Murine Neonatal Oxidant Lung Injury: NRF2-Dependent Predisposition to Adulthood Respiratory Viral Infection and Protection by Maternal Antioxidant
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Time Mating
2.2. Gestational SFN Administration and Placenta Collection
2.3. Neonate Hyperoxia Exposure
2.4. RSV Infection
2.5. Bronchoalveolar Lavage (BAL) Analyses
2.6. Lung Lipid Oxidation Measurement
2.7. Sandwich Enzyme-Linked Immunosorbent Assay (ELISA)
2.8. Lung Histopathology
2.9. Serum Immunoglobulin E (IgE) Measurement
2.10. SFN Metabolites Detection in Urine and Milk Bands
2.11. Protein Extraction and Detection by Western Blot Analysis
2.12. Protein Oxidation Detection
2.13. Nuclear DNA Binding Assay
2.14. Lung DNA Damage Measurement
2.15. Lung RNA Isolation and Reverse Transcriptase-Polymerase Chain Reaction (RT-PCR)
2.16. cDNA Microarray Analyses
2.17. Statistics
3. Results
3.1. Effect of Neonatal Hyperoxia on Adulthood RSV Pathogenesis
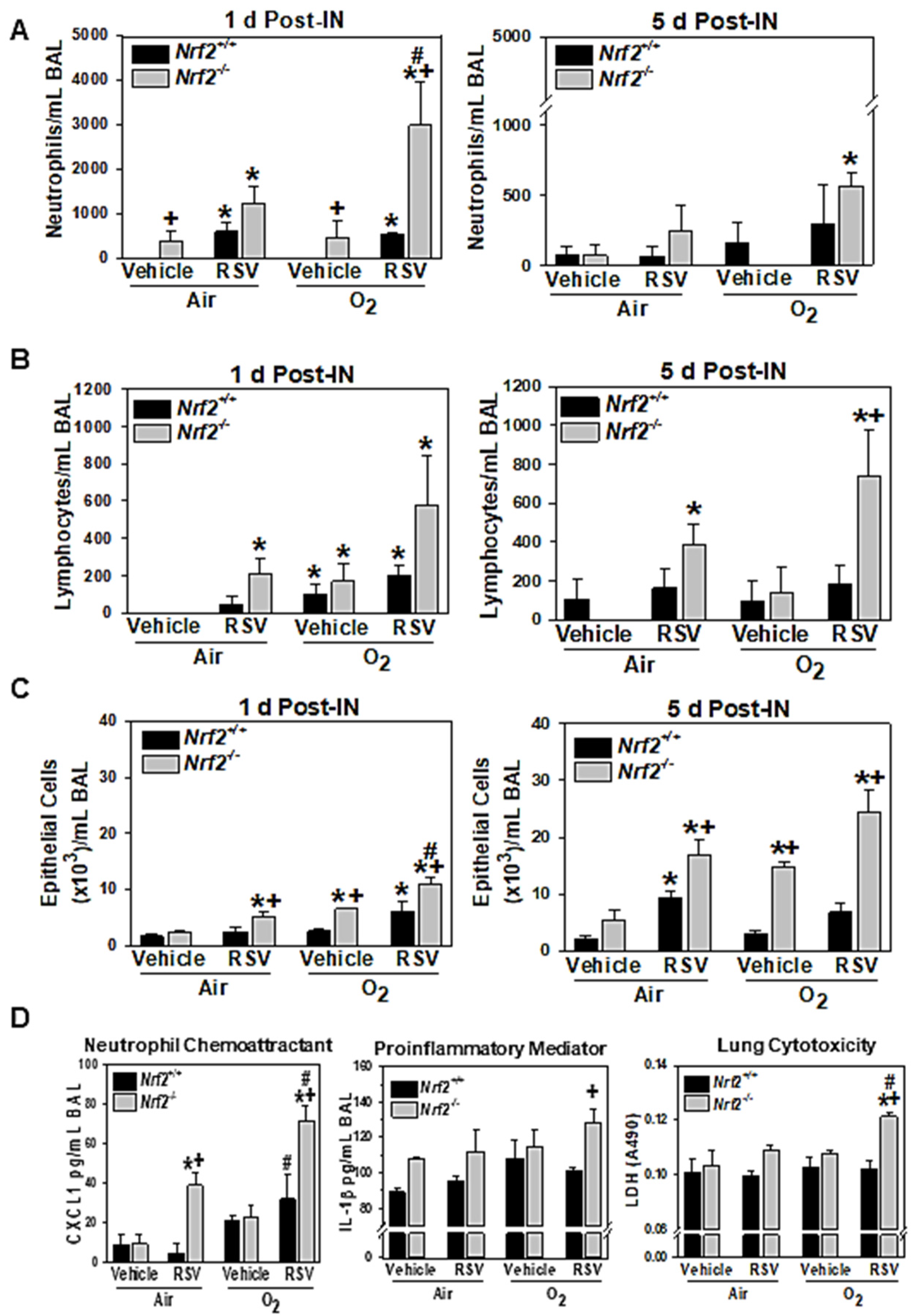
3.1.1. NRF2-Dependent Augmentation of Lung Inflammation and Injury
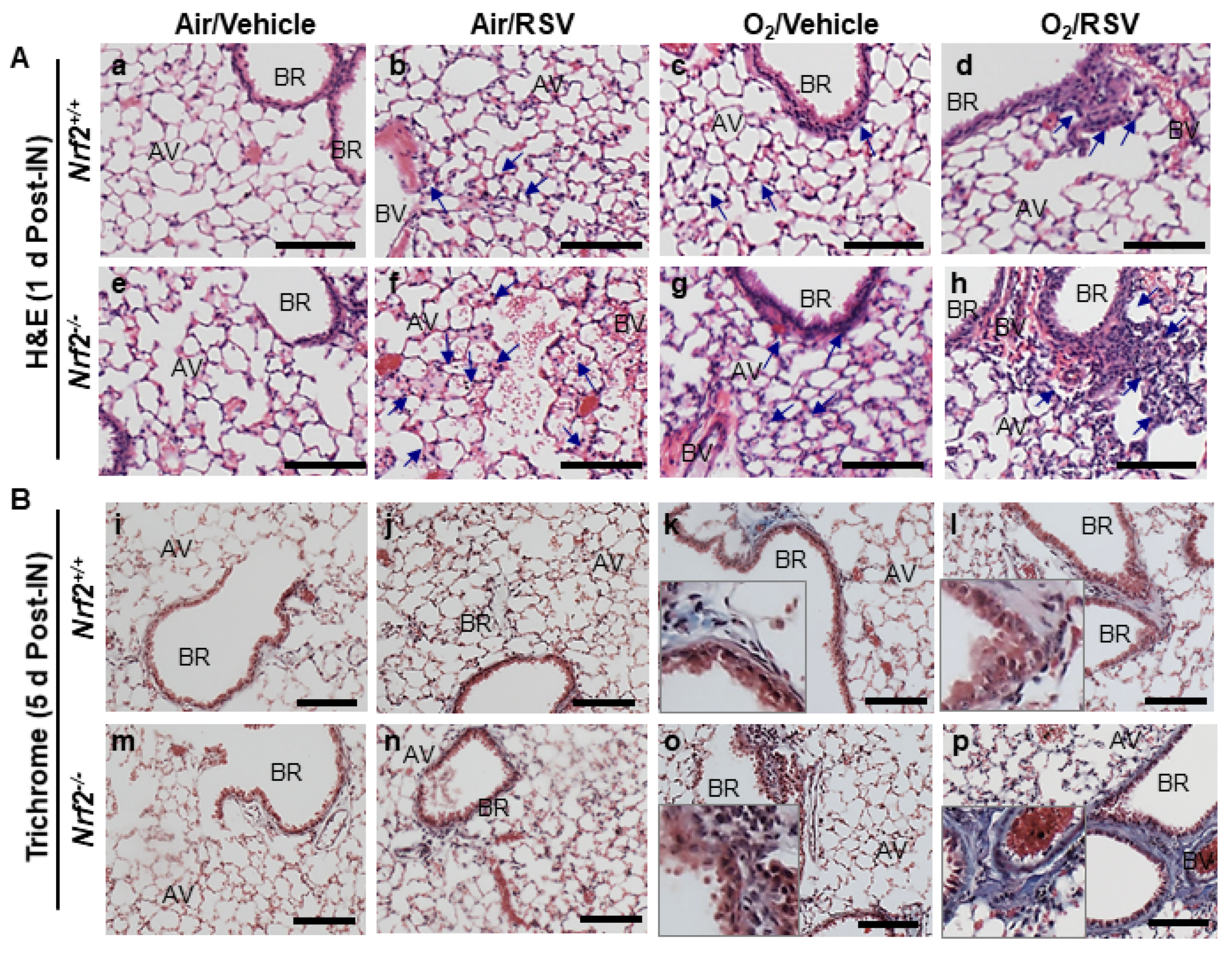
3.1.2. NRF2-Dependent Augmentation of Lung Histopathology
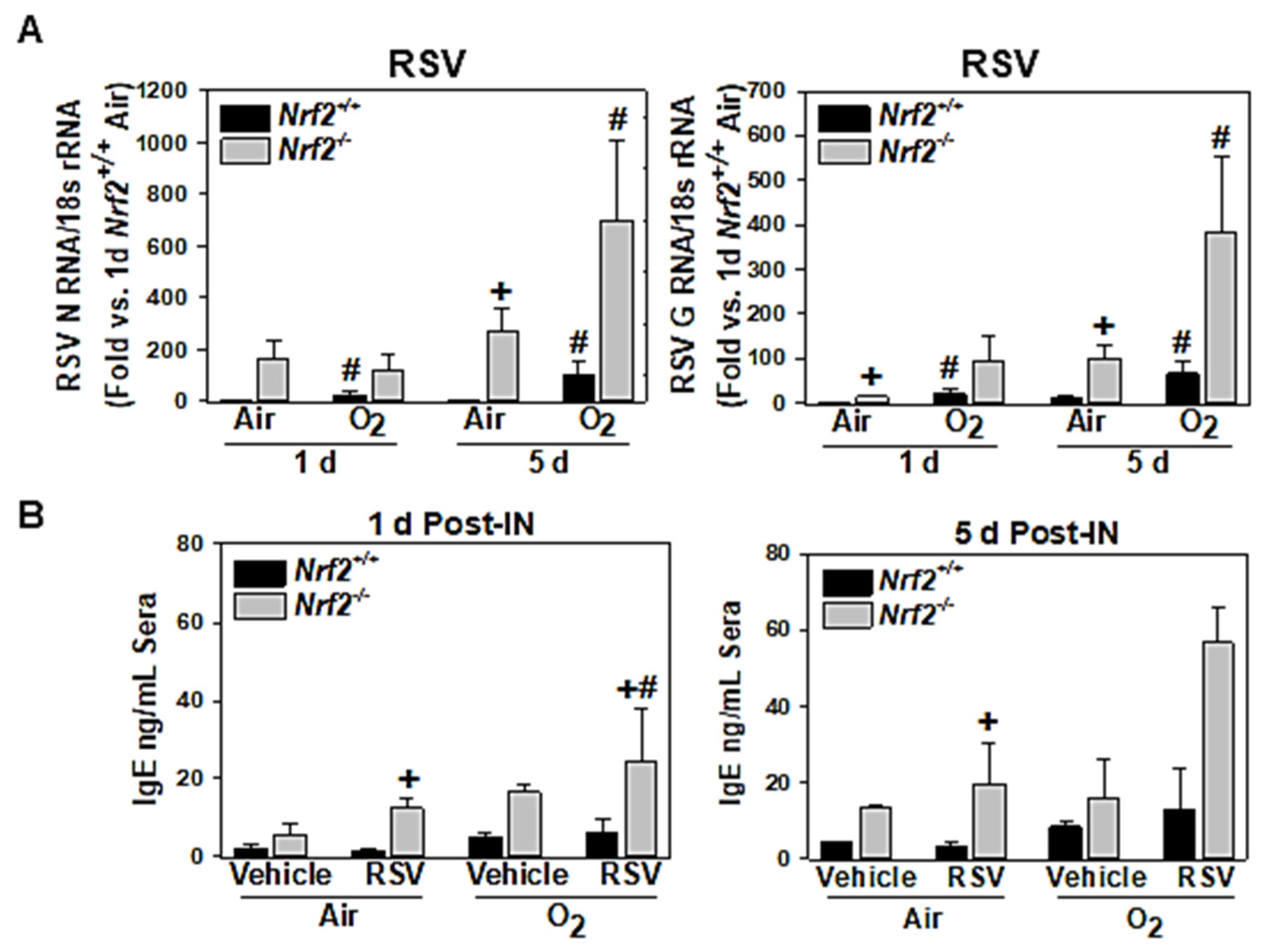
3.1.3. NRF2-Dependent Potentiation of Lung Viral Replication
3.1.4. NRF2-Dependent Potentiation of Serum IgE Increase
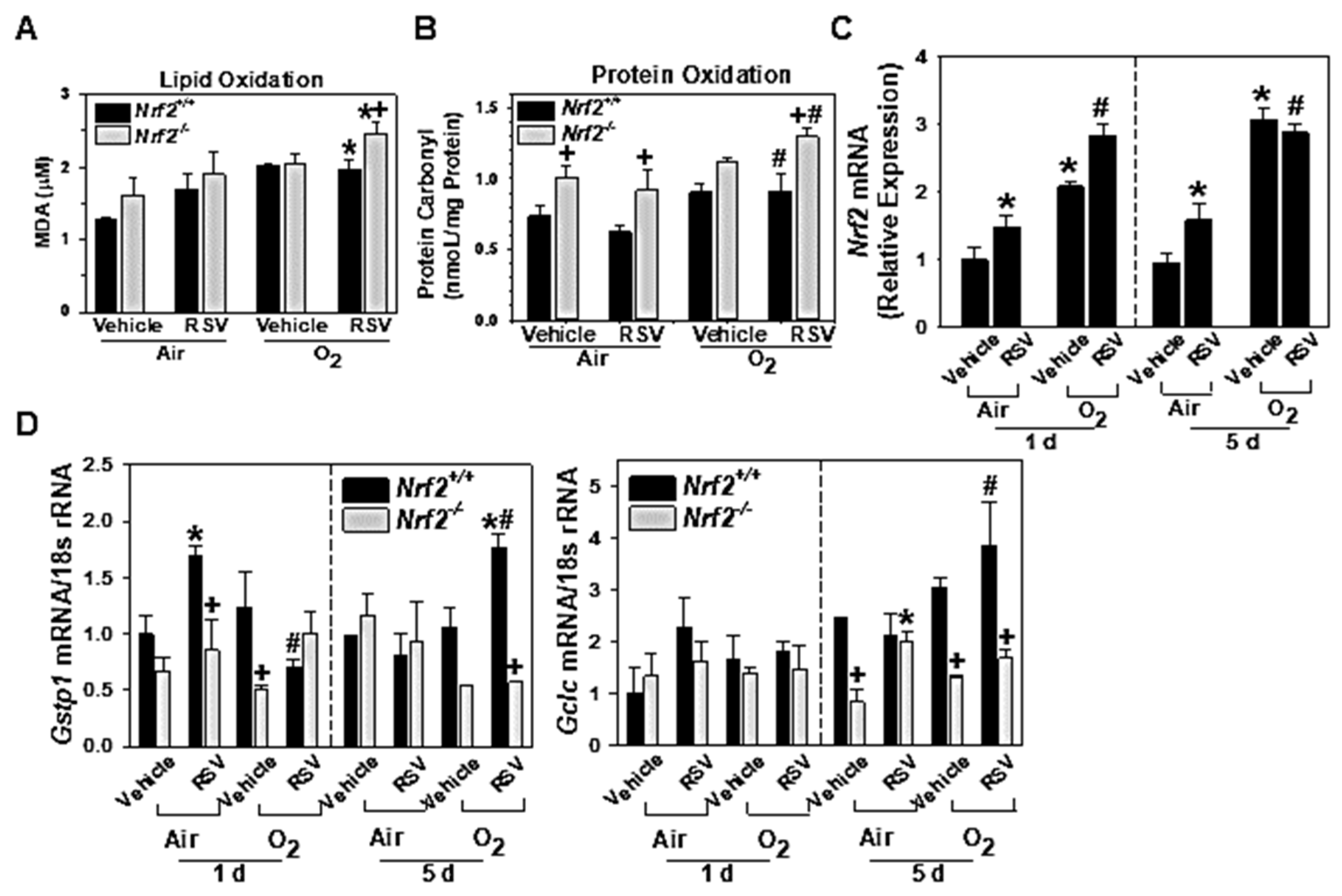
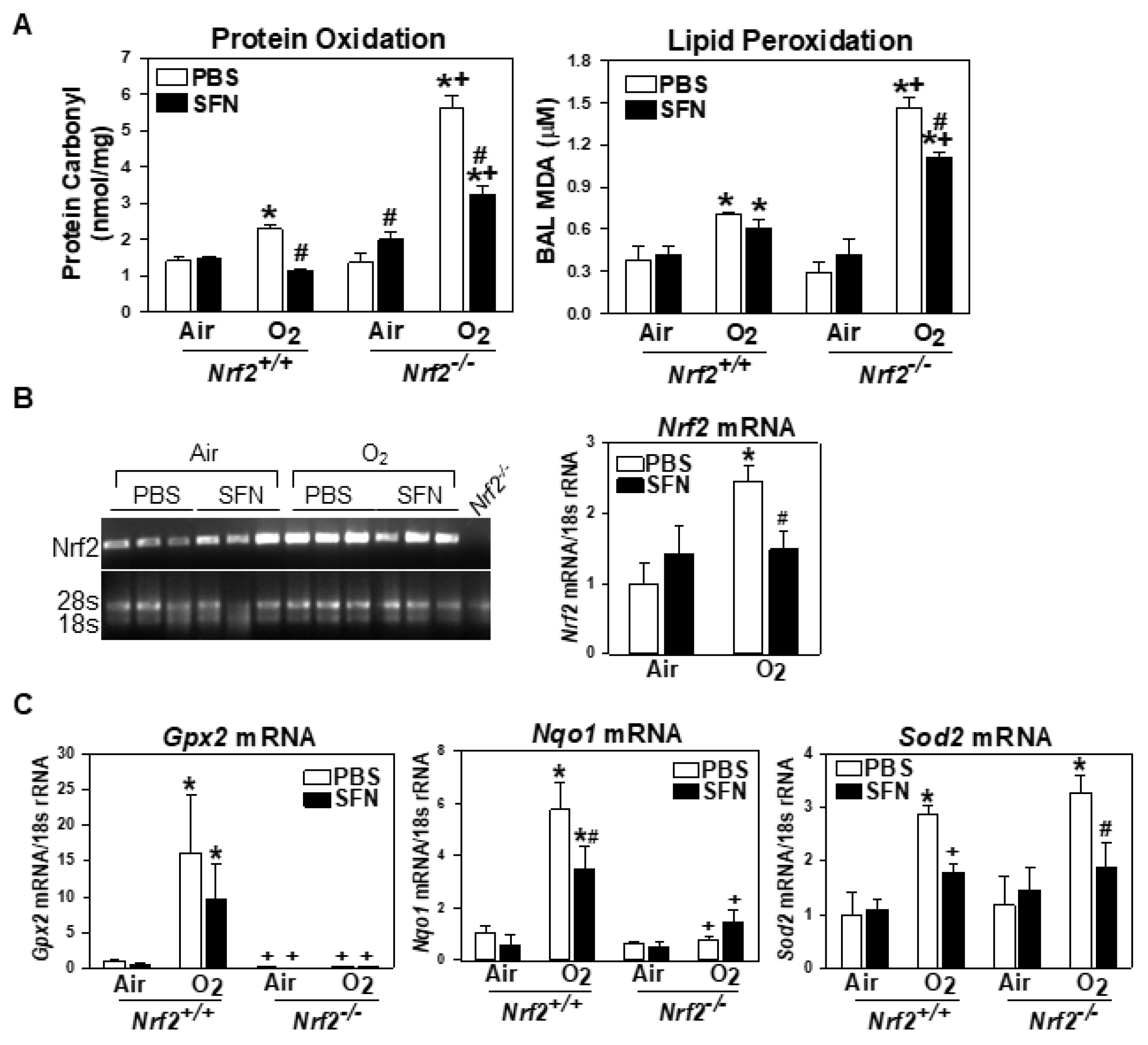
3.1.5. NRF2-Dependent Augmentation of Lung Oxidative Stress
3.1.6. Lung Nrf2 and ARE-Bearing Antioxidant Enzyme Expression
3.2. Effect of Prenatal SFN on Neonatal Hyperoxia-Induced Lung Injury
3.2.1. Decreased Lung Injury in Neonates Supplemented with Prenatal SFN
3.2.2. Detection of Gestational SFN Metabolites in Urine and Milk Bands
3.2.3. Decreased Oxidative Stress and Antioxidant Expression in Neonates by Prenatal SFN
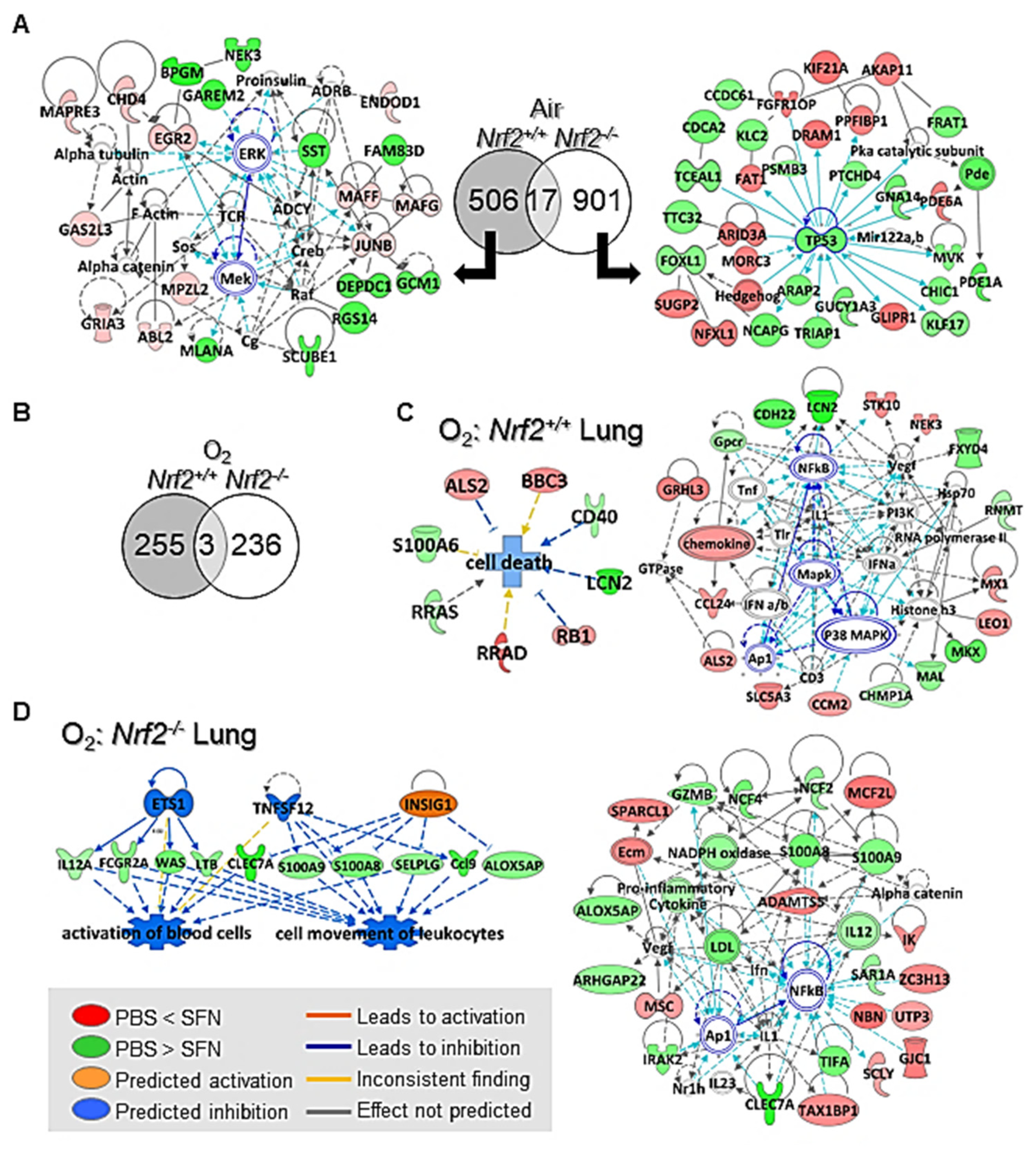
3.2.4. SFN Effects on Lung Transcriptomics
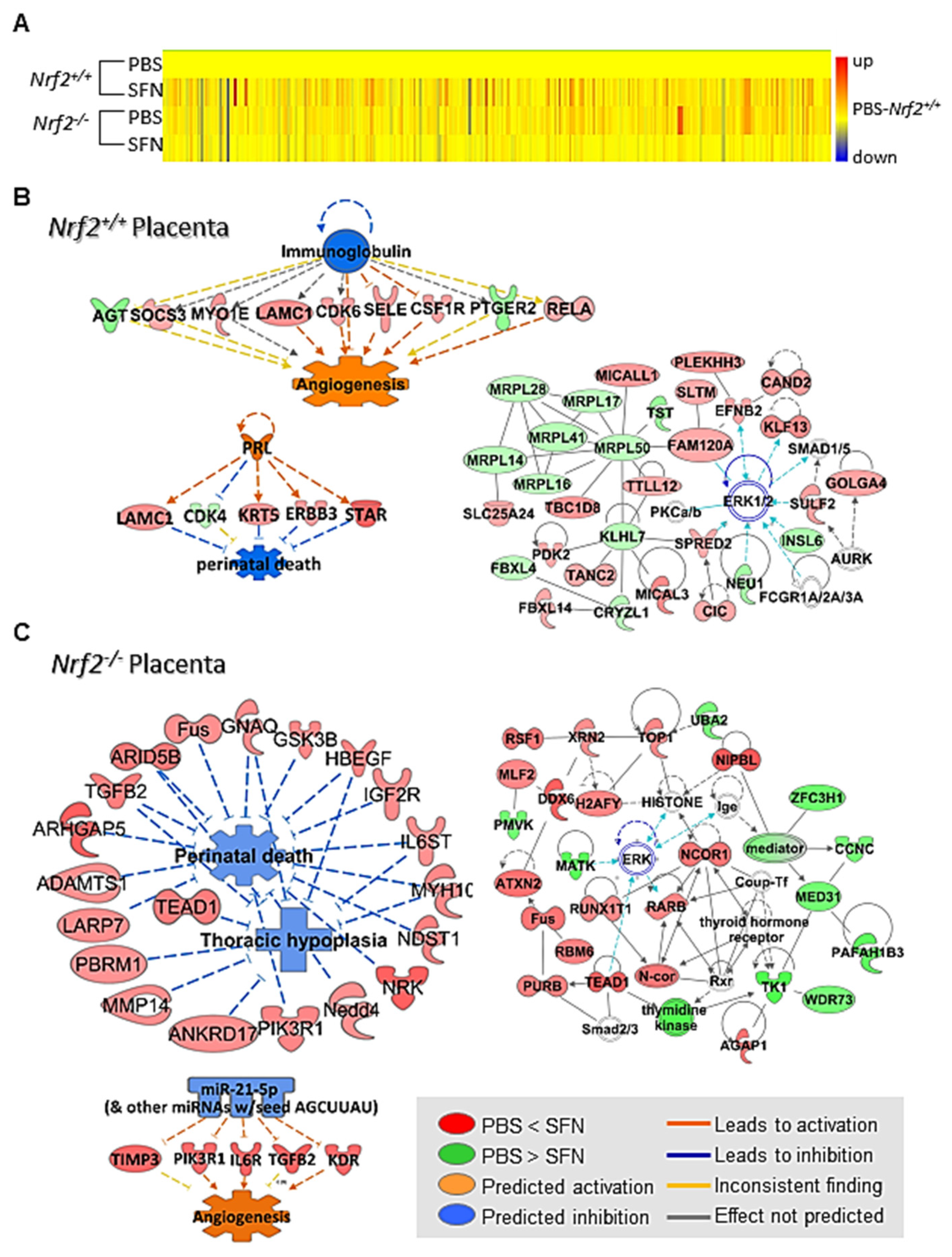
3.2.5. SFN Effects on Placenta Transcriptomics
3.2.6. Validation of Microarray and Pathway Analyses Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Walsh, M.C.; Szefler, S.; Davis, J.; Allen, M.; Van Marter, L.; Abman, S.; Blackmon, L.; Jobe, A. Summary proceedings from the bronchopulmonary dysplasia group. Pediatrics 2006, 117, S52–S56. [Google Scholar] [CrossRef] [PubMed]
- Ruaro, B.; Salton, F.; Braga, L.; Wade, B.; Confalonieri, P.; Volpe, M.C.; Baratella, E.; Maiocchi, S.; Confalonieri, M. The History and Mystery of Alveolar Epithelial Type II Cells: Focus on Their Physiologic and Pathologic Role in Lung. Int. J. Mol. Sci. 2021, 22, 2566. [Google Scholar] [CrossRef] [PubMed]
- Aspal, M.; Zemans, R.L. Mechanisms of ATII-to-ATI Cell Differentiation during Lung Regeneration. Int. J. Mol. Sci. 2020, 21, 3188. [Google Scholar] [CrossRef]
- Baraldi, E.; Filippone, M. Chronic lung disease after premature birth. N. Engl. J. Med. 2007, 357, 1946–1955. [Google Scholar] [CrossRef]
- Jobe, A.H.; Bancalari, E. Bronchopulmonary dysplasia. Am. J. Respir. Crit. Care Med. 2001, 163, 1723–1729. [Google Scholar] [CrossRef] [PubMed]
- Kalikkot Thekkeveedu, R.; Guaman, M.C.; Shivanna, B. Bronchopulmonary dysplasia: A review of pathogenesis and pathophysiology. Respir. Med. 2017, 132, 170–177. [Google Scholar] [CrossRef] [PubMed]
- Skromme, K.; Vollsaeter, M.; Oymar, K.; Markestad, T.; Halvorsen, T. Respiratory morbidity through the first decade of life in a national cohort of children born extremely preterm. BMC Pediatr. 2018, 18, 102. [Google Scholar] [CrossRef]
