Glutathione Is Required for the Early Alert Response and Subsequent Acclimation in Cadmium-Exposed Arabidopsis thaliana Plants
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Cultivation, Cadmium Exposure, and Sampling
2.2. Determination of Cadmium Concentrations in Roots and Leaves
2.3. Gene Expression Analysis
2.4. Determination of Glutathione Concentrations
2.5. Hydrogen Peroxide Measurements
2.6. Determination of Free ACC and Conjugated ACC Concentrations
2.7. Statistical Analysis
3. Results
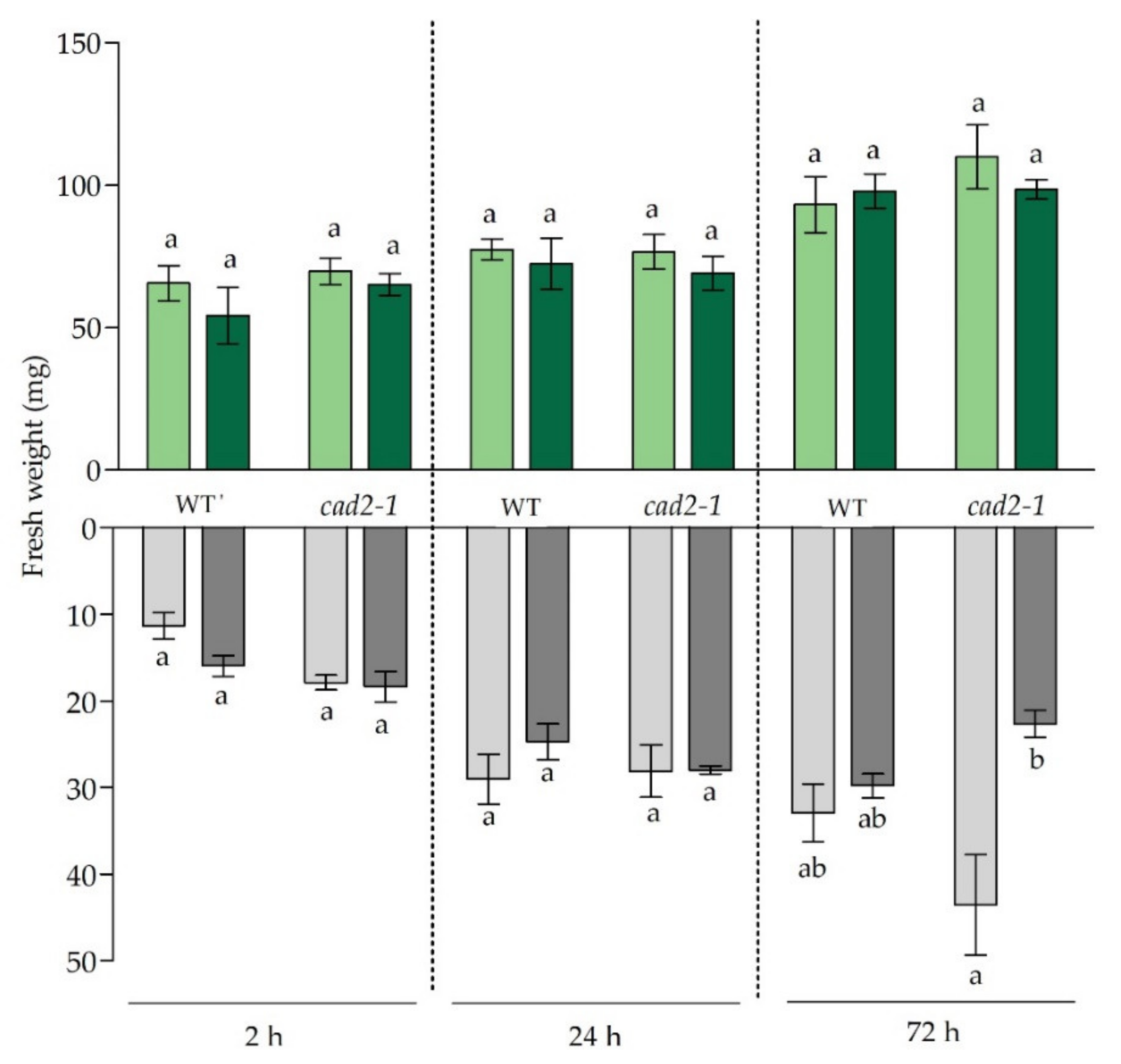
3.1. Growth Responses and Cadmium Concentrations
3.2. The Dual Role of Glutathione as Antioxidant and Chelator under Glutathione-Deficient Conditions
3.3. Balancing the Cadmium-Induced Oxidative Challenge under Glutathione-Limiting Conditions
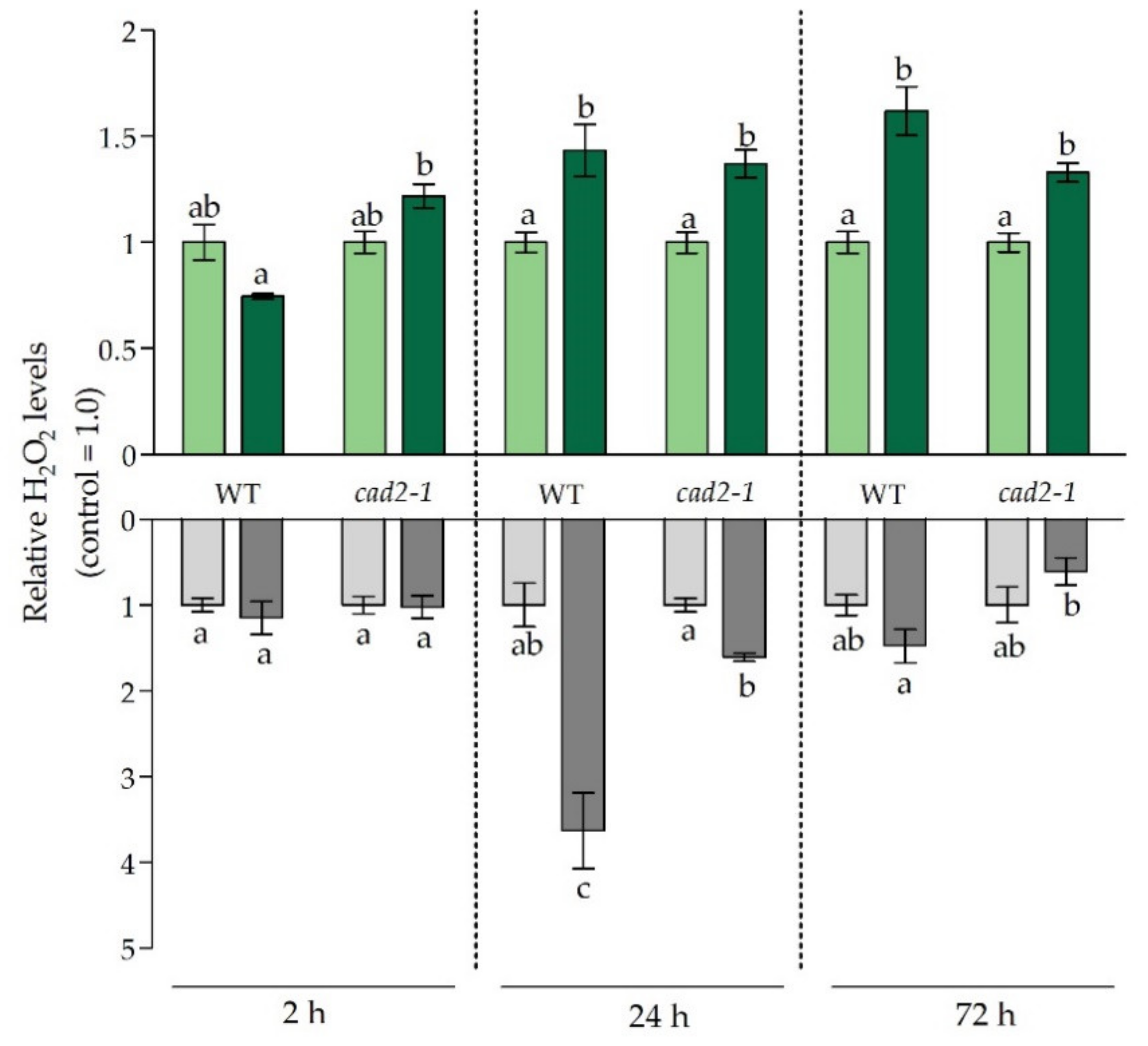
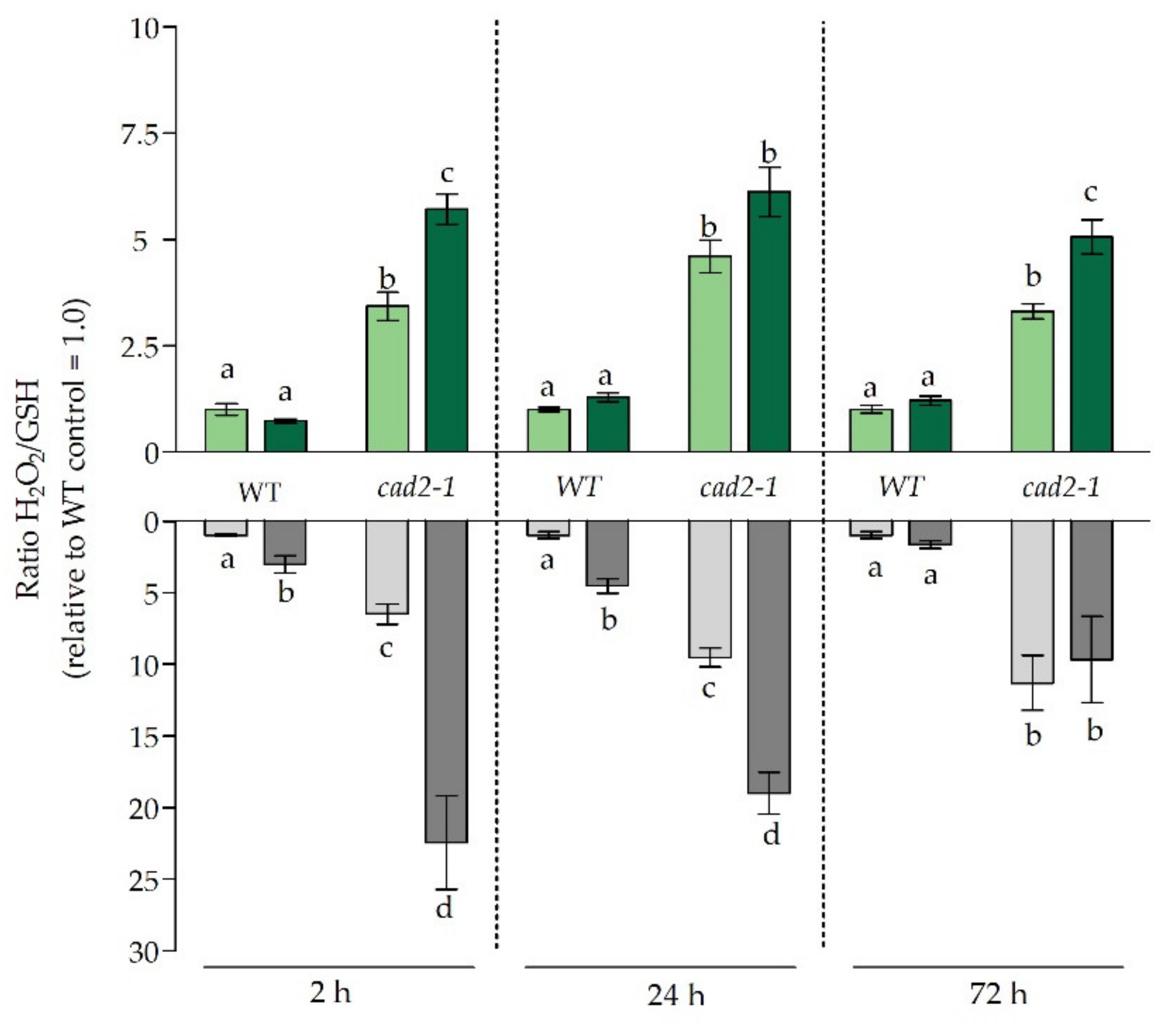
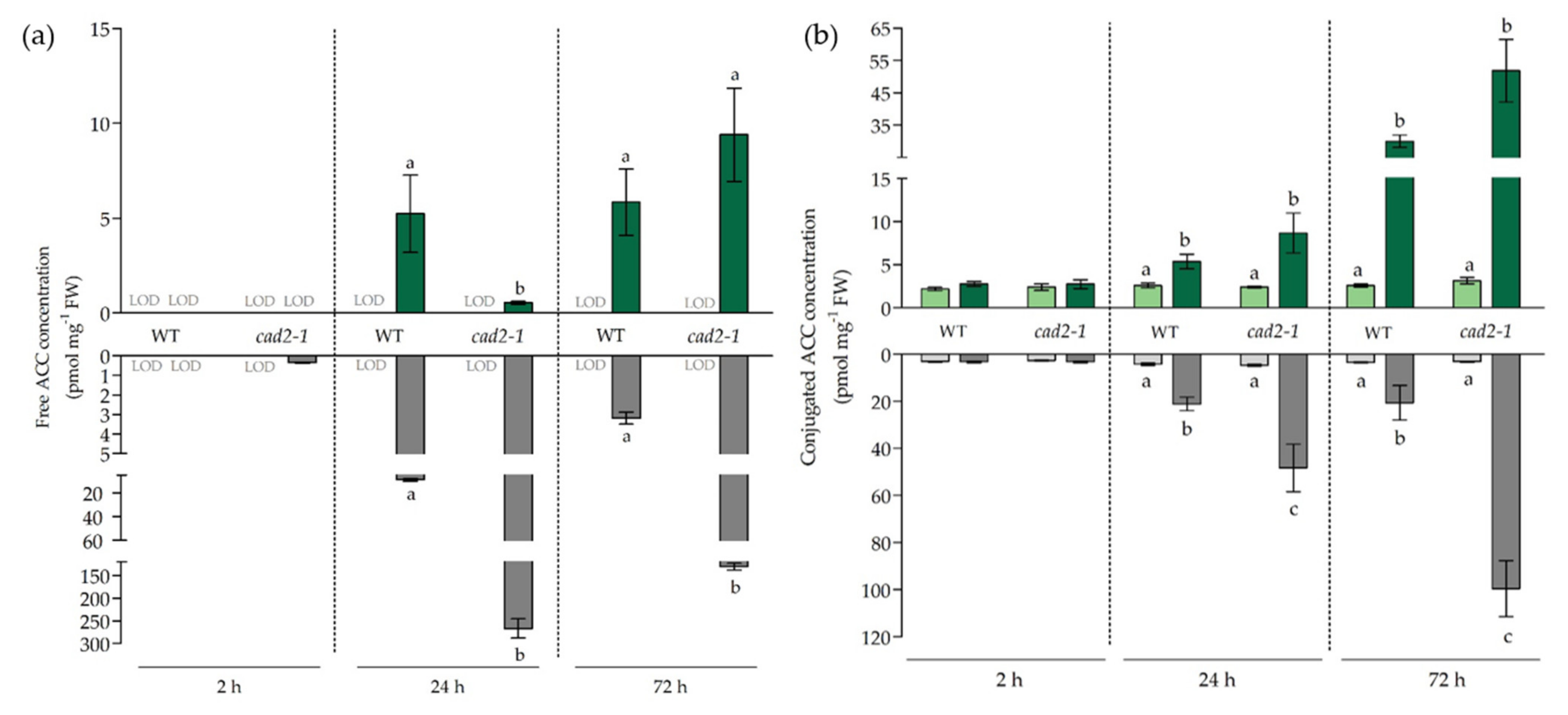
3.4. The Effect of Diminished Glutathione Synthesis on Ethylene-Related Responses under Cadmium Stress
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jarup, L.; Akesson, A. Current status of cadmium as an environmental health problem. Toxicol. Appl. Pharm. 2009, 238, 201–208. [Google Scholar] [CrossRef]
- Khan, M.A.; Khan, S.; Khan, A.; Alam, M. Soil contamination with cadmium, consequences and remediation using organic amendments. Sci. Total Environ. 2017, 601, 1591–1605. [Google Scholar] [CrossRef]
- Verbruggen, N.; Hermans, C.; Schat, H. Mechanisms to cope with arsenic or cadmium excess in plants. Curr. Opin. Plant Biol. 2009, 12, 364–372. [Google Scholar] [CrossRef] [PubMed]
- Clemens, S.; Aarts, M.G.; Thomine, S.; Verbruggen, N. Plant science: The key to preventing slow cadmium poisoning. Trends Plant Sci. 2013, 18, 92–99. [Google Scholar] [CrossRef]
- Jozefczak, M.; Keunen, E.; Schat, H.; Bliek, M.; Hernandez, L.E.; Carleer, R.; Remans, T.; Bohler, S.; Vangronsveld, J.; Cuypers, A. Differential response of Arabidopsis leaves and roots to cadmium: Glutathione-related chelating capacity vs antioxidant capacity. Plant Physiol. Biochem. 2014, 83, 1–9. [Google Scholar] [CrossRef]
- Cuypers, A.; Plusquin, M.; Remans, T.; Jozefczak, M.; Keunen, E.; Gielen, H.; Opdenakker, K.; Nair, A.R.; Munters, E.; Artois, T.J.; et al. Cadmium stress: An oxidative challenge. Biometals 2010, 23, 927–940. [Google Scholar] [CrossRef] [PubMed]
- Gill, S.S.; Anjum, N.A.; Hasanuzzaman, M.; Gill, R.; Trivedi, D.K.; Ahmad, I.; Pereira, E.; Tuteja, N. Glutathione and glutathione reductase: A boon in disguise for plant abiotic stress defense operations. Plant Physiol. Biochem. 2013, 70, 204–212. [Google Scholar] [CrossRef] [PubMed]
- Jozefczak, M.; Remans, T.; Vangronsveld, J.; Cuypers, A. Glutathione is a key player in metal-induced oxidative stress defenses. Int. J. Mol. Sci. 2012, 13, 3145–3175. [Google Scholar] [CrossRef] [Green Version]
- Zagorchev, L.; Seal, C.E.; Kranner, I.; Odjakova, M. A central role for thiols in plant tolerance to abiotic stress. Int. J. Mol. Sci. 2013, 14, 7405–7432. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deckers, J.; Hendrix, S.; Prinsen, E.; Vangronsveld, J.; Cuypers, A. Identifying the Pressure Points of Acute Cadmium Stress Prior to Acclimation in Arabidopsis thaliana. Int. J. Mol. Sci. 2020, 21, 6232. [Google Scholar] [CrossRef] [PubMed]
