Extra Virgin Olive Oil Extracts Modulate the Inflammatory Ability of Murine Dendritic Cells Based on Their Polyphenols Pattern: Correlation between Chemical Composition and Biological Function
Abstract
:1. Introduction
2. Materials and Methods
2.1. EVOO Samples
2.2. Mice
2.3. Culture and Treatment of Murine BMDCs
2.4. Extraction and HPLC-UV-MS/MS Analysis of Polyphenols from EVOO
2.5. ELISA
2.6. Cytofluorimetric Analysis
2.7. Gene Expression Analysis by Real-Time PCR
2.8. Statistical Analysis
3. Results and Discussion
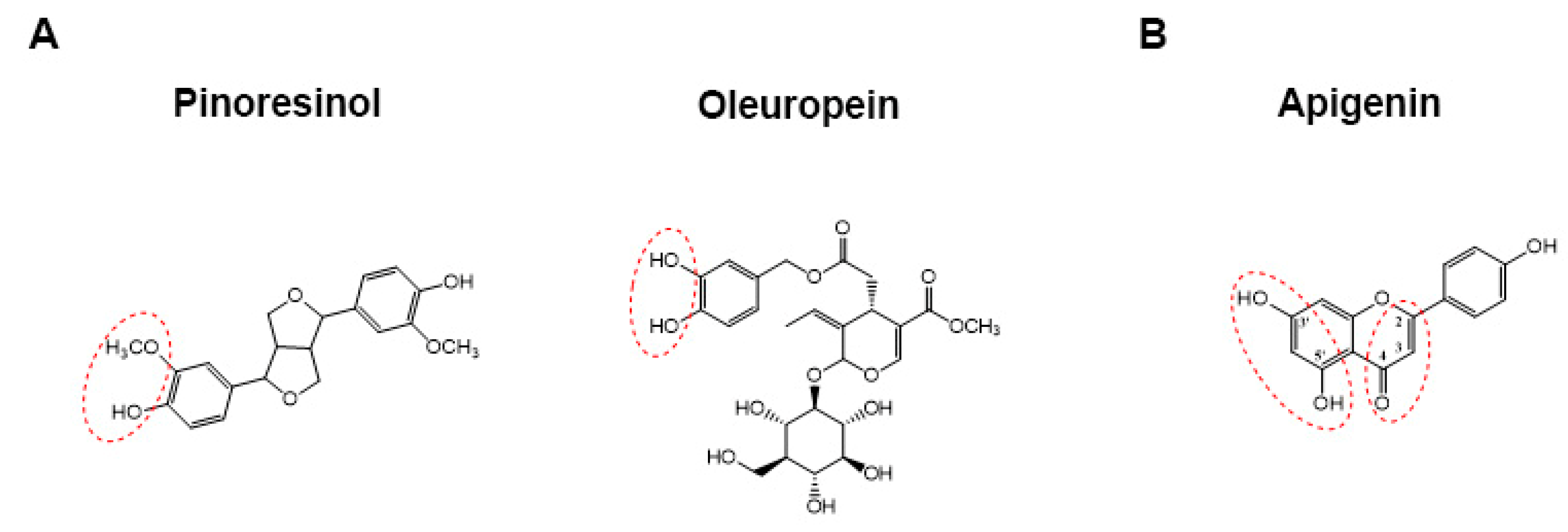
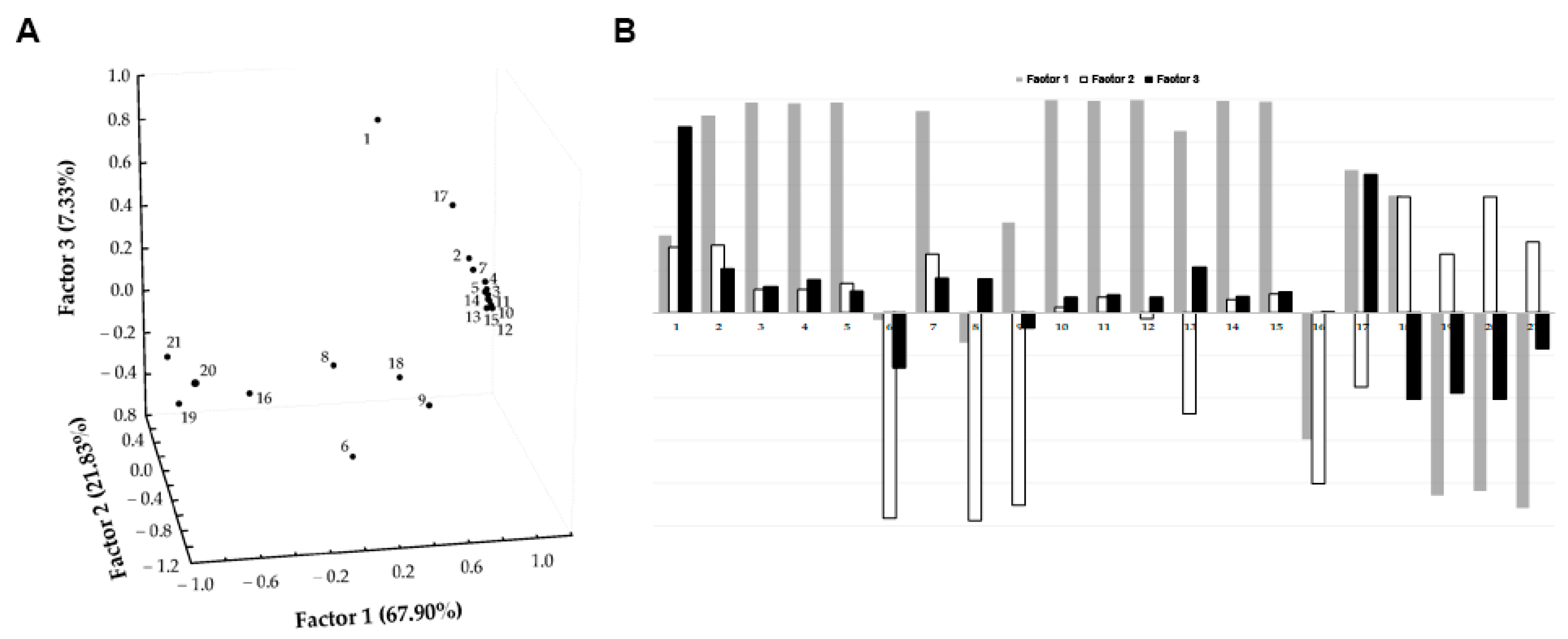
3.1. Chemical Characterization of EVOO Extracts by HPLC-UV-MS/MS Analyses
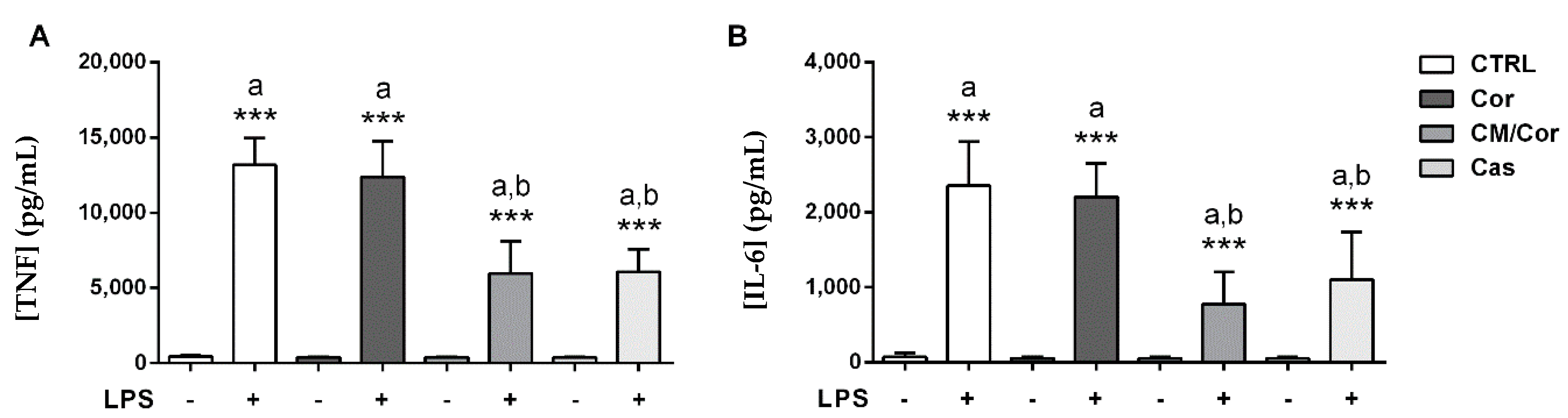
3.2. EVOO Extracts Are Able to Differently Suppress Proinflammatory Cytokine Secretion in LPS-Stimulated BMDCs Based on Their Chemical Composition
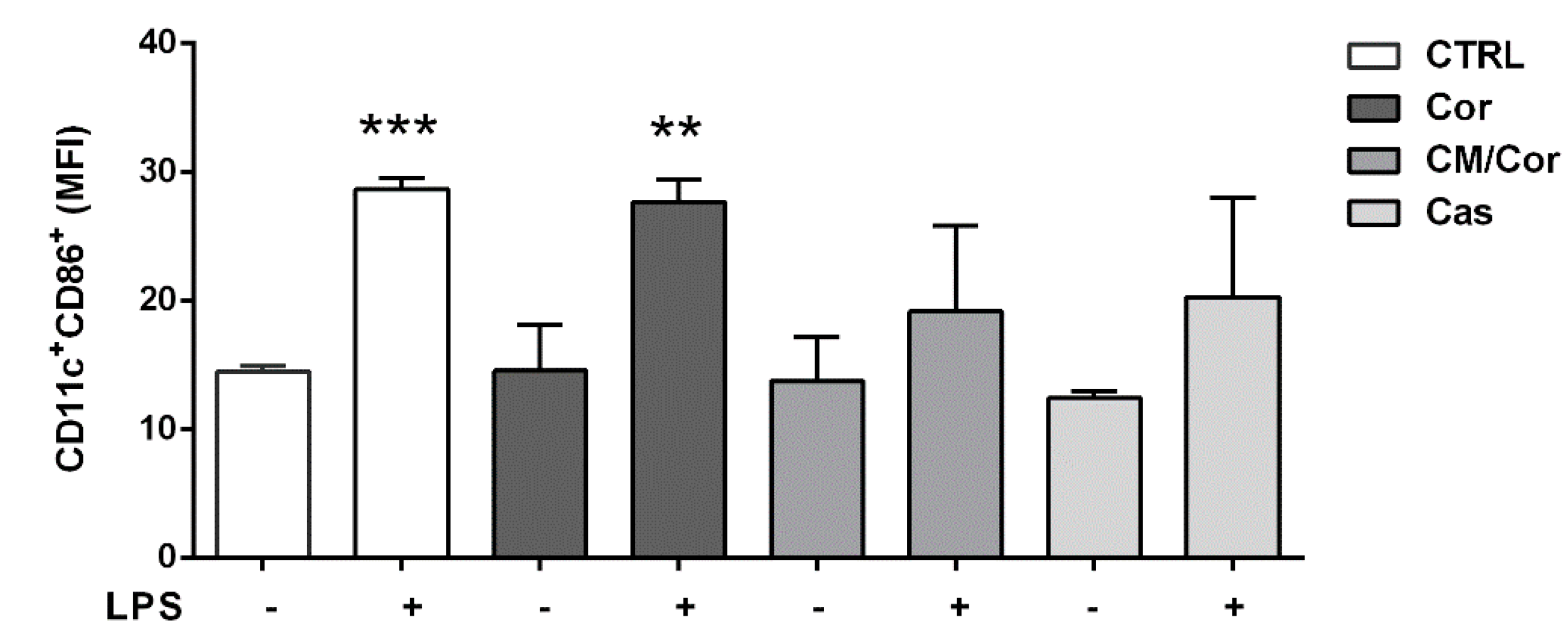