- Davidson, L.M.; Berkelhamer, S.K. Bronchopulmonary Dysplasia: Chronic Lung Disease of Infancy and Long-Term Pulmonary Outcomes. J. Clin. Med. 2017, 6, 4. [Google Scholar] [CrossRef]
- Caskey, S.; Gough, A.; Rowan, S.; Gillespie, S.; Clarke, J.; Riley, M.; Megarry, J.; Nicholls, P.; Patterson, C.; Halliday, H.L.; et al. Structural and Functional Lung Impairment in Adult Survivors of Bronchopulmonary Dysplasia. Ann. Am. Thorac. Soc. 2016, 13, 1262–1270. [Google Scholar] [CrossRef]
- Sillers, L.; Alexiou, S.; Jensen, E.A. Lifelong pulmonary sequelae of bronchopulmonary dysplasia. Curr. Opin. Pediatr. 2020, 32, 252–260. [Google Scholar] [CrossRef]
- Warburton, D.; Gauldie, J.; Bellusci, S.; Shi, W. Lung development and susceptibility to chronic obstructive pulmonary disease. Proc. Am. Thorac. Soc. 2006, 3, 668–672. [Google Scholar] [CrossRef]
- Warner, B.B.; Stuart, L.A.; Papes, R.A.; Wispe, J.R. Functional and pathological effects of prolonged hyperoxia in neonatal mice. Am. J. Physiol. 1998, 275, L110–L117. [Google Scholar] [CrossRef]
- Yee, M.; White, R.J.; Awad, H.A.; Bates, W.A.; McGrath-Morrow, S.A.; O’Reilly, M.A. Neonatal hyperoxia causes pulmonary vascular disease and shortens life span in aging mice. Am. J. Pathol. 2011, 178, 2601–2610. [Google Scholar] [CrossRef] [PubMed]
- McGrath-Morrow, S.A.; Lauer, T.; Collaco, J.M.; Yee, M.; O’Reilly, M.; Mitzner, W.; Neptune, E.; Wise, R.; Biswal, S. Neonatal hyperoxia contributes additively to cigarette smoke-induced chronic obstructive pulmonary disease changes in adult mice. Am. J. Respir. Cell Mol. Biol. 2011, 45, 610–616. [Google Scholar] [CrossRef]
- O’Reilly, M.A.; Marr, S.H.; Yee, M.; McGrath-Morrow, S.A.; Lawrence, B.P. Neonatal hyperoxia enhances the inflammatory response in adult mice infected with influenza A virus. Am. J. Respir. Crit. Care Med. 2008, 177, 1103–1110. [Google Scholar] [CrossRef] [PubMed]
- Cui, T.X.; Maheshwer, B.; Hong, J.Y.; Goldsmith, A.M.; Bentley, J.K.; Popova, A.P. Hyperoxic Exposure of Immature Mice Increases the Inflammatory Response to Subsequent Rhinovirus Infection: Association with Danger Signals. J. Immunol. 2016, 196, 4692–4705. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.Y.; Kleeberger, S.R. Association of Nrf2 with airway pathogenesis: Lessons learned from genetic mouse models. Arch. Toxicol. 2015, 89, 1931–1957. [Google Scholar] [CrossRef]
- McGrath-Morrow, S.; Lauer, T.; Yee, M.; Neptune, E.; Podowski, M.; Thimmulappa, R.K.; O’Reilly, M.; Biswal, S. Nrf2 increases survival and attenuates alveolar growth inhibition in neonatal mice exposed to hyperoxia. Am. J. Physiol. Lung Cell. Mol. Physiol. 2009, 296, L565–L573. [Google Scholar] [CrossRef]
- Cho, H.Y.; van Houten, B.; Wang, X.; Miller-Degraff, L.; Fostel, J.; Gladwell, W.; Perrow, L.; Panduri, V.; Kobzik, L.; Yamamoto, M.; et al. Targeted Deletion of Nrf2 Impairs Lung Development and Oxidant Injury in Neonatal Mice. Antioxid. Redox Signal. 2012, 17, 1066–1082. [Google Scholar] [CrossRef]
- Kensler, T.W.; Egner, P.A.; Agyeman, A.S.; Visvanathan, K.; Groopman, J.D.; Chen, J.G.; Chen, T.Y.; Fahey, J.W.; Talalay, P. Keap1-nrf2 signaling: A target for cancer prevention by sulforaphane. Top. Curr. Chem. 2013, 329, 163–177. [Google Scholar] [CrossRef]
- Dinkova-Kostova, A.T.; Holtzclaw, W.D.; Cole, R.N.; Itoh, K.; Wakabayashi, N.; Katoh, Y.; Yamamoto, M.; Talalay, P. Direct evidence that sulfhydryl groups of Keap1 are the sensors regulating induction of phase 2 enzymes that protect against carcinogens and oxidants. Proc. Natl. Acad. Sci. USA 2002, 99, 11908–11913. [Google Scholar] [CrossRef]
