Efficient SNP Discovery by Combining Microarray and Lab-on-a-Chip Data for Animal Breeding and Selection
Abstract
:1. Introduction
2. Discovery of Novel SNP (Single Nucleotide Polymorphism) for Animal Breeding
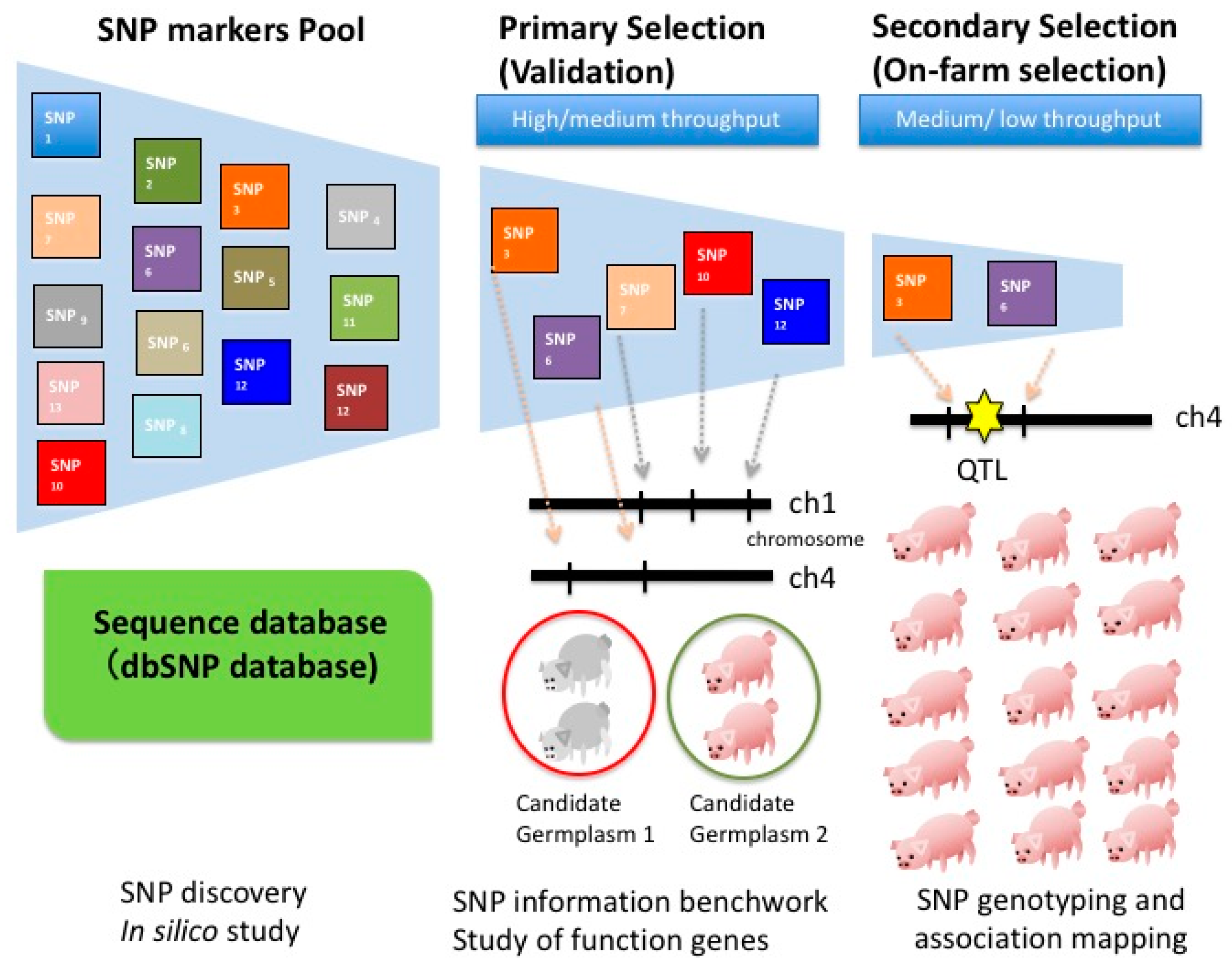
3. Strategies for SNP Discovery
4. SNP Genotyping Technology
| Genotyping Method | Proudct Name | Detection Mechanism | Platform | Throughput | Error Rate | Price | Reference |
|---|---|---|---|---|---|---|---|
| Large Throughput (more than hundreds or thousands SNPs/reaction) | |||||||
| OpenArray® (Applied Biosystems. Foster City, CA, USA) | TaqMAN | Primer extension | Fluorescence | >1000 samples/SNPs | Low | $$$ | [45] |
| Infinium® II (Illumina, San Diego, CA, USA) | Illumina Infinium assay | Primer extension | Fluorescence | >1000 SNPs/sample | Medium to High | $$$$$ | [70] |
| GoldenGate® (Illumina, San Diego, CA, USA) | GoldenGate® | Hybridization | Fluorescence | >1000 SNPs/sample/reaction | Low | $$$$$ | [71] |