3.3. Polyphenols Composition of EVOO Extracts Can Differently Modulate the Expression of CD86 Costimulatory Molecule in BMDCs after LPS Stimulation
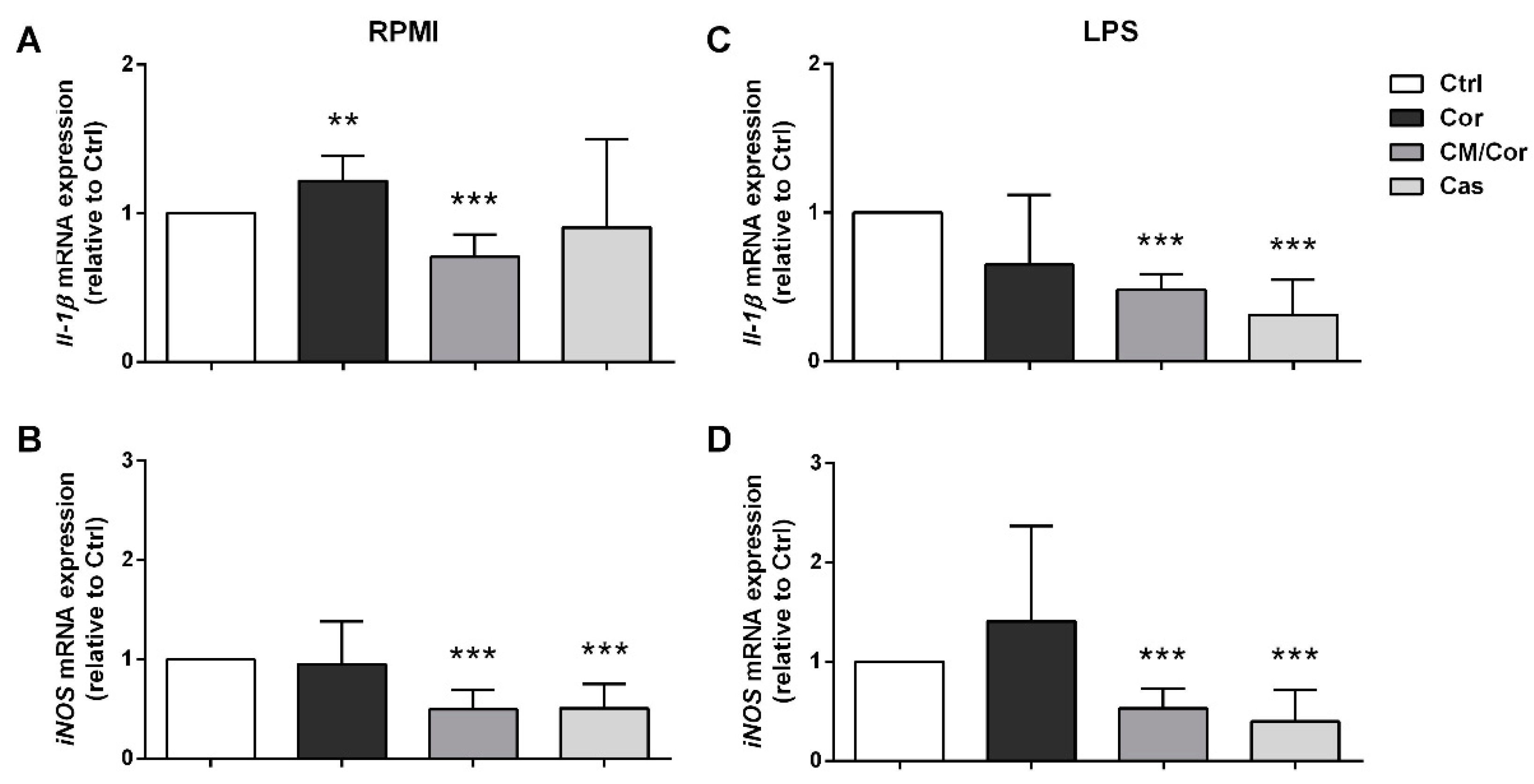
3.4. The Molecular Profile of BMDCs Is Differently Modulated by Specific Combinations of Polyphenols from EVOO Extracts Even without an Inflammatory Stimulus
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- WHO. The Top 10 Causes of Death. 2020. Available online: https://www.who.int/news-room/fact-sheets/detail/the-top-10-causes-of-death (accessed on 30 March 2021).