- Cho, H.Y.; Wang, X.; Li, J.; Bell, D.A.; Kleeberger, S.R. Potential therapeutic targets in Nrf2-dependent protection against neonatal respiratory distress disease predicted by cDNA microarray analysis and bioinformatics tools. Curr. Opin. Toxicol. 2016, 1, 125–133. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Cho, H.Y.; Miller-DeGraff, L.; Blankenship-Paris, T.; Wang, X.; Bell, D.A.; Lih, F.; Deterding, L.; Panduri, V.; Morgan, D.L.; Yamamoto, M.; et al. Sulforaphane enriched transcriptome of lung mitochondrial energy metabolism and provided pulmonary injury protection via Nrf2 in mice. Toxicol. Appl. Pharmacol. 2019, 364, 29–44. [Google Scholar] [CrossRef]
- Wise, R.A.; Holbrook, J.T.; Criner, G.; Sethi, S.; Rayapudi, S.; Sudini, K.R.; Sugar, E.A.; Burke, A.; Thimmulappa, R.; Singh, A.; et al. Lack of Effect of Oral Sulforaphane Administration on Nrf2 Expression in COPD: A Randomized, Double-Blind, Placebo Controlled Trial. PLoS ONE 2016, 11, e0163716. [Google Scholar] [CrossRef]
- Egner, P.A.; Chen, J.G.; Zarth, A.T.; Ng, D.K.; Wang, J.B.; Kensler, K.H.; Jacobson, L.P.; Munoz, A.; Johnson, J.L.; Groopman, J.D.; et al. Rapid and sustainable detoxication of airborne pollutants by broccoli sprout beverage: Results of a randomized clinical trial in China. Cancer Prev. Res. 2014, 7, 813–823. [Google Scholar] [CrossRef] [PubMed]
- Rudan, I.; O’Brien, K.L.; Nair, H.; Liu, L.; Theodoratou, E.; Qazi, S.; Luksic, I.; Fischer Walker, C.L.; Black, R.E.; Campbell, H.; et al. Epidemiology and etiology of childhood pneumonia in 2010: Estimates of incidence, severe morbidity, mortality, underlying risk factors and causative pathogens for 192 countries. J. Glob. Health 2013, 3, 010401. [Google Scholar] [CrossRef] [PubMed]
- Scheltema, N.M.; Gentile, A.; Lucion, F.; Nokes, D.J.; Munywoki, P.K.; Madhi, S.A.; Groome, M.J.; Cohen, C.; Moyes, J.; Thorburn, K.; et al. Global respiratory syncytial virus-associated mortality in young children (RSV GOLD): A retrospective case series. Lancet Glob. Health 2017, 5, e984–e991. [Google Scholar] [CrossRef]
- Paes, B.; Fauroux, B.; Figueras-Aloy, J.; Bont, L.; Checchia, P.A.; Simoes, E.A.; Manzoni, P.; Carbonell-Estrany, X. Defining the Risk and Associated Morbidity and Mortality of Severe Respiratory Syncytial Virus Infection Among Infants with Chronic Lung Disease. Infect. Dis. Ther. 2016, 5, 453–471. [Google Scholar] [CrossRef] [PubMed]
- Chaw, P.S.; Hua, L.; Cunningham, S.; Campbell, H.; Mikolajczyk, R.; Nair, H.; Investigators, R. Respiratory Syncytial Virus-Associated Acute Lower Respiratory Infections in Children With Bronchopulmonary Dysplasia: Systematic Review and Meta-Analysis. J. Infect. Dis. 2019. [Google Scholar] [CrossRef]
- Hall, C.B.; Weinberg, G.A.; Iwane, M.K.; Blumkin, A.K.; Edwards, K.M.; Staat, M.A.; Auinger, P.; Griffin, M.R.; Poehling, K.A.; Erdman, D.; et al. The burden of respiratory syncytial virus infection in young children. N. Engl. J. Med. 2009, 360, 588–598. [Google Scholar] [CrossRef]
- American Academy of Pediatrics Committee on Infectious Diseases; American Academy of Pediatrics Bronchiolitis Guidelines Committee. Updated guidance for palivizumab prophylaxis among infants and young children at increased risk of hospitalization for respiratory syncytial virus infection. Pediatrics 2014, 134, 415–420. [Google Scholar] [CrossRef] [PubMed]
- Acero-Bedoya, S.; Wozniak, P.S.; Sanchez, P.J.; Ramilo, O.; Mejias, A. Recent Trends in RSV Immunoprophylaxis: Clinical Implications for the Infant. Am. J. Perinatol. 2019, 36, S63–S67. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.Y.; Imani, F.; Miller-DeGraff, L.; Walters, D.; Melendi, G.A.; Yamamoto, M.; Polack, F.P.; Kleeberger, S.R. Antiviral activity of Nrf2 in a murine model of respiratory syncytial virus disease. Am. J. Respir. Crit. Care Med. 2009, 179, 138–150. [Google Scholar] [CrossRef]
