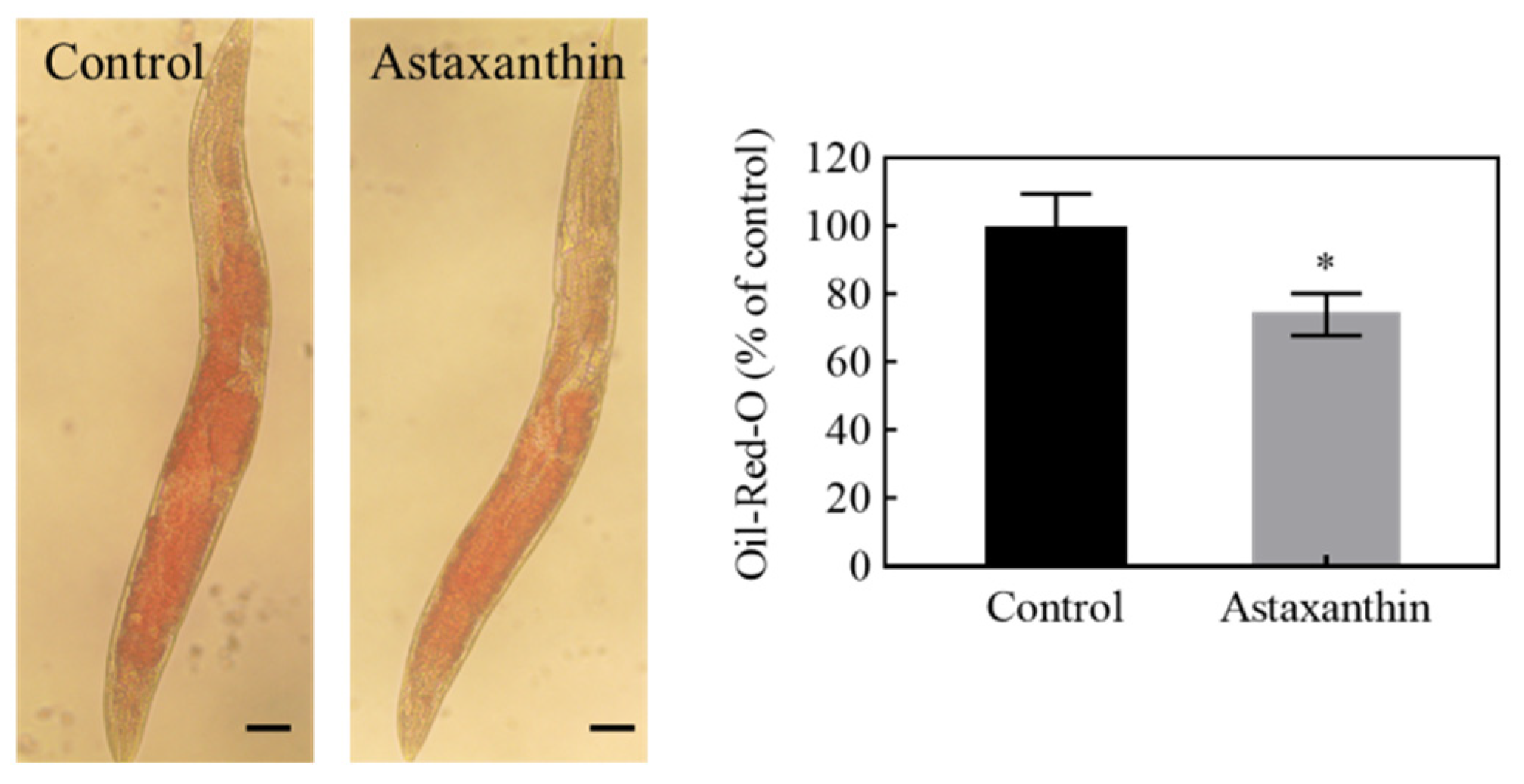

| Conc. (μg/mL) | Mean Lifespan (Days) | Median Lifespan (Days) | Max. Lifespan (Days) |
|---|---|---|---|
| 0 | 15.4 ± 0.3 | 15 ± 0.3 | 21.0 ± 1.0 |
| 5 | 16.3 ± 0.3 * | 16 ± 0.4 | 22.7 ± 1.2 |
| 10 | 16.5 ± 0.4 * | 16 ± 0.4 * | 24.7 ± 0.3 * |
| 20 | 17.3 ± 0.3 * | 17 ± 0.5 * | 23.7 ± 1.6 * |
| 40 | 16.1 ± 0.3 | 16 ± 0.3 | 23.7 ± 0.3 |
| 80 | 15.8 ± 0.3 | 16 ± 0.2 * | 21.0 ± 2.1 |
| Gene Symbols | Description | Fold Change | p-Value |
|---|---|---|---|
| rmd-2 | Regulator of microtubule dynamics | 4.56 | 2.43 × 10−71 |
| T22H9.1 | rRNA biogenesis protein RRP36 | 0.28 | 9.77 × 10−4 |
| F47E1.2 | Solute carrier organic anion transporter family member | 0.49 | 1.09 × 10−3 |
| lex-1 | Tat-binding homolog 7 | 0.46 | 4.66 × 10−3 |
| kvs-5 | K (potassium) voltage-sensitive channel subunit 5 | 0.47 | 0.01 |
| clec-78 | C-type lectin | 0.33 | 0.01 |
| mxl-3 | Max-like | 2.09 | 0.01 |
| phf-34 | PHD finger family | 0.13 | 0.01 |
| drr-1 | Dietary restriction response | 0.35 | 0.01 |
| his-28 | Histone 28 | 0.44 | 0.01 |
| hpo-36 | Hypersensitive to pore-forming toxin 36 | 0.25 | 0.01 |
| oat-1 | Organic anion transporter | 0.44 | 0.02 |
| rpl-37 | 60S ribosomal protein L37 | 0.35 | 0.02 |
| F02D8.4 | Peptidase_M14 domain-containing protein | 0.47 | 0.02 |
| nlp-15 | Neuropeptide-like protein | 0.44 | 0.02 |
| F28H7.8 | CRAL-TRIO domain-containing protein | 0.26 | 0.02 |
| H06I04.6 | CX domain-containing protein | 0.48 | 0.03 |
| cyp-14A3 | Cytochrome P450 family | 0.38 | 0.03 |
| linc-60 | Long intervening non-coding RNA 60 | 0.38 | 0.03 |