| Genome Wide SNP Array (Affymetrix, Santa Clara, CA, USA) | Affymetrix® | Hybridization | Fluorescence | >40 K SNPs/sample/reaction | Low | $$$$$ | [72] |
| RAD sequencing | Illumina | Sequencing | Capillary electrophoresis | >13 K SNPs/sample/reaction | Low | $$$ | [73] |
| Pyrosequencing | Pyrosequencing™ | Sequencing | Pyrophosphate | >96 samples | Low | $$$ | [74] |
| Medium Throughput (hundreds to low thousands SNPs/reaction) | |||||||
| TaqMan assay (Applied Biosystems. Foster City, CA, USA) | TaqMan | Primer extension | Fluorescence | Up to 384 samples/SNP | Low | $$ | [45] |
| MassARRAY® system (Agena Bioscience, San Diego, CA, USA) | iPLEX | Primer extension | Mass spectrometer | 60 SNPs/sample/reaction | Low | $$$ | [75] |
| SNPstream genotyping system (Beckman Coulter, Brea, CA, USA) | 48-plex GenomeLab SNPstream | Primer extension | Fluorescence | 48 SNPs/sample/reaction | N.A. | $$$ | [76] |
| SNaPshot® multiplex system (Applied Biosystems) | SNaPshot | Primer extension | Capillary electrophoresis | 10 SNPs/sample/reaction | Low | $$$ | [77] |
| PCR-APEX | Genorama® | Primer extension | Fluorescene | Up to 384 samples/SNP | Low | $$$ | [78,79] |
| Luminex xMAP technology (Luminex, Austin, TX, USA ) | Luminex100™ | Ligation | Flow cytometer | Up to 100 samples/SNP | Medium to high | $$$ | [80] |
| Small Throughput (less than hundred SNPs/reaction) | |||||||
| Invader assay | Laboratory use | Endonuclease cleavage | Fluorescence | 1 SNP/sample/reaction | Low | $ | [18] |
| PCR-RFLP | Laboratory use | Restriction enzyme | Gel electrophoresis | 1 SNP/sample/reaction | Low | $ | [51] |
| DASH | Laboratory use | Hybridization | Fluorescence | 1 SNP/sample/reaction | Low | $ | [81] |
4.1. Large Throughput Methods
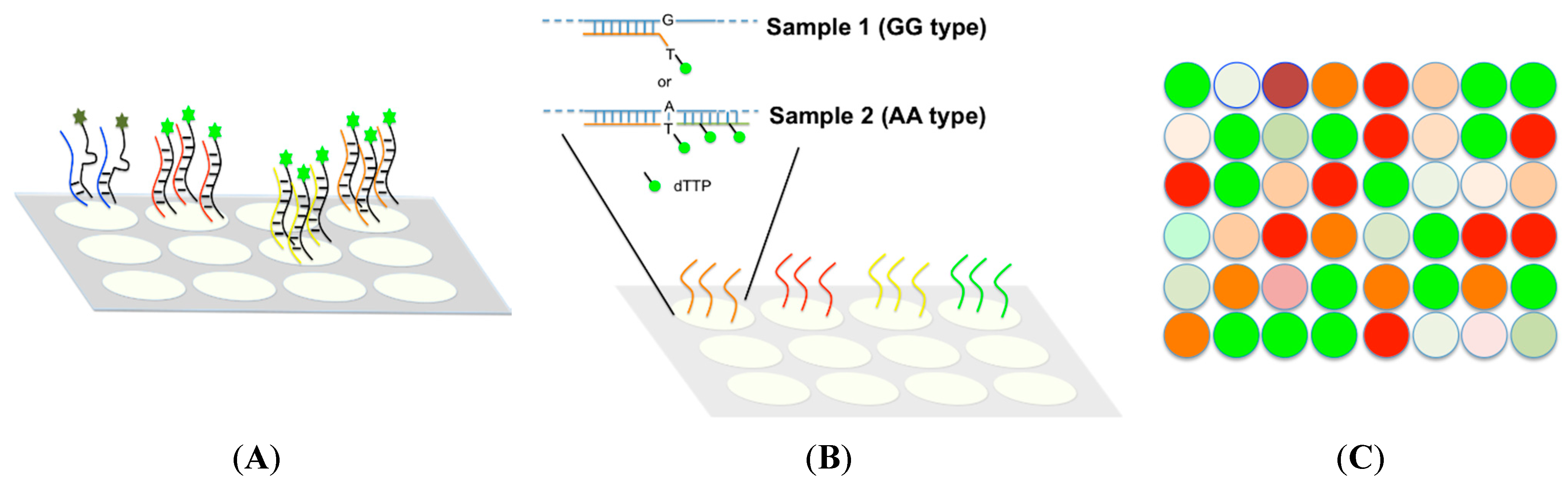
4.2. Medium Throughput Methods
4.3. Small Throughput Methods
5. Miniaturizing Platforms of SNP Genotyping
5.1. Lab-on-a-Chip Platform
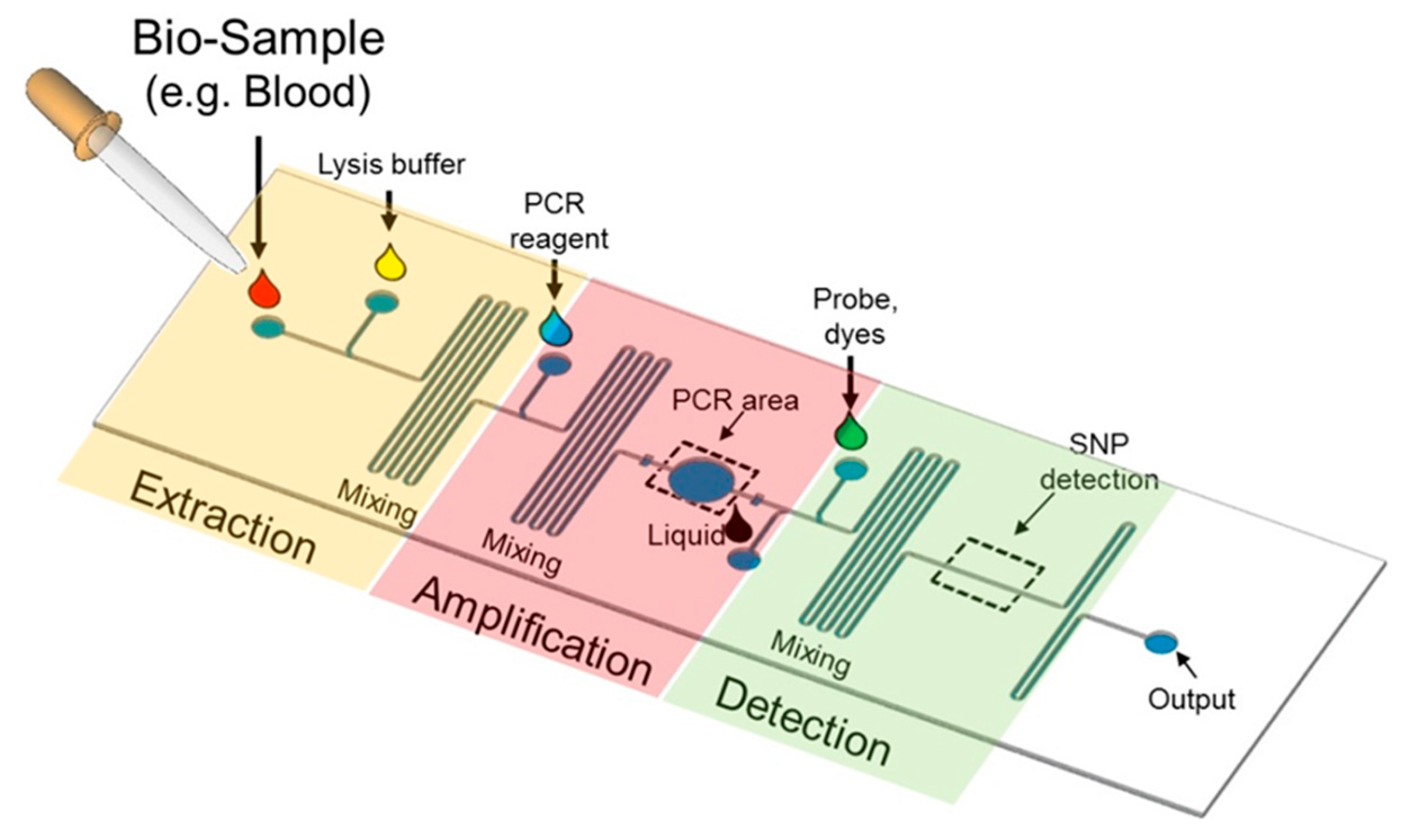