- Cobbett, C.S.; May, M.J.; Howden, R.; Rolls, B. The glutathione-deficient, cadmium-sensitive mutant, cad2-1, of Arabidopsis thaliana is deficient in gamma-glutamylcysteine synthetase. Plant J. 1998, 16, 73–78. [Google Scholar] [CrossRef]
- Hothorn, M.; Wachter, A.; Gromes, R.; Stuwe, T.; Rausch, T.; Scheffzek, K. Structural basis for the redox control of plant glutamate cysteine ligase. J Biol. Chem. 2006, 281, 27557–27565. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keunen, E.; Schellingen, K.; Vangronsveld, J.; Cuypers, A. Ethylene and metal stress: Small molecule, big impact. Front. Plant Sci. 2016, 7, 23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Argueso, C.T.; Hansen, M.; Kieber, J.J. Regulation of ethylene biosynthesis. J. Plant Growth Regul. 2007, 26, 92–105. [Google Scholar] [CrossRef]
- Vanderstraeten, L.; Van Der Straeten, D. Accumulation and Transport of 1-Aminocyclopropane-1-Carboxylic Acid (ACC) in Plants: Current Status, Considerations for Future Research and Agronomic Applications. Front. Plant Sci. 2017, 8, 38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van de Poel, B.; Van Der Straeten, D. 1-aminocyclopropane-1-carboxylic acid (ACC) in plants: More than just the precursor of ethylene! Front. Plant Sci. 2014, 5, 640. [Google Scholar] [CrossRef] [Green Version]
- Schellingen, K.; Van Der Straeten, D.; Vandenbussche, F.; Prinsen, E.; Remans, T.; Vangronsveld, J.; Cuypers, A. Cadmium-induced ethylene production and responses in Arabidopsis thaliana rely on ACS2 and ACS6 gene expression. BMC Plant Biol. 2014, 14, 214. [Google Scholar] [CrossRef] [Green Version]
- Opdenakker, K.; Remans, T.; Keunen, E.; Vangronsveld, J.; Cuypers, A. Exposure of Arabidopsis thaliana to Cd or Cu excess leads to oxidative stress mediated alterations in MAPKinase transcript levels. Environ. Exp. Bot. 2012, 83, 53–61. [Google Scholar] [CrossRef]
- Rentel, M.C.; Lecourieux, D.; Ouaked, F.; Usher, S.L.; Petersen, L.; Okamoto, H.; Knight, H.; Peck, S.C.; Grierson, C.S.; Hirt, H.; et al. OXI1 kinase is necessary for oxidative burst-mediated signalling in Arabidopsis. Nature 2004, 427, 858–861. [Google Scholar] [CrossRef] [PubMed]
- Remans, T.; Opdenakker, K.; Smeets, K.; Mathijsen, D.; Vangronsveld, J.; Cuypers, A. Metal-specific and NADPH oxidase dependent changes in lipoxygenase and NADPH oxidase gene expression in Arabidopsis thaliana exposed to cadmium or excess copper. Funct. Plant Biol. 2010, 37, 532–544. [Google Scholar] [CrossRef]
- Schellingen, K.; Van Der Straeten, D.; Remans, T.; Vangronsveld, J.; Keunen, E.; Cuypers, A. Ethylene signalling is mediating the early cadmium-induced oxidative challenge in Arabidopsis thaliana. Plant Sci 2015, 239, 137–146. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, S.; Tamaoki, M.; Ioki, M.; Ogawa, D.; Sato, Y.; Aono, M.; Kubo, A.; Saji, S.; Saji, H.; Satoh, S.; et al. Ethylene and salicylic acid control glutathione biosynthesis in ozone-exposed Arabidopsis thaliana. Physiol. Plant. 2009, 136, 284–298. [Google Scholar] [CrossRef]