- Castro, S.M.; Guerrero-Plata, A.; Suarez-Real, G.; Adegboyega, P.A.; Colasurdo, G.N.; Khan, A.M.; Garofalo, R.P.; Casola, A. Antioxidant treatment ameliorates respiratory syncytial virus-induced disease and lung inflammation. Am. J. Respir. Crit. Care Med. 2006, 174, 1361–1369. [Google Scholar] [CrossRef] [PubMed]
- Kerns, M.L.; DePianto, D.; Dinkova-Kostova, A.T.; Talalay, P.; Coulombe, P.A. Reprogramming of keratin biosynthesis by sulforaphane restores skin integrity in epidermolysis bullosa simplex. Proc. Natl. Acad. Sci. USA 2007, 104, 14460–14465. [Google Scholar] [CrossRef]
- Ward, J.M.; Elmore, S.A.; Foley, J.F. Pathology methods for the evaluation of embryonic and perinatal developmental defects and lethality in genetically engineered mice. Vet. Pathol. 2012, 49, 71–84. [Google Scholar] [CrossRef]
- Cho, H.Y.; Jedlicka, A.E.; Reddy, S.P.; Kensler, T.W.; Yamamoto, M.; Zhang, L.Y.; Kleeberger, S.R. Role of NRF2 in protection against hyperoxic lung injury in mice. Am. J. Respir. Cell Mol. Biol. 2002, 26, 175–182. [Google Scholar] [CrossRef]
- Cho, H.Y.; Morgan, D.L.; Bauer, A.K.; Kleeberger, S.R. Signal transduction pathways of tumor necrosis factor--mediated lung injury induced by ozone in mice. Am. J. Respir. Crit. Care Med. 2007, 175, 829–839. [Google Scholar] [CrossRef]
- Farraj, A.K.; Harkema, J.R.; Kaminski, N.E. Allergic rhinitis induced by intranasal sensitization and challenge with trimellitic anhydride but not with dinitrochlorobenzene or oxazolone in A/J mice. Toxicol. Sci. 2004, 79, 315–325. [Google Scholar] [CrossRef]
- Santos, J.H.; Meyer, J.N.; Mandavilli, B.S.; Van Houten, B. Quantitative PCR-based measurement of nuclear and mitochondrial DNA damage and repair in mammalian cells. Meth. Mol. Biol. 2006, 314, 183–199. [Google Scholar]
- Cho, H.Y.; Gladwell, W.; Yamamoto, M.; Kleeberger, S.R. Exacerbated airway toxicity of environmental oxidant ozone in mice deficient in Nrf2. Oxid. Med. Cell. Longev. 2013, 2013, 254069. [Google Scholar] [CrossRef] [PubMed]
- Maitre, N.L.; Ballard, R.A.; Ellenberg, J.H.; Davis, S.D.; Greenberg, J.M.; Hamvas, A.; Pryhuber, G.S.; Prematurity and Respiratory Outcomes Program. Respiratory consequences of prematurity: Evolution of a diagnosis and development of a comprehensive approach. J. Perinatol. 2015, 35, 313–321. [Google Scholar] [CrossRef]
- Doyle, L.W.; Ford, G.; Davis, N. Health and hospitalistions after discharge in extremely low birth weight infants. Semin. Neonatol. 2003, 8, 137–145. [Google Scholar] [CrossRef]
- Bogdan, R.D.; Rusu, L.; Toma, A.I.; Nastase, L. Respiratory Outcome of the Former Premature Infants. J. Med. Life 2019, 12, 381–394. [Google Scholar] [CrossRef]
- Benitez-Guerra, D.; Pina-Flores, C.; Zamora-Lopez, M.; Escalante-Padron, F.; Lima-Rogel, V.; Gonzalez-Ortiz, A.M.; Guevara-Tovar, M.; Bernal-Silva, S.; Benito-Cruz, B.; Castillo-Martinez, F.; et al. Respiratory syncytial virus acute respiratory infection-associated hospitalizations in preterm Mexican infants: A cohort study. Influenza Other Respir. Viruses 2020, 14, 182–188. [Google Scholar] [CrossRef]
- Noyan-Ashraf, M.H.; Wu, L.; Wang, R.; Juurlink, B.H. Dietary approaches to positively influence fetal determinants of adult health. FASEB J. 2006, 20, 371–373. [Google Scholar] [CrossRef]