5.2. Microfluidics for SNP Detection
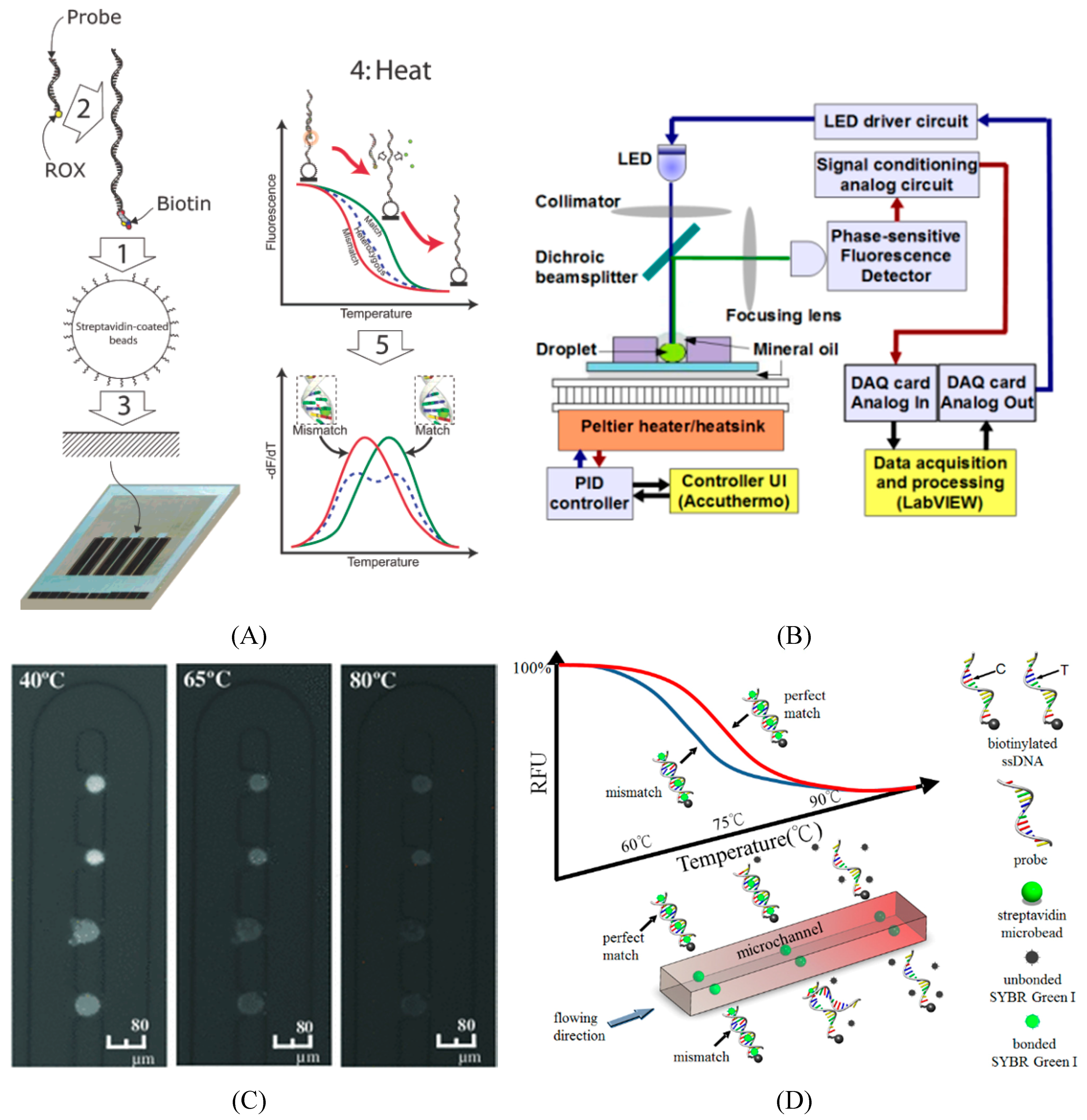
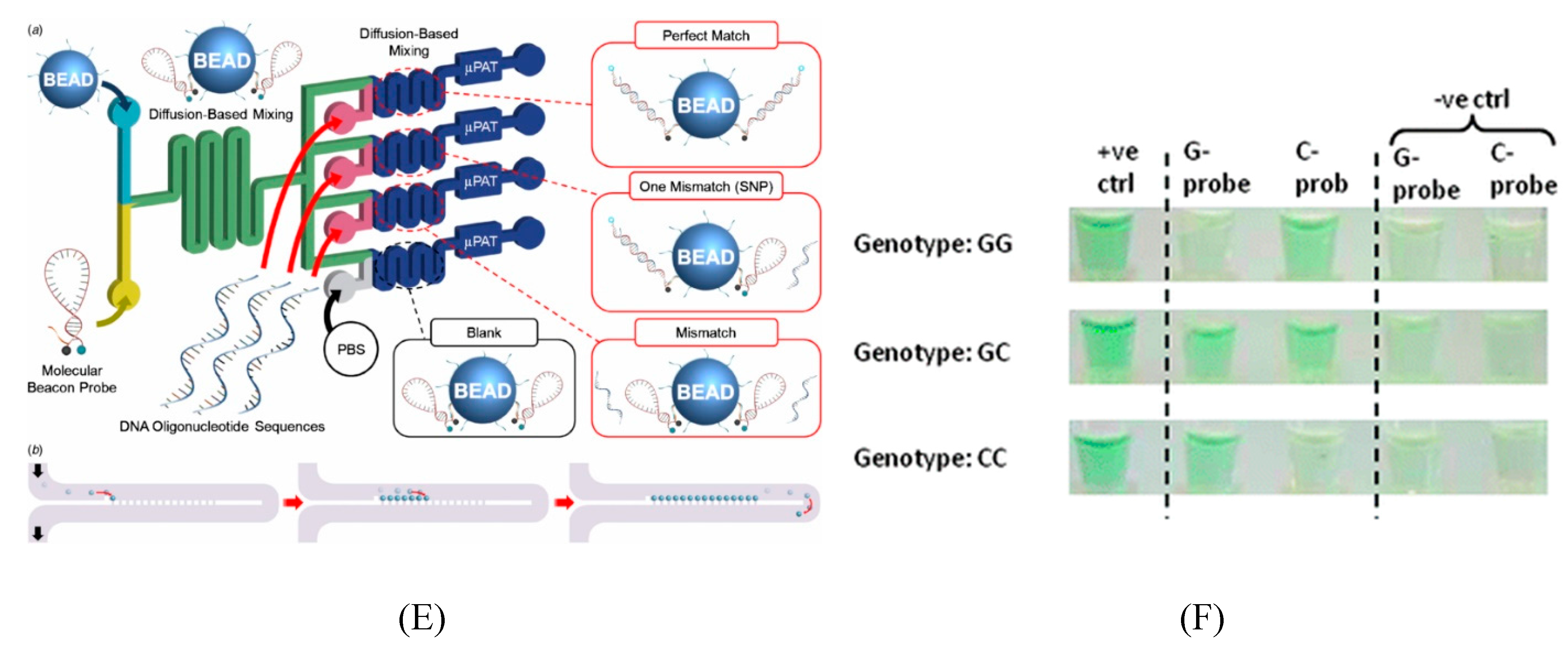
6. Application of Trait Selection in Farm Animals by Using SNPs
6.1. SNP Discovery in Swine
6.1.1. Reproduction
6.1.2. Meat Quality Traits
6.2. Poultry
6.2.1. Reproduction
6.2.2. Growth
| Species | Traits | Gene 1 | Chromosome Location | Putative Functions | Refs. |
|---|---|---|---|---|---|
| Swine | Reproduction | ATM | 9 | Morula development | [103] |
| Reproduction | ESR | 1 | Effect of follicular growth and litter size | [104,105]. | |
| Reproduction | PRLR | 16 | Control luteal and follicular steroidogenesis | [107] | |
| Meat Quality | MSTN | 15 | Negative regulator for muscle mass | [4,45,113] | |
| Swine | Meat Quality | IGF-2 | 2 | Growth-promoting peptidesStructurally homologous with insulin Producing uniformity of pork leanness | [114,115] |
| Meat Quality | RYR1 | 6 | Known as Halothane gene, Ryanodine receptor causing Ca2+ release | [118] | |
| Meat Quality | Myf5, Myf6 | 5 | Transcription regulator of skeleton muscle development and increase of meat mass | [118,119,120] | |
| Poultry | Reproduction | LRP8 | 1 | Cholesterol supply for steroid biosynthesis, which enables folliculogenesis, melatonin in ovarian | [124] |
| Reproduction | MTNR family | 8 | MTNR binds melatonin, affecting growth and reproduction | [125,126] | |
| Reproduction (geese) | (MAGI-1), KIAA1462, ARHGAP21, ACSF2, ASTN2 | MAGI-1: 12 KIAA1462: 2 ARHGAP21: 2 ACSF2: 18 ASTN2: 17 | MAGI-1: cell proliferation and apoptosis KIAA1462: Meiotic recombination ARHGAP21: cell-cell adhesion formation and cellular migration ACSF2: fatty acid synthesis ASTN2: cell adhesion | [127] | |
| Growth | PIT1 | 4 | Secretion of growth hormone, prolactin and thyroid-stimulating hormone | [128,129] | |
| Growth | MC4R | 18 | Appetite, growth and weight gain | [130] |
7. Prospective
8. Summary
- Animal breeding by using phenotypic selection is insufficient.