- Chen, H.J.; Huang, C.S.; Huang, G.J.; Chow, T.J.; Lin, Y.H. NADPH oxidase inhibitor diphenyleneiodonium and reduced glutathione mitigate ethephon-mediated leaf senescence, H2O2 elevation and senescence-associated gene expression in sweet potato (Ipomoea batatas). J. Plant Physiol. 2013, 170, 1471–1483. [Google Scholar] [CrossRef]
- Zhang, Y.; He, Q.; Zhao, S.; Huang, L.; Hao, L. Arabidopsis ein2-1 and npr1-1 response to Al stress. Bull. Environ. Contam. Toxicol. 2014, 93, 78–83. [Google Scholar] [CrossRef]
- Smeets, K.; Ruytinx, J.; Van Belleghem, F.; Semane, B.; Lin, D.; Vangronsveld, J.; Cuypers, A. Critical evaluation and statistical validation of a hydroponic culture system for Arabidopsis thaliana. Plant Physiol. Biochem. 2008, 46, 212–218. [Google Scholar] [CrossRef] [PubMed]
- Keunen, E.; Truyens, S.; Bruckers, L.; Remans, T.; Vangronsveld, J.; Cuypers, A. Survival of Cd-exposed Arabidopsis thaliana: Are these plants reproductively challenged? Plant Physiol. Biochem. 2011, 49, 1084–1091. [Google Scholar] [CrossRef] [PubMed]
- Remans, T.; Keunen, E.; Bex, G.J.; Smeets, K.; Vangronsveld, J.; Cuypers, A. Reliable Gene Expression Analysis by Reverse Transcription-Quantitative PCR: Reporting and Minimizing the Uncertainty in Data Accuracy. Plant Cell 2014, 26, 3829–3837. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tausz, M.; Šircelj, H.; Grill, D. The glutathione system as a stress marker in plant ecophysiology: Is a stress-response concept valid? J. Exp. Bot. 2004, 55, 1955–1962. [Google Scholar] [CrossRef]
- Krznaric, E.; Verbruggen, N.; Wevers, J.H.L.; Carleer, R.; Vangronsveld, J.; Colpaert, J.V. Cd-tolerant Suillus luteus: A fungal insurance for pines exposed to Cd. Environ. Pollut. 2009, 157, 1581–1588. [Google Scholar] [CrossRef] [PubMed]
- Jozefczak, M.; Bohler, S.; Schat, H.; Horemans, N.; Guisez, Y.; Remans, T.; Vangronsveld, J.; Cuypers, A. Both the concentration and redox state of glutathione and ascorbate influence the sensitivity of Arabidopsis to cadmium. Annals of Botany 2015. [Google Scholar] [CrossRef] [PubMed]
- Jozefczak, M. Deficiency in Ascorbate is Compensated by Glutathione in Cadmium-Exposed Arabidopsis Mutants but Glutathione Deficiency Demands for Multiple Alternatives. Ph.D. Thesis, Hasselt University, Hasselt, Belgium, 2014. Chapter 4. [Google Scholar]
- Cuypers, A.; Hendrix, S.; dos Reis, R.A.; De Smet, S.; Deckers, J.; Gielen, H.; Jozefczak, M.; Loix, C.; Vercampt, H.; Vangronsveld, J.; et al. Hydrogen peroxide, signaling in disguise during metal phytotoxicity. Front. Plant Sci. 2016, 7, 470. [Google Scholar] [CrossRef] [Green Version]