- Furman, D.; Campisi, J.; Verdin, E.; Carrera-Bastos, P.; Targ, S.; Franceschi, C.; Ferrucci, L.; Gilroy, D.W.; Fasano, A.; Miller, G.W.; et al. Chronic inflammation in the etiology of disease across the life span. Nat. Med. 2019, 25, 1822–1832. [Google Scholar] [CrossRef] [PubMed]
- Ojo, O. Nutrition and Chronic Conditions. Nutrients 2019, 11, 459. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buckland, G.; Gonzalez, C.A. The role of olive oil in disease prevention: A focus on the recent epidemiological evidence from cohort studies and dietary intervention trials. Br. J. Nutr. 2015, 113, S94–S101. [Google Scholar] [CrossRef] [PubMed]
- De Santis, S.; Cariello, M.; Piccinin, E.; Sabba, C.; Moschetta, A. Extra Virgin Olive Oil: Lesson from Nutrigenomics. Nutrients 2019, 11, 2085. [Google Scholar] [CrossRef] [Green Version]
- Tripoli, E.; Giammanco, M.; Tabacchi, G.; Di Majo, D.; Giammanco, S.; La Guardia, M. The phenolic compounds of olive oil: Structure, biological activity and beneficial effects on human health. Nutr. Res. Rev. 2005, 18, 98–112. [Google Scholar] [CrossRef]
- Boskou DTMBG. Olive Oil: Chemistry and Technology, 2nd ed.; AOCS Press: Champaign, IL, USA, 2006; pp. 73–92. [Google Scholar]
- Abbattista, R.; Losito, I.; Castellaneta, A.; De Ceglie, C.; Calvano, C.D.; Cataldi, T.R.I. Insight into the Storage-Related Oxidative/Hydrolytic Degradation of Olive Oil Secoiridoids by Liquid Chromatography and High-Resolution Fourier Transform Mass Spectrometry. J. Agric. Food Chem. 2020, 68, 12310–12325. [Google Scholar] [CrossRef]
- Borges, T.H.; Serna, A.; López, L.C.; Lara, L.; Nieto, R.; Seiquer, I. Composition and Antioxidant Properties of Spanish Extra Virgin Olive Oil Regarding Cultivar, Harvest Year and Crop Stage. Antioxidants 2019, 8, 217. [Google Scholar] [CrossRef] [Green Version]
- Cariello, M.; Contursi, A.; Gadaleta, R.M.; Piccinin, E.; De Santis, S.; Piglionica, M.; Spaziante, A.F.; Sabbà, C.; Villani, G.; Moschetta, A. Extra-Virgin Olive Oil from Apulian Cultivars and Intestinal Inflammation. Nutrients 2020, 12, 1084. [Google Scholar] [CrossRef] [Green Version]
- Clodoveo, M.L.; Dipalmo, T.; Crupi, P.; Durante, V.; Pesce, V.; Maiellaro, I.; Lovece, A.; Mercurio, A.; Laghezza, A.; Corbo, F.; et al. Comparison Between Different Flavored Olive Oil Production Techniques: Healthy Value and Process Efficiency. Plant Foods Hum. Nutr. 2016, 71, 81–87. [Google Scholar] [CrossRef]
- Clodoveo, M.L.; Moramarco, V.; Paduano, A.; Sacchi, R.; Di Palmo, T.; Crupi, P.; Corbo, F.; Pesce, V.; Distaso, E.; Tamburrano, P.; et al. Engineering design and prototype development of a full scale ultrasound system for virgin olive oil by means of numerical and experimental analysis. Ultrason. Sonochem. 2017, 37, 169–181. [Google Scholar] [CrossRef]
- Cerretani, L. Phenolic Fraction of Virgin Olive Oil: An Overview on Identified Compounds and Analytical Methods for their Determination. Funct. Plant Sci. Biotechnol. 2009, 3, 69–80. [Google Scholar]
- Ansary, J.; Cianciosi, D. Natural antioxidants: Is the research going in the right direction? Mediterr. J. Nutr. Metab. 2020, 13, 187–191. [Google Scholar] [CrossRef]
- De Santis, S.; Clodoveo, M.L.; Cariello, M.; D’Amato, G.; Franchini, C.; Faienza, M.F.; Corbo, F. Polyphenols and obesity prevention: Critical insights on molecular regulation, bioavailability and dose in preclinical and clinical settings. Crit. Rev. Food Sci. Nutr. 2021, 61, 1804–1826. [Google Scholar]
- Corona, G.; Deiana, M.; Incani, A.; Vauzour, D.; Dessi, M.A.; Spencer, J.P. Hydroxytyrosol inhibits the proliferation of human colon adenocarcinoma cells through inhibition of ERK1/2 and cyclin D1. Mol. Nutr. Food Res. 2009, 53, 897–903. [Google Scholar] [CrossRef]
- Cusimano, A.; Balasus, D.; Azzolina, A.; Augello, G.; Emma, M.R.; Di Sano, C.; Gramignoli, R.; Strom, S.C.; Mccubrey, J.A.; Montalto, G.; et al. Oleocanthal exerts antitumor effects on human liver and colon cancer cells through ROS generation. Int. J. Oncol. 2017, 51, 533–544. [Google Scholar] [CrossRef] [Green Version]
- Pei, T.; Meng, Q.; Han, J.; Sun, H.; Li, L.; Song, R.; Sun, B.; Pan, S.; Liang, D.; Liu, L. (−)-Oleocanthal inhibits growth and metastasis by blocking activation of STAT3 in human hepatocellular carcinoma. Oncotarget 2016, 7, 43475–43491. [Google Scholar] [CrossRef] [Green Version]