- Li, Y.; Buckhaults, P.; Li, S.; Tollefsbol, T. Temporal Efficacy of a Sulforaphane-Based Broccoli Sprout Diet in Prevention of Breast Cancer through Modulation of Epigenetic Mechanisms. Cancer Prev. Res. 2018, 11, 451–464. [Google Scholar] [CrossRef] [PubMed]
- Yuan, F.; Chen, X.; Liu, J.; Feng, W.; Cai, L.; Wu, X.; Chen, S.Y. Sulforaphane restores acetyl-histone H3 binding to Bcl-2 promoter and prevents apoptosis in ethanol-exposed neural crest cells and mouse embryos. Exp. Neurol. 2018, 300, 60–66. [Google Scholar] [CrossRef]
- Zhang, P.; Li, T.; Liu, C.; Sindi, M.; Cheng, X.; Qi, S.; Liu, X.; Yan, Y.; Bao, Y.; Brand-Saberi, B.; et al. Nano-sulforaphane attenuates PhIP-induced early abnormal embryonic neuro-development. Ann. Anat. 2021, 233, 151617. [Google Scholar] [CrossRef]
- Heiss, E.; Herhaus, C.; Klimo, K.; Bartsch, H.; Gerhauser, C. Nuclear factor kappa B is a molecular target for sulforaphane-mediated anti-inflammatory mechanisms. J. Biol. Chem. 2001, 276, 32008–32015. [Google Scholar] [CrossRef]
- Moon, D.O.; Kim, M.O.; Kang, S.H.; Choi, Y.H.; Kim, G.Y. Sulforaphane suppresses TNF-alpha-mediated activation of NF-kappaB and induces apoptosis through activation of reactive oxygen species-dependent caspase-3. Cancer Lett. 2009, 274, 132–142. [Google Scholar] [CrossRef]
- Jo, C.; Kim, S.; Cho, S.J.; Choi, K.J.; Yun, S.M.; Koh, Y.H.; Johnson, G.V.; Park, S.I. Sulforaphane induces autophagy through ERK activation in neuronal cells. FEBS Lett. 2014, 588, 3081–3088. [Google Scholar] [CrossRef] [PubMed]
- Keum, Y.S.; Yu, S.; Chang, P.P.; Yuan, X.; Kim, J.H.; Xu, C.; Han, J.; Agarwal, A.; Kong, A.N. Mechanism of action of sulforaphane: Inhibition of p38 mitogen-activated protein kinase isoforms contributing to the induction of antioxidant response element-mediated heme oxygenase-1 in human hepatoma HepG2 cells. Cancer Res. 2006, 66, 8804–8813. [Google Scholar] [CrossRef]
- Ho, E.; Clarke, J.D.; Dashwood, R.H. Dietary sulforaphane, a histone deacetylase inhibitor for cancer prevention. J. Nutr. 2009, 139, 2393–2396. [Google Scholar] [CrossRef] [PubMed]
- Cuadrado, A.; Manda, G.; Hassan, A.; Alcaraz, M.J.; Barbas, C.; Daiber, A.; Ghezzi, P.; Leon, R.; Lopez, M.G.; Oliva, B.; et al. Transcription Factor NRF2 as a Therapeutic Target for Chronic Diseases: A Systems Medicine Approach. Pharmacol. Rev. 2018, 70, 348–383. [Google Scholar] [CrossRef]
- Robledinos-Anton, N.; Fernandez-Gines, R.; Manda, G.; Cuadrado, A. Activators and Inhibitors of NRF2: A Review of Their Potential for Clinical Development. Oxid. Med. Cell. Longev. 2019, 2019, 9372182. [Google Scholar] [CrossRef] [PubMed]
- Barker, G.F.; Manzo, N.D.; Cotich, K.L.; Shone, R.K.; Waxman, A.B. DNA damage induced by hyperoxia: Quantitation and correlation with lung injury. Am. J. Respir. Cell Mol. Biol. 2006, 35, 277–288. [Google Scholar] [CrossRef]
- Yue, X.; Fu, J.; Xue, X.; Gao, H.; Liu, D.; Zong, Z.; Wang, W.; Li, H.; Yuan, Z. Detection of p16 promoter methylation in premature rats with chronic lung disease induced by hyperoxia. Pediatr. Int. 2010, 52, 520–526. [Google Scholar] [CrossRef]
- Panayiotidis, M.I.; Rancourt, R.C.; Pappa, A.; White, C.W. Effect of cell cycle growth arrest on global DNA methylation status in human lung epithelial-like (A549) cells. In Vivo 2006, 20, 861–865. [Google Scholar]
- Kweider, N.; Huppertz, B.; Rath, W.; Lambertz, J.; Caspers, R.; ElMoursi, M.; Pecks, U.; Kadyrov, M.; Fragoulis, A.; Pufe, T.; et al. The effects of Nrf2 deletion on placental morphology and exchange capacity in the mouse. J. Matern. Fetal Neonatal. Med. 2017, 30, 2068–2073. [Google Scholar] [CrossRef]