- Gadjev, I.; Vanderauwera, S.; Gechev, T.S.; Laloi, C.; Minkov, I.N.; Shulaev, V.; Apel, K.; Inze, D.; Mittler, R.; Van Breusegem, F. Transcriptomic footprints disclose specificity of reactive oxygen species signaling in Arabidopsis. Plant Physiol. 2006, 141, 436–445. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dubreuil-Maurizi, C.; Vitecek, J.; Marty, L.; Branciard, L.; Frettinger, P.; Wendehenne, D.; Meyer, A.J.; Mauch, F.; Poinssot, B. Glutathione Deficiency of the Arabidopsis Mutant pad2-1 Affects Oxidative Stress-Related Events, Defense Gene Expression, and the Hypersensitive Response. Plant Physiol. 2011, 157, 2000–2012. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, Y.; Chaouch, S.; Mhamdi, A.; Queval, G.; Zechmann, B.; Noctor, G. Functional analysis of Arabidopsis mutants points to novel roles for glutathione in coupling H2O2 to activation of salicylic acid accumulation and signaling. Antioxid. Redox Signal. 2013, 18, 2106–2121. [Google Scholar] [CrossRef] [Green Version]
- Mhamdi, A.; Hager, J.; Chaouch, S.; Queval, G.; Han, Y.; Taconnat, L.; Saindrenan, P.; Gouia, H.; Issakidis-Bourguet, E.; Renou, J.P.; et al. Arabidopsis GLUTATHIONE REDUCTASE1 plays a crucial role in leaf responses to intracellular hydrogen peroxide and in ensuring appropriate gene expression through both salicylic acid and jasmonic acid signaling pathways. Plant Physiol. 2010, 153, 1144–1160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.; Yang, L.; Yan, X.; Liu, Y.; Wang, R.; Fan, T.; Ren, Y.; Tang, X.; Xiao, F.; Liu, Y.; et al. Zinc-Finger Transcription Factor ZAT6 Positively Regulates Cadmium Tolerance through the Glutathione-Dependent Pathway in Arabidopsis. Plant Physiol. 2016, 171, 707–719. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.M.; Nguyen, X.C.; Kim, K.E.; Han, H.J.; Yoo, J.; Lee, K.; Kim, M.C.; Yun, D.J.; Chung, W.S. Phosphorylation of the zinc finger transcriptional regulator ZAT6 by MPK6 regulates Arabidopsis seed germination under salt and osmotic stress. Biochem. Biophys. Res. Commun. 2013, 430, 1054–1059. [Google Scholar] [CrossRef]
- Rizhsky, L.; Davletova, S.; Liang, H.J.; Mittler, R. The zinc finger protein Zat12 is required for cytosolic ascorbate peroxidase 1 expression during oxidative stress in Arabidopsis. J. Biol. Chem. 2004, 279, 11736–11743. [Google Scholar] [CrossRef] [Green Version]
- Han, G.L.; Lu, C.X.; Guo, J.R.; Qiao, Z.Q.; Sui, N.; Qiu, N.W.; Wang, B.S. C2H2 Zinc Finger Proteins: Master Regulators of Abiotic Stress Responses in Plants. Front. Plant Sci. 2020, 11, 115. [Google Scholar] [CrossRef] [Green Version]
- Schellingen, K.; Van Der Straeten, D.; Remans, T.; Loix, C.; Vangronsveld, J.; Cuypers, A. Ethylene biosynthesis is involved in the early oxidative challenge induced by moderate Cd exposure in Arabidopsis thaliana. Environ. Exp. Bot. 2015, 117, 1–11. [Google Scholar] [CrossRef]
- Datta, R.; Kumar, D.; Sultana, A.; Hazra, S.; Bhattacharyya, D.; Chattopadhyay, S. Glutathione Regulates 1-Aminocyclopropane-1-Carboxylate Synthase Transcription via WRKY33 and 1-Aminocyclopropane-1-Carboxylate Oxidase by Modulating Messenger RNA Stability to Induce Ethylene Synthesis during Stress. Plant Physiol. 2015, 169, 2963–2981. [Google Scholar] [CrossRef] [PubMed]
- Montero-Palmero, M.B.; Martín-Barranco, A.; Escobar, C.; Hernández, L.E. Early transcriptional responses to mercury: A role for ethylene in mercury-induced stress. New Phytol. 2014, 201, 116–130. [Google Scholar] [CrossRef] [PubMed]