| nhr-3 | Nuclear hormone receptor family member nhr-3 | 0.49 | 0.03 |
| ugt-19 | UDP-glucuronosyltransferase | 0.44 | 0.04 |
| F55A3.6 | FACT complex subunit spt-16 | 0.46 | 0.05 |
| miRNAs | Target Sequence (5′ to 3′) | Accession Number | Fold Change | p-Value |
|---|---|---|---|---|
| cel-miR-4814-5p | TTCTCAACCAACTTTGGCCACT | OP709781 | inf | 0.0036 |
| cel-miR-86-3p | CTGGGCTCAGATTCGCTTAGGC | OP709782 | 6.78 | 0.018 |
| cbn-miR-75_L+4 | GATCTTAAAGCTACCAACCGGCTTCA | OP709783 | 3.76 | 0.018 |
| cel-miR-82-5p | CGGTTTTCTCTGTGATCTACAGA | OP709784 | inf | 0.043 |
| cbn-miR-60 | TATTATGCACATTTTCTAGTCCA | OP709785 | 0.45 | 0.044 |
| cel-miR-230-5p | ACTTGGTCGGCGATTTAATATT | OP709786 | 2.89 | 0.047 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ding, F.; Zhao, Y. Astaxanthin Induces Transcriptomic Responses Associated with Lifespan Extension in Caenorhabditis elegans. Antioxidants 2022, 11, 2115. https://doi.org/10.3390/antiox11112115
Ding F, Zhao Y. Astaxanthin Induces Transcriptomic Responses Associated with Lifespan Extension in Caenorhabditis elegans. Antioxidants. 2022; 11(11):2115. https://doi.org/10.3390/antiox11112115
Chicago/Turabian StyleDing, Feng, and Yan Zhao. 2022. "Astaxanthin Induces Transcriptomic Responses Associated with Lifespan Extension in Caenorhabditis elegans" Antioxidants 11, no. 11: 2115. https://doi.org/10.3390/antiox11112115
APA StyleDing, F., & Zhao, Y. (2022). Astaxanthin Induces Transcriptomic Responses Associated with Lifespan Extension in Caenorhabditis elegans. Antioxidants, 11(11), 2115. https://doi.org/10.3390/antiox11112115