- Huebner, A.J.; Dai, D.; Morasso, M.; Schmidt, E.E.; Schafer, M.; Werner, S.; Roop, D.R. Amniotic fluid activates the nrf2/keap1 pathway to repair an epidermal barrier defect in utero. Dev. Cell 2012, 23, 1238–1246. [Google Scholar] [CrossRef] [PubMed]



| Genotype | † FC | Gene Symbol | Gene Title | Gene Ontology |
|---|---|---|---|---|
| Nrf2+/+ | 6.85 | Ibsp | integrin binding sialoprotein | cell adhesion |
| 5.31 | Ctsk | cathepsin K | proteolysis | |
| 2.07 | Calcb | calcitonin-related polypeptide, beta | vasodilation | |
| 2.05 | Lilrb4 | leukocyte immunoglobulin-like receptor, subfamily B, member 4 | immune system process | |
| 2.01 | Dnm3os | dynamin 3, opposite strand (Mir214) | skeletal system development | |
| 1.69 | Egr2 | early growth response 2 | transcription | |
| 1.69 | Myof | Myoferlin | vascular endothelial growth factor receptor signaling | |
| 1.53 | Aldh3a1 | aldehyde dehydrogenase family 3, subfamily A1 | response to hypoxia | |
| 1.52 | Ereg | Epiregulin | angiogenesis | |
| 1.49 | Igfbp5 | insulin-like growth factor binding protein 5 | cell growth, glucose metabolism | |
| 1.42 | Maff | v-maf musculoaponeurotic fibrosarcoma oncogene family, protein F | embryonic development, transcription regulation | |
| −1.64 | Gpr165 | G protein-coupled receptor 165 | signal transduction | |
| −1.60 | Tspo2 | translocator protein 2 | transport | |
| −1.56 | Bpgm | 2,3-bisphosphoglycerate mutase | glycolytic process | |
| −1.52 | Lce1i | late cornified envelope 1I | epidermal development | |
| −1.48 | Il20 | interleukin 20 | hemopoiesis | |
| −1.42 | Nkx2-9 | NK2 homeobox 9 | respiratory tube development | |
| 2.73 | Olfm4 | olfactomedin 4 | cell adhesion | |
| Nrf2−/− | 2.66 | Neat1 | nuclear paraspeckle assembly transcript 1 | cellular component maintenance |
| 2.46 | Meg3 | maternally expressed 3 | in utero embryonic development | |
| 2.33 | Stfa1 | stefin A1 | negative regulation of peptidase activity | |
| 1.98 | Amy1 | amylase 1, salivary | carbohydrate metabolism | |
| 1.65 | Ptprc | protein tyrosine phosphatase, receptor type, C | MAPK activation | |
| 1.55 | Cd14 | CD14 antigen | immune system process | |
| 1.50 | Sema4a | sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A | angiogenesis | |
| 1.30 | Vegfa | vascular endothelial growth factor A | angiogenesis | |
| −2.48 | Cda | cytidine deaminase | negative regulation of cell growth | |
| −2.44 | St6gal2 | beta galactoside alpha 2,6 sialyltransferase 2 | carbohydrate metabolism | |
| −2.18 | Arsk | arylsulfatase K | metabolic process | |
| −2.15 | Igfbp2 | insulin-like growth factor binding protein 2 | cell growth, response to stress | |
| −1.67 | Cdc25c | cell division cycle 25C | mitotic cell cycle | |
| −1.61 | Igf1 | insulin-like growth factor 1 | cell activation, vessel remodeling | |
| −1.41 | Nox4 | NADPH oxidase 4 | oxidation–reduction process pppprocessremodeling |
| Genotype | * FC by O2 | † FD PBS:SFN | Gene Symbol | Gene Title | Gene Ontology |
|---|---|---|---|---|---|
| Nrf2+/+ | −2.22 | 2.32 | Dkk2 | dickkopf homolog 2 | transcriptional regulation |
| −4.35 | 1.89 | Prss35 | protease, serine 35 | proteolysis, metabolism | |
| −1.70 | 1.89 | Frem1 | Fras1 related extracellular matrix protein 1 | cell communication | |
| −2.33 | 1.86 | Rrad | Ras-related associated with diabetes | negative regulation of cell growth | |
| −1.04 | 1.78 | Klhdc7a | kelch domain containing 7A (microRNA 2139) | protein binding | |
| −1.46 | 1.61 | Fam26e | family with sequence similarity 26, member E | transport | |
| −1.55 | 1.52 | Cep128 | centrosomal protein 128 | microtubule organization | |