- Skottke, K.R.; Yoon, G.M.; Kieber, J.J.; DeLong, A. Protein Phosphatase 2A Controls Ethylene Biosynthesis by Differentially Regulating the Turnover of ACC Synthase Isoforms. PLoS Genet. 2011, 7, e1001370. [Google Scholar] [CrossRef]
- Li, G.; Meng, X.; Wang, R.; Mao, G.; Han, L.; Liu, Y.; Zhang, S. Dual-Level Regulation of ACC Synthase Activity by MPK3/MPK6 Cascade and Its Downstream WRKY Transcription Factor during Ethylene Induction in Arabidopsis. PLoS Genet. 2012, 8, e1002767. [Google Scholar] [CrossRef] [PubMed]
- Yoo, S.D.; Cho, Y.H.; Sheen, J. Emerging connections in the ethylene signaling network. Trends Plant Sci. 2009, 14, 270–279. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Zhang, S. Phosphorylation of 1-Aminocyclopropane-1-Carboxylic Acid Synthase by MPK6, a Stress-Responsive Mitogen-Activated Protein Kinase, Induces Ethylene Biosynthesis in Arabidopsis. Plant Cell 2004, 16, 3386–3399. [Google Scholar] [CrossRef] [Green Version]
- Mersmann, S.; Bourdais, G.; Rietz, S.; Robatzek, S. Ethylene Signaling Regulates Accumulation of the FLS2 Receptor and Is Required for the Oxidative Burst Contributing to Plant Immunity. Plant Physiol. 2010, 154, 391–400. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hendrix, S.; Jozefczak, M.; Wojcik, M.; Deckers, J.; Vangronsveld, J.; Cuypers, A. Glutathione: A key player in metal chelation, nutrient homeostasis, cell cycle regulation and the DNA damage response in cadmium-exposed Arabidopsis thaliana. Plant Physiol. Biochem. 2020, 154, 498–507. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, S.; Yoshidomi, T.; Yoshimura, E. Cadmium(II)-stimulated enzyme activation of Arabidopsis thaliana phytochelatin synthase 1. J. Inorg. Biochem. 2011, 105, 111–117. [Google Scholar] [CrossRef]

| Cd Concentration (mg kg−1 DW) | |||||||
| 2 h | 24 h | 72 h | |||||
| [CdSO4] | WT | cad2-1 | WT | cad2-1 | WT | cad2-1 | |
| Leaf | 5 µM | 1.75 ± 0.12 | 8.71 ± 1.09 * | 1001.02 ± 15.36 | 275.20 ± 14.53 * | 1492.14 ± 24.95 | 584.86 ± 17.23 * |
| Root | 5 µM | 1009.09 ± 28.66 | 1054.35 ± 20.62 | 2168.99 ± 52.39 | 3070.92 ± 52.39 * | 2680.88 ± 92.24 | 3209.21 ± 88.32 * |
| Translocation factor | |||||||
| 2 h | 24 h | 72 h | |||||
| [CdSO4] | WT | cad2-1 | WT | cad2-1 | WT | cad2-1 | |
| 5 µM | 0.17 ± 0.01 | 0.82 ± 0.01 * | 46.21 ± 0.78 | 8.97 ± 0.49 * | 55.89 ± 1.99 | 18.29 ± 0.81 * | |
| GSH-Related Genes | |||||||
|---|---|---|---|---|---|---|---|
| 2 h | 24 h | 72 h | |||||
| Gene | CdSO4 | WT | cad2-1 | WT | cad2-1 | WT | cad2-1 |
| Leaf | |||||||
| GSH1 | 0 µM | 1.00 ± 0.03 | 1.00 ± 0.06 | 1.00 ± 0.14 | 1.00 ± 0.10 | 1.00 ± 0.12 | 1.00 ± 0.07 |
| 5 µM | 1.01 ± 0.07 | 0.92 ± 0.14 | 2.34 ± 0.12 | 2.30 ± 0.57 | 1.23 ± 0.09 | 0.84 ± 0.04 * | |
| GSH2 | 0 µM | 1.00 ± 0.06 | 1.00 ± 0.07 | 1.00 ± 0.14 | 1.00 ± 0.11 | 1.00 ± 0.10 | 1.00 ± 0.16 |
| 5 µM | 1.21 ± 0.03 | 1.06 ± 0.05 | 3.39 ± 0.31 | 3.14 ± 0.77 | 1.48 ± 0.05 | 2.07 ± 0.56 | |
| GR1 | 0 µM | 1.00 ± 0.05 | 1.00 ± 0.07 | 1.00 ± 0.09 | 1.00 ± 0.12 | 1.00 ± 0.10 | 1.00 ± 0.09 |
| 5 µM | 0.89 ± 0.04 | 0.78 ± 0.02 | 3.00 ± 0.15 | 1.65 ± 0.29 * | 1.23 ± 0.05 | 1.02 ± 0.05 | |