- Zhao, B.; Ma, Y.; Xu, Z.; Wang, J.; Wang, F.; Wang, D.; Pan, S.; Wu, Y.; Pan, H.; Xu, D.; et al. Hydroxytyrosol, a natural molecule from olive oil, suppresses the growth of human hepatocellular carcinoma cells via inactivating AKT and nuclear factor-kappa B pathways. Cancer Lett. 2014, 347, 79–87. [Google Scholar] [CrossRef]
- Dell’Agli, M.; Fagnani, R.; Galli, G.V.; Maschi, O.; Gilardi, F.; Bellosta, S.; Crestani, M.; Bosisio, E.; De Fabiani, E.; Caruso, D. Olive oil phenols modulate the expression of metalloproteinase 9 in THP-1 cells by acting on nuclear factor-kappaB signaling. J. Agric. Food Chem. 2010, 58, 2246–2252. [Google Scholar] [CrossRef]
- Bigagli, E.; Cinci, L.; Paccosi, S.; Parenti, A.; D’Ambrosio, M.; Luceri, C. Nutritionally relevant concentrations of resveratrol and hydroxytyrosol mitigate oxidative burst of human granulocytes and monocytes and the production of pro-inflammatory mediators in LPS-stimulated RAW 264.7 macrophages. Int. Immunopharmacol. 2017, 43, 147–155. [Google Scholar] [CrossRef]
- Maiuri, M.C.; De Stefano, D.; Di Meglio, P.; Irace, C.; Savarese, M.; Sacchi, R.; Cinelli, M.P.; Carnuccio, R. Hydroxytyrosol, a phenolic compound from virgin olive oil, prevents macrophage activation. Naunyn-Schmiedeberg’s Arch. Pharmacol. 2005, 371, 457–465. [Google Scholar] [CrossRef]
- Kim, J.S.; Jobin, C. The flavonoid luteolin prevents lipopolysaccharide-induced NF-kappaB signalling and gene expression by blocking IkappaB kinase activity in intestinal epithelial cells and bone-marrow derived dendritic cells. Immunology 2005, 115, 375–387. [Google Scholar] [CrossRef]
- Yoon, M.-S.; Lee, J.S.; Choi, B.-M.; Jeong, Y.-I.; Lee, C.-M.; Park, J.-H.; Moon, Y.; Sung, S.-C.; Lee, S.K.; Chang, Y.H.; et al. Apigenin inhibits immunostimulatory function of dendritic cells: Implication of Immunotherapeutic adjuvant. Mol. Pharmacol. 2006, 70, 1033–1044. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Banchereau, J.; Steinman, R.M. Dendritic cells and the control of immunity. Nature 1998, 392, 245–252. [Google Scholar] [CrossRef]
- Eri, R.; Chieppa, M. Messages from the Inside. The Dynamic Environment that Favors Intestinal Homeostasis. Front. Immunol. 2013, 4, 323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reis e Sousa, C. Activation of dendritic cells: Translating innate into adaptive immunity. Curr. Opin. Immunol. 2004, 16, 21–25. [Google Scholar] [CrossRef] [PubMed]
- Rescigno, M.; Martino, M.; Sutherland, C.L.; Gold, M.; Ricciardi-Castagnoli, P.; Williams, N.S.; Moore, T.A.; Schatzle, J.D.; Puzanov, I.J.; Sivakumar, P.; et al. Dendritic cell survival and maturation are regulated by different signaling pathways. J. Exp. Med. 1998, 188, 2175–2180. [Google Scholar] [CrossRef]
- Joffre, O.; Nolte, M.A.; Spörri, R.; E Sousa, C.R. Inflammatory signals in dendritic cell activation and the induction of adaptive immunity. Immunol. Rev. 2009, 227, 234–247. [Google Scholar] [CrossRef] [PubMed]
- Cavalcanti, E.; Vadrucci, E.; Delvecchio, F.R.; Addabbo, F.; Bettini, S.; Liou, R.; Monsurrò, V.; Huang, A.Y.-C.; Pizarro, T.T.; Santino, A.; et al. Administration of reconstituted polyphenol oil bodies efficiently suppresses dendritic cell inflammatory pathways and acute intestinal inflammation. PLoS ONE 2014, 9, e88898. [Google Scholar] [CrossRef] [Green Version]
- De Santis, S.; Galleggiante, V.; Scandiffio, L.; Liso, M.; Sommella, E.; Sobolewski, A.; Spilotro, V.; Pinto, A.; Campiglia, P.; Serino, G.; et al. Secretory Leukoprotease Inhibitor (Slpi) Expression Is Required for Educating Murine Dendritic Cells Inflammatory Response Following Quercetin Exposure. Nutrients 2017, 9, 706. [Google Scholar] [CrossRef] [Green Version]
- De Santis, S.; Kunde, D.; Serino, G.; Galleggiante, V.; Caruso, M.L.; Mastronardi, M.; Cavalcanti, E.; Ranson, N.; Pinto, A.; Campiglia, P.; et al. Secretory leukoprotease inhibitor is required for efficient quercetin-mediated suppression of TNFalpha secretion. Oncotarget 2016, 7, 75800–75809. [Google Scholar] [CrossRef] [Green Version]
- Delvecchio, F.R.; Vadrucci, E.; Cavalcanti, E.; De Santis, S.; Kunde, D.; Vacca, M.; Myers, J.; Allen, F.; Bianco, G.; Huang, A.Y.; et al. Polyphenol administration impairs T-cell proliferation by imprinting a distinct dendritic cell maturational profile. Eur. J. Immunol. 2015, 45, 2638–2649. [Google Scholar] [CrossRef]