| −1.29 | 1.28 | Ggt7 | gamma-glutamyltransferase 7 | glutathione metabolism | |
| 3.50 | 1.19 | Ephx1 | epoxide hydrolase 1, microsomal | response to toxicant | |
| 4.39 | −3.07 | Lcn2 | lipocalin 2 | immune system process | |
| 1.10 | −2.33 | H2-D1 | histocompatibility 2, D region locus 1 | T cell mediated cytotoxicity | |
| 1.82 | −2.29 | Cdh22 | cadherin 22 | cell adhesion | |
| 2.13 | −2.28 | Mkx | mohawk homeobox | negative regulation of transcription | |
| 3.03 | −2.19 | Msln | mesothelin | cell adhesion | |
| 1.88 | −1.75 | Tgm1 | transglutaminase 1, K polypeptide | organ morphogenesis | |
| 2.75 | −1.31 | Liph | lipase, member H | lipid metabolism | |
| 1.98 | −1.28 | Ctsh | cathepsin H | T cell mediated cytotoxicity | |
| Nrf2−/− | −1.69 | 3.10 | Amy1 | amylase 1, salivary | carbohydrate metabolism |
| −2.03 | 2.24 | Scara5 | scavenger receptor class A, member 5 | transport | |
| −1.80 | 1.79 | Fap | fibroblast activation protein | proteolysis | |
| −1.73 | 1.73 | Cep128 | centrosomal protein 128 | microtubule organization | |
| −1.98 | 1.65 | Cc2d2a | coiled-coil and C2 domain containing 2A | cilium assembly | |
| −1.71 | 1.64 | Uimc1 | ubiquitin interaction motif containing 1 | DNA repair | |
| −1.37 | 1.57 | Cep83 | centrosomal protein 83 | cilium assembly | |
| −1.37 | 1.56 | Nbn | nibrin | DNA damage checkpoint | |
| −2.11 | 1.51 | Smc4 | structural maintenance of chromosomes 4 | DNA repair | |
| 4.51 | −2.77 | Clec7a | C-type lectin domain family 7, member a | pattern recognition receptor signaling | |
| 1.89 | −2.40 | Cd52 | CD52 antigen | immune response | |
| 4.53 | −2.26 | Ccl9 | chemokine (C-C motif) ligand 9 | chemotaxis | |
| 1.67 | −1.67 | Ncf4 | neutrophil cytosolic factor 4 | cell communication | |
| 1.66 | −1.67 | Tifa | TRAF-interacting protein with forkhead-associated domain | NF-kappa B signaling | |
| 1.74 | −1.52 | Gzmb | granzyme B | T cell mediated cytotoxicity | |
| 2.11 | −1.46 | Csf2rb | colony stimulating factor 2 receptor, beta, low-affinity (granulocyte-macrophage) | cytokine signaling | |
| 2.16 | −1.46 | Cyp3a13 | cytochrome P450, family 3, subfamily a, polypeptide 13 | oxidation–reduction process | |
| 1.46 | −1.45 | Ltb | lymphotoxin B | immune response | |
| 1.58 | −1.40 | Selplg | selectin, platelet (p-selectin) ligand | leukocyte adhesion |
| Genotype | † FC | Gene Symbol | Gene Title | Gene Ontology |
|---|---|---|---|---|
| Nrf2+/+ | 7.33 | Stfa3 | stefin A3 | endopeptidase inhibitor |
| 4.33 | Tmem178 | transmembrane protein 178 | structural molecule activity | |
| 3.69 | Uchl1 | ubiquitin carboxy-terminal hydrolase L1 | axonogenesis | |
| 3.04 | Lce1d | late cornified envelope 1D | epidermal development | |
| 2.92 | Col8a2 | collagen, type VIII, alpha 2 | morphogenesis | |
| 2.88 | Slc6a2 | solute carrier family 6 (neurotransmitter transporter, noradrenalin), member 2 | monoamine transport | |
| 2.77 | Lor | loricrin | keratinocyte differentiation | |
| 2.57 | Pdgfrb | platelet derived growth factor receptor, beta polypeptide | cell proliferation | |
| 2.51 | Dnmt3a | DNA methyltransferase 3A | DNA methylation | |
| 2.19 | Sema3g | sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3G | organ development | |
| 1.97 | Pik3r1 | phosphatidylinositol 3-kinase, regulatory subunit, polypeptide 1 | insulin-like growth factor receptor signaling pathway | |
| 1.92 | Med13l | mediator complex subunit 13-like | transcription regulation | |
| 1.55 | Aldh1l2 | aldehyde dehydrogenase 1 family, member L2 | oxidation reduction | |
| −109.27 | Adamdec1 | ADAM-like, decysin 1 | proteolysis | |