| GGT1 | 0 µM | 1.00 ± 0.08 | 1.00 ± 0.06 | 1.00 ± 0.09 | 1.00 ± 0.19 | 1.00 ± 0.06 | 1.00 ± 0.17 |
| 5 µM | 0.86 ± 0.06 | 1.12 ± 0.07 | 2.98 ± 0.20 | 0.79 ± 0.12 * | 1.76 ± 0.32 | 0.98 ± 0.10 | |
| ZAT6 | 0 µM | 1.00 ± 0.16 | 1.00 ± 0.26 | 1.00 ± 0.27 | 1.00 ± 0.21 | 1.00 ± 0.04 | 1.00 ± 0.20 |
| 5 µM | 1.41 ± 0.34 | 2.10 ± 0.26 | 3.93 ± 0.71 | 1.85 ± 0.48 | 1.73 ± 0.66 | 1.13 ± 0.13 | |
| Root | |||||||
| GSH1 | 0 µM | 1.00 ± 0.05 | 1.00 ± 0.11 | 1.00 ± 0.02 | 1.00 ± 0.03 | 1.00 ± 0.09 | 1.00 ± 0.08 |
| 5 µM | 1.11 ± 0.03 | 1.25 ± 0.03 | 1.66 ± 0.12 | 2.15 ± 0.11 * | 0.85 ± 0.05 | 0.96 ± 0.14 | |
| GSH2 | 0 µM | 1.00 ± 0.09 | 1.00 ± 0.11 | 1.00 ± 0.04 | 1.00 ± 0.02 | 1.00 ± 0.08 | 1.00 ± 0.08 |
| 5 µM | 1.04 ± 0.03 | 1.39 ± 0.00 | 2.83 ± 0.08 | 2.67 ± 0.13 | 1.39 ± 0.23 | 1.78 ± 0.26 | |
| GR1 | 0 µM | 1.00 ± 0.03 | 1.00 ± 0.14 | 1.00 ± 0.08 | 1.00 ± 0.03 | 1.00 ± 0.10 | 1.00 ± 0.04 |
| 5 µM | 0.98 ± 0.07 | 1.29 ± 0.02 * | 2.17 ± 0.08 | 1.99 ± 0.05 | 1.74 ± 0.33 | 1.16 ± 0.12 | |
| GGT1 | 0 µM | 1.00 ± 0.05 | 1.00 ± 0.01 | 1.00 ± 0.04 | 1.00 ± 0.05 | 1.00 ± 0.16 | 1.00 ± 0.10 |
| 5 µM | 1.43 ± 0.15 | 1.35 ± 0.03 | 2.85 ± 0.20 | 1.33 ± 0.05 * | 1.08 ± 0.05 | 1.08 ± 0.09 | |
| ZAT6 | 0 µM | 1.00 ± 0.13 | 1.00 ± 0.25 | 1.00 ± 0.17 | 1.00 ± 0.14 | 1.00 ± 0.14 | 1.00 ± 0.05 |
| 5 µM | 3.97 ± 0.48 | 23.05 ± 0.92 * | 7.54 ± 0.81 | 10.89 ± 0.37 | 1.62 ± 0.22 | 9.48 ± 1.05 * | |
| Oxidative Challenge-Related Genes | |||||||
|---|---|---|---|---|---|---|---|
| 2 h | 24 h | 72 h | |||||
| Gene | CdSO4 | WT | cad2-1 | WT | cad2-1 | WT | cad2-1 |
| Leaf | |||||||
| AT1G05340 | 0 µM | 1.00 ± 0.50 | 1.00 ± 0.29 | 1.00 ± 0.29 | 1.00 ± 0.34 | 1.00 ± 0.24 | 1.00 ± 0.12 |
| 5 µM | 0.41 ± 0.12 | 0.15 ± 0.04 | 32.29 ± 4.11 | 1.07 ± 0.42 * | 1.17 ± 0.51 | 1.03 ± 0.21 | |
| AT1G19020 | 0 µM | 1.00 ± 0.21 | 1.00 ± 0.27 | 1.00 ± 0.29 | 1.00 ± 0.34 | 1.00 ± 0.17 | 1.00 ± 0.11 |
| 5 µM | 0.92 ± 0.07 | 1.15 ± 0.08 | 9.45 ± 0.52 | 2.41 ± 0.59 * | 2.00 ± 0.40 | 1.98 ± 0.48 | |
| AT1G57630 | 0 µM | 1.00 ± 0.31 | 1.00 ± 0.30 | 1.00 ± 0.31 | 1.00 ± 0.38 | 1.00 ± 0.25 | 1.00 ± 0.16 |
| 5 µM | 0.57 ± 0.16 | 0.14 ± 0.05 | 9.88 ± 0.42 | 1.16 ± 0.22 | 2.26 ± 0.46 | 1.04 ± 0.23 | |
| AT2G21640 | 0 µM | 1.00 ± 0.10 | 1.00 ± 0.08 | 1.00 ± 0.02 | 1.00 ± 0.32 | 1.00 ± 0.06 | 1.00 ± 0.20 |
| 5 µM | 1.11 ± 0.07 | 1.17 ± 0.05 | 12.72 ± 2.12 | 2.55 ± 0.38 * | 2.68 ± 0.58 | 1.43 ± 0.08 | |
| AT2G43510 | 0 µM | 1.00 ± 0.25 | 1.00 ± 0.18 | 1.00 ± 0.48 | 1.00 ± 0.18 | 1.00 ± 0.11 | 1.00 ± 0.19 |
| 5 µM | 1.21 ± 0.26 | 0.68 ± 0.03 | 20.04 ± 0.43 | 10.81 ± 3.70 | 3.19 ± 0.43 | 2.83 ± 0.40 | |
| ZAT12 | 0 µM | 1.00 ± 0.22 | 1.00 ± 0.28 | 1.00 ± 0.35 | 1.00 ± 0.28 | 1.00 ± 0.25 | 1.00 ± 0.16 |
| 5 µM | 0.98 ± 0.21 | 1.86 ± 0.33 | 5.47 ± 0.58 | 2.07 ± 0.52 * | 1.39 ± 0.37 | 1.31 ± 0.20 | |
| RRTF1 | 0 µM | 1.00 ± 0.41 | 1.00 ± 0.61 | 1.00 ± 0.57 | 1.00 ± 0.35 | 1.00 ± 0.32 | 1.00 ± 0.24 |
| 5 µM | 0.62 ± 0.22 | 6.96 ± 1.57 * | 0.39 ± 0.11 | 0.32 ± 0.12 | 1.71 ± 0.66 | 0.56 ± 0.27 | |
| Root | |||||||
| AT1G05340 | 0 µM | 1.00 ± 0.13 | 1.00 ± 0.14 | 1.00 ± 0.07 | 1.00 ± 0.06 | 1.00 ± 0.16 | 1.00 ± 0.12 |