- del Corno, M.; Scazzocchio, B.; Masella, R.; Gessani, S. Regulation of Dendritic Cell Function by Dietary Polyphenols. Crit. Rev. Food Sci. Nutr. 2016, 56, 737–747. [Google Scholar] [CrossRef]
- Santangelo, C.; Varì, R.; Scazzocchio, B.; De Sancti, P.; Giovannini, C.; D’Archivio, M.; Masella, R. Anti-inflammatory Activity of Extra Virgin Olive Oil Polyphenols: Which Role in the Prevention and Treatment of Immune-Mediated Inflammatory Diseases? Endocr. Metab. Immune Disord. Drug Targets 2017, 18, 36–50. [Google Scholar] [CrossRef]
- de la Torre-Carbot, K.; Jauregui, O.; Gimeno, E.; Castellote, A.I.; Lamuela-Raventos, R.M.; Lopez-Sabater, M.C. Characterization and quantification of phenolic compounds in olive oils by solid-phase extraction, HPLC-DAD, and HPLC-MS/MS. J. Agric. Food Chem. 2005, 53, 4331–4340. [Google Scholar] [CrossRef]
- Negro, C.; Aprile, A.; Luvisi, A.; Nicolì, F.; Nutricati, E.; Vergine, M.; Miceli, A.; Blando, F.; Sabella, E.; De Bellis, L. Phenolic Profile and Antioxidant Activity of Italian Monovarietal Extra Virgin Olive Oils. Antioxidants 2019, 8, 161. [Google Scholar] [CrossRef] [Green Version]
- De Nino, A.; Mazzotti, F.; Perri, E.; Procopio, A.; Raffaelli, A.; Sindona, G. Virtual freezing of the hemiacetal-aldehyde equilibrium of the aglycones of oleuropein and ligstroside present in olive oils from Carolea and Coratina cultivars by ionspray ionization tandem mass spectrometry. J. Mass Spectrom. 2000, 35, 461–467. [Google Scholar] [CrossRef]
- Lenschow, D.J.; Walunas, T.L.; Bluestone, J.A. CD28/B7 system of T cell costimulation. Annu. Rev. Immunol. 1996, 14, 233–258. [Google Scholar] [CrossRef]
- Xue, Q.; Yan, Y.; Zhang, R.; Xiong, H. Regulation of iNOS on Immune Cells and Its Role in Diseases. Int. J. Mol. Sci. 2018, 19, 3805. [Google Scholar] [CrossRef] [Green Version]
- Scalavino, V.; Liso, M.; Cavalcanti, E.; Gigante, I.; Lippolis, A.; Mastronardi, M.; Chieppa, M.; Serino, G. miR-369-3p modulates inducible nitric oxide synthase and is involved in regulation of chronic inflammatory response. Sci. Rep. 2020, 10, 15942. [Google Scholar] [CrossRef]
- Lu, J.; Huang, G.; Wang, Z.; Zhuang, S.; Xu, L.; Song, B.; Xiong, Y.; Guan, S. Tyrosol exhibits negative regulatory effects on LPS response and endotoxemia. Food Chem. Toxicol. 2013, 62, 172–178. [Google Scholar] [CrossRef]
- Richard, N.; Arnold, S.; Hoeller, U.; Kilpert, C.; Wertz, K.; Schwager, J. Hydroxytyrosol is the major anti-inflammatory compound in aqueous olive extracts and impairs cytokine and chemokine production in macrophages. Planta Med. 2011, 77, 1890–1897. [Google Scholar] [CrossRef] [Green Version]
- Rodríguez-López, P.; Lozano-Sanchez, J.; Borrás-Linares, I.; Emanuelli, T.; Menéndez, J.A.; Segura-Carretero, A. Structure–Biological Activity Relationships of Extra-Virgin Olive Oil Phenolic Compounds: Health Properties and Bioavailability. Antioxidants 2020, 9, 685. [Google Scholar] [CrossRef]
- García-Villalba, R.; Carrasco-Pancorbo, A.; Oliveras-Ferraros, C.; Menendez, J.A.; Segura-Carretero, A.; Fernández-Gutiérrez, A. Uptake and metabolism of olive oil polyphenols in human breast cancer cells using nano-liquid chromatography coupled to electrospray ionization–time of flight-mass spectrometry. J. Chromatogr. B 2012, 898, 69–77. [Google Scholar] [CrossRef]
- García-Villalba, R.; Larrosa, M.; Possemiers, S.; Tomás-Barberán, F.A.; Espín, J.C. Bioavailability of phenolics from an oleuropein-rich olive (Olea europaea) leaf extract and its acute effect on plasma antioxidant status: Comparison between pre- and postmenopausal women. Eur. J. Nutr. 2014, 53, 1015–1027. [Google Scholar] [CrossRef]
- Koudoufio, M.; Desjardins, Y.; Feldman, F.; Spahis, S.; Delvin, E.; Levy, E. Insight into Polyphenol and Gut Microbiota Crosstalk: Are Their Metabolites the Key to Understand Protective Effects Against Metabolic Disorders? Antioxidants 2020, 9, 982. [Google Scholar] [CrossRef]
- Kawabata, K.; Yoshioka, Y.; Terao, J. Role of Intestinal Microbiota in the Bioavailability and Physiological Functions of Dietary Polyphenols. Molecules 2019, 24, 370. [Google Scholar] [CrossRef] [Green Version]
- Vamanu, E.; Gatea, F. Correlations between Microbiota Bioactivity and Bioavailability of Functional Compounds: A Mini-Review. Biomedicines 2020, 8, 39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Renaud, J.; Martinoli, M.-G. Considerations for the Use of Polyphenols as Therapies in Neurodegenerative Diseases. Int. J. Mol. Sci. 2019, 20, 1883. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mileo, A.M.; Nisticò, P.; Miccadei, S. Polyphenols: Immunomodulatory and Therapeutic Implication in Colorectal Cancer. Front. Immunol. 2019, 10, 729. [Google Scholar] [CrossRef] [PubMed]