| −16.14 | H2-D1 | histocompatibility 2, D region locus 1 | antigen processing and presentation | |
| −7.84 | Lect1 | leukocyte cell derived chemotaxin 1 | organ development | |
| −5.37 | Hao3 | hydroxyacid oxidase (glycolate oxidase) 3 | oxidation reduction | |
| −4.43 | Gstt1/t2 | glutathione S-transferase, theta 1/2 | glutathione metabolism | |
| −4.11 | Gzmd | granzyme D | cytolysis | |
| −3.86 | Igfbp1 | insulin-like growth factor binding protein 1 | cell growth regulation | |
| −1.80 | Ccnc | cyclin C | transcription regulation | |
| −1.78 | Ndufaf1 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, assembly factor 1 | ubiquinone) activity | |
| −1.69 | Irak1bp1 | Interleukin-1 receptor-associated kinase 1 binding protein 1 | NF-kappa B cascade | |
| −1.68 | Prdx5 | peroxiredoxin 5 | oxidation reduction | |
| Nrf2−/− | 4.71 | Lect1 | leukocyte cell derived chemotaxin 1 | organism development |
| 2.19 | Uchl1 | ubiquitin carboxy-terminal hydrolase L1 | response to ischemia | |
| 2.16 | Ppargc1b | peroxisome proliferative activated receptor, gamma, coactivator 1 beta | transcription regulation | |
| 1.95 | Arid5b | AT rich interactive domain 5B | development | |
| 1.74 | Hbegf | heparin-binding EGF-like growth factor | angiogenesis | |
| 1.74 | Timp3 | tissue inhibitor of metalloproteinase 3 | neurotransmitter secretion | |
| 1.72 | Kdr | kinase insert domain protein receptor | angiogenesis | |
| 1.70 | Figf | c-fos induced growth factor | angiogenesis | |
| 1.63 | Igf2r | insulin-like growth factor 2 receptor | post-embryonic development | |
| −3.13 | Uty | ubiquitously transcribed tetratricopeptide repeat gene, Y chromosome | in utero embryonic development | |
| −1.96 | Ccne2 | cyclin E2 | mitotic cell cycle | |
| −1.73 | Tk1 | thymidine kinase 1 | DNA replication | |
| −1.62 | Cox7a1 | cytochrome c oxidase subunit VIIa 1 | oxidation–reduction process | |
| −1.59 | Cyp1a1 | cytochrome P450, family 1, subfamily a, polypeptide 1 | response to hypoxia | |
| −1.59 | Mir17hg | Mir17 host gene 1 | in utero embryonic development | |
| −1.58 | Pafah1b3 | platelet-activating factor acetylhydrolase, isoform 1b, subunit 3 | lipid metabolic process |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cho, H.-Y.; Miller-DeGraff, L.; Perrow, L.A.; Gladwell, W.; Panduri, V.; Lih, F.B.; Kleeberger, S.R. Murine Neonatal Oxidant Lung Injury: NRF2-Dependent Predisposition to Adulthood Respiratory Viral Infection and Protection by Maternal Antioxidant. Antioxidants 2021, 10, 1874. https://doi.org/10.3390/antiox10121874
Cho H-Y, Miller-DeGraff L, Perrow LA, Gladwell W, Panduri V, Lih FB, Kleeberger SR. Murine Neonatal Oxidant Lung Injury: NRF2-Dependent Predisposition to Adulthood Respiratory Viral Infection and Protection by Maternal Antioxidant. Antioxidants. 2021; 10(12):1874. https://doi.org/10.3390/antiox10121874
Chicago/Turabian StyleCho, Hye-Youn, Laura Miller-DeGraff, Ligon A. Perrow, Wesley Gladwell, Vijayalakshmi Panduri, Fred B. Lih, and Steven R. Kleeberger. 2021. "Murine Neonatal Oxidant Lung Injury: NRF2-Dependent Predisposition to Adulthood Respiratory Viral Infection and Protection by Maternal Antioxidant" Antioxidants 10, no. 12: 1874. https://doi.org/10.3390/antiox10121874
APA StyleCho, H.-Y., Miller-DeGraff, L., Perrow, L. A., Gladwell, W., Panduri, V., Lih, F. B., & Kleeberger, S. R. (2021). Murine Neonatal Oxidant Lung Injury: NRF2-Dependent Predisposition to Adulthood Respiratory Viral Infection and Protection by Maternal Antioxidant. Antioxidants, 10(12), 1874. https://doi.org/10.3390/antiox10121874