| 5 µM | 1.35 ± 0.19 | 1.16 ± 0.10 | 1.54 ± 0.04 | 2.84 ± 0.19 * | 1.09 ± 0.10 | 2.02 ± 0.22 * | |
| AT1G19020 | 0 µM | 1.00 ± 0.09 | 1.00 ± 0.18 | 1.00 ± 0.19 | 1.00 ± 0.29 | 1.00 ± 0.14 | 1.00 ± 0.21 |
| 5 µM | 6.66 ± 0.54 | 30.54 ± 2.30 * | 21.94 ± 1.10 | 8.79 ± 0.68 * | 4.34 ± 0.61 | 12.98 ± 1.05 * | |
| AT1G57630 | 0 µM | 1.00 ± 0.17 | 1.00 ± 0.02 | 1.00 ± 0.19 | 1.00 ± 0.19 | 1.00 ± 0.17 | 1.00 ± 0.17 |
| 5 µM | 1.00 ± 0.02 | 2.96 ± 0.21 * | 39.49 ± 3.52 | 14.65 ± 0.97 * | 6.80 ± 0.70 | 16.10 ± 1.14 * | |
| AT2G21640 | 0 µM | 1.00 ± 0.11 | 1.00 ± 0.14 | 1.00 ± 0.06 | 1.00 ± 0.08 | 1.00 ± 0.12 | 1.00 ± 0.13 |
| 5 µM | 1.13 ± 0.05 | 1.47 ± 0.04 * | 6.49 ± 0.49 | 10.27 ± 1.10 | 1.40 ± 0.24 | 6.05 ± 0.47 * | |
| AT2G43510 | 0 µM | 1.00 ± 0.25 | 1.00 ± 0.25 | 1.00 ± 0.24 | 1.00 ± 0.12 | 1.00 ± 0.30 | 1.00 ± 0.21 |
| 5 µM | 2.48 ± 0.33 | 1.47 ± 0.08 | 25.89 ± 3.21 | 18.84 ± 1.68 | 158.69 ± 9.91 | 138.00 ± 16.19 | |
| ZAT12 | 0 µM | 1.00 ± 0.21 | 1.00 ± 0.37 | 1.00 ± 0.24 | 1.00 ± 0.24 | 1.00 ± 0.15 | 1.00 ± 0.16 |
| 5 µM | 2.98 ± 0.29 | 12.88 ± 0.81 * | 14.83 ± 0.04 | 10.04 ± 0.40 | 3.27 ± 0.30 | 36.59 ± 1.12 * | |
| RRTF1 | 0 µM | 1.00 ± 0.25 | 1.00 ± 0.60 | 1.00 ± 0.60 | 1.00 ± 0.42 | 1.00 ± 0.61 | 1.00 ± 0.49 |
| 5 µM | 1.22 ± 0.34 | 0.46 ± 0.10 | 6.67 ± 0.86 | 0.22 ± 0.02 * | 1.18 ± 0.37 | 4.33 ± 0.31 * | |
| Ethylene-Related Genes | |||||||
|---|---|---|---|---|---|---|---|
| 2 h | 24 h | 72 h | |||||
| Gene | CdSO4 | WT | cad2-1 | WT | cad2-1 | WT | cad2-1 |
| Leaf | |||||||
| ACS2 | 0 µM | 1.00 ± 0.38 | 1.00 ± 0.34 | 1.00 ± 0.32 | 1.00 ± 0.35 | 1.00 ± 0.23 | 1.00 ± 0.18 |
| 5 µM | 0.59 ± 0.21 | 0.34 ± 0.07 | 113.06 ± 13.61 | 16.22 ± 5.99 * | 0.62 ± 0.21 | 1.52 ± 0.31 | |
| ACS6 | 0 µM | 1.00 ± 0.34 | 1.00 ± 0.19 | 1.00 ± 0.14 | 1.00 ± 0.22 | 1.00 ± 0.16 | 1.00 ± 0.13 |
| 5 µM | 0.81 ± 0.44 | 2.70 ± 0.29 * | 6.74 ± 1.07 | 1.17 ± 0.36 * | 1.84 ± 0.27 | 2.80 ± 0.74 * | |
| ACO2 | 0 µM | 1.00 ± 0.03 | 1.00 ± 0.01 | 1.00 ± 0.08 | 1.00 ± 0.07 | 1.00 ± 0.14 | 1.00 ± 0.09 |
| 5 µM | 0.91 ± 0.05 | 0.96 ± 0.05 | 4.09 ± 0.77 | 6.25 ± 1.16 | 1.47 ± 0.14 | 1.39 ± 0.02 | |
| ACO4 | 0 µM | 1.00 ± 0.13 | 1.00 ± 0.15 | 1.00 ± 0.08 | 1.00 ± 0.61 | 1.00 ± 0.09 | 1.00 ± 0.11 |
| 5 µM | 0.96 ± 0.03 | 0.80 ± 0.05 | 6.49 ± 0.49 | 2.18 ± 0.44 * | 2.37 ± 0.15 | 1.66 ± 0.19 | |
| ERF1 | 0 µM | 1.00 ± 0.19 | 1.00 ± 0.23 | 1.00 ± 0.15 | 1.00 ± 0.29 | 1.00 ± 0.24 | 1.00 ± 0.13 |
| 5 µM | 1.57 ± 0.15 | 1.23 ± 0.21 | 38.31 ± 6.76 | 9.18 ± 2.49 * | 2.99 ± 0.55 | 4.53 ± 0.77 | |
| OXI1 | 0 µM | 1.00 ± 0.23 | 1.00 ± 0.20 | 1.00 ± 0.35 | 1.00 ± 0.22 | 1.00 ± 0.17 | 1.00 ± 0.08 |
| 5 µM | 1.22 ± 0.14 | 1.10 ± 0.26 | 32.46 ± 4.95 | 3.92 ± 0.89 * | 1.70 ± 0.54 | 1.87 ± 0.09 | |
| MPK3 | 0 µM | 1.00 ± 0.12 | 1.00 ± 0.15 | 1.00 ± 0.18 | 1.00 ± 0.24 | 1.00 ± 0.13 | 1.00 ± 0.10 |
| 5 µM | 1.08 ± 0.01 | 0.99 ± 0.09 | 1.80 ± 0.07 | 0.54 ± 0.04 * | 2.01 ± 0.33 | 1.20 ± 0.16 | |
| MPK6 | 0 µM | 1.00 ± 0.07 | 1.00 ± 0.11 | 1.00 ± 0.11 | 1.00 ± 0.90 | 1.00 ± 0.03 | 1.00 ± 0.15 |
| 5 µM | 0.88 ± 0.05 | 0.86 ± 0.03 | 2.26 ± 0.23 | 0.98 ± 0.12 * | 1.21 ± 0.15 | 0.96 ± 0.11 | |
| WRKY33 | 0 µM | 1.00 ± 0.34 | 1.00 ± 0.24 | 1.00 ± 0.30 | 1.00 ± 0.26 | 1.00 ± 0.11 | 1.00 ± 0.25 |