- De Angelis, M.; Ferrocino, I.; Calabrese, F.M.; De Filippis, F.; Cavallo, N.; Siragusa, S.; Rampelli, S.; Di Cagno, R.; Rantsiou, K.; Vannini, L.; et al. Diet influences the functions of the human intestinal microbiome. Sci. Rep. 2020, 10, 4247. [Google Scholar] [CrossRef] [Green Version]
- Hachimura, S.; Totsuka, M.; Hosono, A. Immunomodulation by food: Impact on gut immunity and immune cell function. Biosci. Biotechnol. Biochem. 2018, 82, 584–599. [Google Scholar] [CrossRef]
- Vitetta, L.; Vitetta, G.; Hall, S. Immunological Tolerance and Function: Associations Between Intestinal Bacteria, Probiotics, Prebiotics, and Phages. Front. Immunol. 2018, 9, 2240. [Google Scholar] [CrossRef] [Green Version]
- Chieppa, M.; Galleggiante, V.; Serino, G.; Massaro, M.; Santino, A. Iron Chelators Dictate Immune Cells Inflammatory Ability: Potential Adjuvant Therapy for IBD. Curr. Pharm. Des. 2017, 23, 2289–2298. [Google Scholar] [CrossRef]
- Chieppa, M.; Giannelli, G. Immune Cells and Microbiota Response to Iron Starvation. Front. Med. 2018, 5, 109. [Google Scholar] [CrossRef] [Green Version]
- EFSA. SCIENTIFIC OPINION—EFSA Panel on Dietetic Products, Nutrition and Allergies (NDA). EFSA J. 2011, 9, 2033. [Google Scholar]
| Compound | RT (min) | [M-H]−(m/z) | MS/MS Experiments m/z (% Base Peak) | Cor | CM/Cor | Cas |
|---|---|---|---|---|---|---|
| 3-hydroxytyrosol a | 2.918 ± 0.012 | 153.0 | 123.4 (100), 95.4 (7) | 53 (5) a,f,g | 4.4 (0.4) b | 1.37 (0.14) b |
| Decarboxymethyl-elenolic acid derivative b | 3.512 ± 0.014 | 185.1 | 111.5 (100), 95.2 (66), 69.2 (8) | 1.56 (0.15) a | 0.026 (0.003) b | n.d. |
| Decarboxymethyl elenolic acid b | 4.19 ± 0.02 | 199.0 | 111.4 (91), 95.1 (28), 85.2 (56), 69.4 (81), 59.2 (100) | 7.8 (0.8) a | 0.065 (0.006) b | n.d. |
| Tyrosol b | 4.63 ± 0.03 | 137.0 | 119.2 (100) | 0.020 (0.002) | n.d. | n.d. |
| Oleacinic acid Open Forms II a | 15.418 ± 0.014 | 335.1 | 199.6 (100), 155.3 (14), 111.3 (11), 59.4 (6) | 10.8 (1.1) a | 10.1 (1.0) a | 2.3 (0.2) b |
| Oleuropein aglycone carboxylic acid Open Forms I or II a | 15.46 ± 0.04 | 393.2 | 257.5 (27), 169.4 (100), 111.4 (66) | 0.33 (0.03) a | 0.172 (0.017) b | 0.170 (0.017) b |
| Oleuropein isomer 1 a | 15.51 ± 0.02 | 539.1 | 113.2 (100) | 1.28 (0.13) a | 0.65 (0.06) b | 0.47 (0.05) b |
| Pinoresinol c | 15.53 ± 0.02 | 357.0 | 221.6 (100) | 0.53 (0.05) a | 0.62 (0.06) a | 0.125 (0.013) b |
| Oleuropein aglycone enolic-aldehydic Open Form I a | 15.75 ± 0.04 | 377.1 | 275.7 (27), 149.4 (22), 139.3 (100), 121.4 (9), 111.1 (20), 101.2 (8), 95.2 (41) | 3.6 (0.4) b | 3.6 (0.4) b | 5.8 (0.6) a |
| Oleuropein aglycone enolic-aldehydic Open Form I a | 16.50 ± 0.04 | 377.1 | 275.8 (15), 149.4 (50), 139.3 (100), 121.3 (9), 111.2 (25), 101.2 (10), 95.2 (75) | 10.8 (1.1) b | 10.0 (1.0) b | 15.1 (1.5) a |
| Oleuropein aglycone carboxylic acid Open Forms I or II a | 16.60 ± 0.03 | 393.2 | 257.5 (77), 169.4 (100), 111.4 (66) | 0.30 (0.03) a | 0.174 (0.017) b | 0.153 (0.015) b |
| Oleuropein isomer 2 a | 16.650 ± 0.019 | 539.1 | 113.2 (100) | 0.77 (0.08) a | 0.43 (0.04) b | 0.45 (0.04) b |
| Oleocanthalic acid Open Forms II b | 18.48 ± 0.05 | 319.1 | 199.6 (87), 181.7(10), 155.4(11), 139.3(7), 121.4 (14), 111.3 (100), 85.2 (9) | 7.5 (0.7) a | 7.4 (0.7) a | 1.59 (0.16) b |
| Oleuropein aglycone dialdehydic Open Form I a | 18.64 ± 0.04 | 377.1 | 275.6 (16), 149.3 (43), 139.5 (100), 111.2 (16), 101.3 (13), 95.4 (37) | 2.6 (0.3) a | 1.28 (0.13) b | 1.64 (0.16) b |
| Ligstroside aglycone enolic-aldehydic Open Form I b | 18.69 ± 0.04 | 361.0 | 291.8 (12), 171.3 (21), 139.3 (57), 127.2 (36), 101.3(100), 69.2 (18) | 5.0 (0.5) a,b | 4.5 (0.4) b | 6.2 (0.6) a |
| Luteolin d | 18.99 ± 0.05 | 285.2 | 217.1 (8), 199.0 (39), 175.0 (18), 133.2 (100) | 1.32 (0.13) b | 1.35 (0.13) b | 1.83 (0.18) a |
| Oleuropein aglycone carboxylic acid Open Forms I or II a | 19.30 ± 0.04 | 393.2 | 257.5 (77), 169.4 (100), 111.4 (66) | 0.29 (0.03) a | 0.109 (0.011) b | 0.139 (0.014) b |
| Acetoxy pinoresinol c | 19.43 ± 0.03 | 415.1 | 264.9 (43), 204.9 (18), 136.2 (100) | 0.21 (0.02) b | 0.29 (0.03) b | 0.70 (0.07) a |
| Hydroxy-methyl decarboxymethyl ligstroside aglycone isomer 1 b | 19.50 ± 0.03 | 333.0 | 181.5 (31), 111.3 (100), 99.1 (50), 94.9 (34), 69.4 (33) | 2.2 (0.2) a,b | 1.69 (0.17) b | 2.5 (0.2) a |
| Ligstroside aglycone enolic-aldehydic Open Form I b | 19.50 ± 0.03 | 361.0 | 291.7 (40), 259.7 (12), 171.4 (17), 139.3 (33), 127.4 (37), 101.3 (100), 69.2 (10) | 7.8 (0.8) a,b | 6.6 (0.7) b | 9.0 (0.9) a |
| Oleuropein aglycone carboxylic acid Open Forms I or II a | 19.80 ± 0.03 | 393.2 | 257.5 (77), 169.4 (100), 111.4 (66) | n.q. | n.q. | n.q. |
| Hydroxy-methyl decarboxymethyl ligstroside aglycone isomer 2 b | 19.92 ± 0.04 | 333.0 | 181.6 (28), 153.4 (8), 111.3 (100), 99.1 (50), 94.9 (34), 69.4 (33) | 0.30 (0.03) b | 0.47 (0.05) a | 0.34 (0.03) b |
| Oleuropein aglycone dialdehydic Open Form I a | 20.14 ± 0.03 | 377.1 | 275.7 (19), 149.4 (50), 139.4 (100), 111.2 (23), 101.2 (19), 95.4 (43) | 6.2 (0.6) a | 3.4 (0.3) b | 5.4 (0.5) a |
| Hydroxy-methyl decarboxymethyl ligstroside aglycone isomer 3 b | 20.28 ± 0.04 | 333.0 | 181.6 (28), 153.4 (8), 111.3 (100), 99.1 (50), 94.9 (34), 69.4 (33) | 0.22 (0.02) b | 0.42 (0.04) a | 0.24 (0.02) b |
| Oleuropein aglycone dialdehydic Open Form I a | 20.86 ± 0.02 | 377.1 | 275.7 (28), 149.4 (52), 139.4 (100), 111.3 (41), 101.2 (18), 95.4 (64) | 16.5 (1.6) a | 11.9 (1.2) b | 17.8 (1.8) a |
| Ligstroside aglycone dialdehydic Open Form I b | 21.00 ± 0.02 | 361.1 | 291.8 (37), 259.6 (21), 139.4 (47), 127.3 (46), 101.3 (100), 69.2 (12) | 1.86 (0.19) a | 0.90 (0.09) b | 0.56 (0.06) c |
| Apigenin d | 21.23 ± 0.04 | 269.2 | 150.8 (12), 117.0 (100), 107.1 (35) | 0.54 (0.05) b | 0.66 (0.06) b | 1.02 (0.10) a |
| Methoxyluteolin d | 21.58 ± 0.03 | 299.1 | 227.0 (100), 199.1 (13) | 0.39 (0.04) b | 0.24 (0.02) c | 0.55 (0.05) a |
| Mono enolic-aldehydic oleuropein aglycone Closed FormI a | 21.94 ± 0.03 | 377.1 | 275.7 (30), 149.4 (43), 139.4 (100), 111.2 (44), 101.2 (16), 95.4 (69) | 95 (9) a | 69 (7) b | 96 (10) a |
| Ligstroside aglycone dialdehydic Open Form I b | 22.07 ± 0.07 | 361.1 | 291.7 (44), 171.3 (19), 139.3 (56), 127.3 (41), 101.2 (100), 69.1 (35) | 7.3 (0.7) | 6.4 (0.6) | 6.5 (0.6) |
| Ligstroside aglycone dialdehydic Open Form I b | 22.80 ± 0.02 | 361.1 | 291.7 (45), 171.3 (19), 139.3 (42), 127.3 (63), 101.2 (100), 69.5 (12) | 4.8 (0.5) a | 3.6 (0.4) b | 4.4 (0.4) a,b |
| Mono enolic-aldehydic dihydropyranic ligstroside aglycone Closed FormI b | 24.08 ± 0.03 | 361.1 | 291.7 (32), 259.6 (10), 171.5 (12), 139.4 (38), 127.3 (55), 101.3 (100), 69.3 (15) | 27 (3) a | 18.1 (1.8) b | 16.2 (1.6) b |
| Total polyphenols e | 280 (30) a | 169 (17) b | 200 (20) a,b |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
De Santis, S.; Liso, M.; Verna, G.; Curci, F.; Milani, G.; Faienza, M.F.; Franchini, C.; Moschetta, A.; Chieppa, M.; Clodoveo, M.L.; et al. Extra Virgin Olive Oil Extracts Modulate the Inflammatory Ability of Murine Dendritic Cells Based on Their Polyphenols Pattern: Correlation between Chemical Composition and Biological Function. Antioxidants 2021, 10, 1016. https://doi.org/10.3390/antiox10071016
De Santis S, Liso M, Verna G, Curci F, Milani G, Faienza MF, Franchini C, Moschetta A, Chieppa M, Clodoveo ML, et al. Extra Virgin Olive Oil Extracts Modulate the Inflammatory Ability of Murine Dendritic Cells Based on Their Polyphenols Pattern: Correlation between Chemical Composition and Biological Function. Antioxidants. 2021; 10(7):1016. https://doi.org/10.3390/antiox10071016
Chicago/Turabian StyleDe Santis, Stefania, Marina Liso, Giulio Verna, Francesca Curci, Gualtiero Milani, Maria Felicia Faienza, Carlo Franchini, Antonio Moschetta, Marcello Chieppa, Maria Lisa Clodoveo, and et al. 2021. "Extra Virgin Olive Oil Extracts Modulate the Inflammatory Ability of Murine Dendritic Cells Based on Their Polyphenols Pattern: Correlation between Chemical Composition and Biological Function" Antioxidants 10, no. 7: 1016. https://doi.org/10.3390/antiox10071016
APA StyleDe Santis, S., Liso, M., Verna, G., Curci, F., Milani, G., Faienza, M. F., Franchini, C., Moschetta, A., Chieppa, M., Clodoveo, M. L., Crupi, P., & Corbo, F. (2021). Extra Virgin Olive Oil Extracts Modulate the Inflammatory Ability of Murine Dendritic Cells Based on Their Polyphenols Pattern: Correlation between Chemical Composition and Biological Function. Antioxidants, 10(7), 1016. https://doi.org/10.3390/antiox10071016