| 5 µM | 1.41 ± 0.34 | 1.25 ± 0.41 | 3.69 ± 0.39 | 0.93 ± 0.25 * | 1.19 ± 0.28 | 1.42 ± 0.25 | |
| Root | |||||||
| ACS2 | 0 µM | 1.00 ± 0.10 | 1.00 ± 0.12 | 1.00 ± 0.18 | 1.00 ± 0.24 | 1.00 ± 0.09 | 1.00 ± 0.13 |
| 5 µM | 1.29 ± 0.10 | 1.25 ± 0.08 | 14.22 ± 1.63 | 108.08 ± 0.74 * | 6.29 ± 0.1 | 23.74 ± 1.68 * | |
| ACS6 | 0 µM | 1.00 ± 0.18 | 1.00 ± 0.23 | 1.00 ± 0.18 | 1.00 ± 0.24 | 1.00 ± 0.14 | 1.00 ± 0.08 |
| 5 µM | 3.22 ± 0.77 | 10.08 ± 1.14 * | 14.22 ± 1.63 | 13.02 ± 0.74 | 3.98 ± 0.52 | 12.22 ± 1.58 * | |
| ACO2 | 0 µM | 1.00 ± 0.12 | 1.00 ± 0.09 | 1.00 ± 0.07 | 1.00 ± 0.03 | 1.00 ± 0.10 | 1.00 ± 0.05 |
| 5 µM | 0.88 ± 0.09 | 1.28 ± 0.12 | 2.07 ± 0.21 | 1.98 ± 0.06 | 1.08 ± 0.10 | 0.96 ± 0.13 | |
| ACO4 | 0 µM | 1.00 ± 0.10 | 1.00 ± 0.15 | 1.00 ± 0.05 | 1.00 ± 0.05 | 1.00 ± 0.00 | 1.00 ± 0.06 |
| 5 µM | 2.61 ± 0.14 | 3.71 ± 0.26 | 6.99 ± 0.39 | 6.26 ± 0.22 | 3.07 ± 0.29 | 4.13 ± 0.33 | |
| ERF1 | 0 µM | 1.00 ± 0.13 | 1.00 ± 0.18 | 1.00 ± 0.15 | 1.00 ± 0.07 | 1.00 ± 0.12 | 1.00 ± 0.09 |
| 5 µM | 3.37 ± 0.27 | 39.93 ± 2.37 * | 137.31 ± 19.52 | 69.26 ± 5.58 * | 13.55 ± 2.44 | 79.92 ± 7.98 * | |
| OXI1 | 0 µM | 1.00 ± 0.10 | 1.00 ± 0.19 | 1.00 ± 0.06 | 1.00 ± 0.22 | 1.00 ± 0.14 | 1.00 ± 0.13 |
| 5 µM | 3.44 ± 0.35 | 16.77 ± 1.46 * | 7.75 ± 1.12 | 12.54 ± 0.10 | 1.81 ± 0.19 | 11.55 ± 0.84 * | |
| MPK3 | 0 µM | 1.00 ± 0.03 | 1.00 ± 0.03 | 1.00 ± 0.05 | 1.00 ± 0.12 | 1.00 ± 0.16 | 1.00 ± 0.02 |
| 5 µM | 2.28 ± 0.09 | 6.41 ± 0.36 * | 3.50 ± 0.9 | 5.82 ± 0.27 * | 2.00 ± 0.24 | 3.06 ± 0.23 | |
| MPK6 | 0 µM | 1.00 ± 0.03 | 1.00 ± 0.13 | 1.00 ± 0.05 | 1.00 ± 0.01 | 1.00 ± 0.12 | 1.00 ± 0.12 |
| 5 µM | 1.81 ± 0.38 | 2.46 ± 0.02 | 2.23 ± 0.03 | 1.49 ± 0.27 * | 0.79 ± 0.05 | 0.82 ± 0.07 | |
| WRKY33 | 0 µM | 1.00 ± 0.11 | 1.00 ± 0.17 | 1.00 ± 0.09 | 1.00 ± 0.22 | 1.00 ± 0.23 | 1.00 ± 0.17 |
| 5 µM | 5.01 ± 0.84 | 19.71 ± 1.02 * | 8.47 ± 0.59 | 21.07 ± 1.24 * | 2.77 ± 0.32 | 7.16 ± 0.76 * | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Deckers, J.; Hendrix, S.; Prinsen, E.; Vangronsveld, J.; Cuypers, A. Glutathione Is Required for the Early Alert Response and Subsequent Acclimation in Cadmium-Exposed Arabidopsis thaliana Plants. Antioxidants 2022, 11, 6. https://doi.org/10.3390/antiox11010006
Deckers J, Hendrix S, Prinsen E, Vangronsveld J, Cuypers A. Glutathione Is Required for the Early Alert Response and Subsequent Acclimation in Cadmium-Exposed Arabidopsis thaliana Plants. Antioxidants. 2022; 11(1):6. https://doi.org/10.3390/antiox11010006
Chicago/Turabian StyleDeckers, Jana, Sophie Hendrix, Els Prinsen, Jaco Vangronsveld, and Ann Cuypers. 2022. "Glutathione Is Required for the Early Alert Response and Subsequent Acclimation in Cadmium-Exposed Arabidopsis thaliana Plants" Antioxidants 11, no. 1: 6. https://doi.org/10.3390/antiox11010006
APA StyleDeckers, J., Hendrix, S., Prinsen, E., Vangronsveld, J., & Cuypers, A. (2022). Glutathione Is Required for the Early Alert Response and Subsequent Acclimation in Cadmium-Exposed Arabidopsis thaliana Plants. Antioxidants, 11(1), 6. https://doi.org/10.3390/antiox11010006