- Utilization of genetic markers for breeding has become mainstream in the livestock industry.
- In particular, breeding using SNP markers is an effective and economical approach as it possesses the advantages of abundance and wide distribution in genome, and is easy to analyze.
- SNP markers can be discovered by using sequence-dependent and sequence-independent methods. The former one is large throughput but expensive, while the latter one is small throughput but robust.
- Appropriate SNP genotyping techniques for animal breeding will be adapted addressing concerns regarding throughput, reliability, associated expense, and procedure complexity.
- A combination of whole-genome sequencing for a small population of individuals and a SNP chip for the genotyping of the majority of individuals will be a feasible and economical approach for animal breeding.
Acknowledgments
Conflicts of Interest
References
- Lande, R.; Thompson, R. Efficiency of marker-assisted selection in the improvement of quantitative traits. Genetics 1990, 124, 743–756. [Google Scholar] [PubMed]
- Collard, B.C.Y.; Jahufer, M.Z.Z.; Brouwer, J.B.; Pang, E.C.K. An introduction to markers, quantitative trait loci (QTL) mapping and marker-assisted selection for crop improvement: The basic concepts. Euphytica 2005, 142, 169–196. [Google Scholar] [CrossRef]
- Revilla, M.; Ramayo-Caldas, Y.; Castelló, A.; Corominas, J.; Puig-Oliveras, A.; Ibáñez-Escriche, N.; Muñoz, M.; Ballester, M.; Folch, J.M. New insight into the SSC8 genetic determination of fatty acid composition in pigs. Genet. Sel. Evol. 2014, 46. [Google Scholar] [CrossRef] [PubMed]
- Tu, P.-A.; Shiau, J.-W.; Ding, S.-T.; Lin, E.-C.; Wu, M.-C.; Wang, P.-H. The association of genetic variations in the promoter region of myostatin gene with growth traits in Duroc pigs. Anim. Biotechnol. 2012, 23, 291–298. [Google Scholar] [CrossRef] [PubMed]
- Dekkers, J.C. Commercial application of marker-and gene-assisted selection in livestock: Strategies and lessons. J. Anim. Sci. 2004, 82, E313–E328. [Google Scholar] [PubMed]
- Vos, P.; Hogers, R.; Bleeker, M.; Reijans, M.; van de Lee, T.; Hornes, M.; Friters, A.; Pot, J.; Paleman, J.; Kuiper, M.; et al. AFLP: A new technique for DNA fingerprinting. Nucleic Acids Res. 1995, 23, 4407–4414. [Google Scholar] [CrossRef] [PubMed]
- Williams, J.G.; Kubelik, A.R.; Livak, K.J.; Rafalski, J.A.; Tingey, S.V. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 1990, 18, 6531–6535. [Google Scholar] [CrossRef] [PubMed]
- Tautz, D. Hypervariability of simple sequences as a general source for polymorphic DNA markers. Nucleic Acids Res. 1989, 17, 6463–6471. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Kang, X.; Yang, Q.; Lin, Y.; Fang, M. Review on the development of genotyping methods for assessing farm animal diversity. J. Anim. Sci. Biotechnol. 2013, 4. [Google Scholar] [CrossRef] [PubMed]
- Hayes, B.J.; Lewin, H.A.; Goddard, M.E. The future of livestock breeding: Genomic selection for efficiency, reduced emissions intensity, and adaptation. Trends Genet. 2013, 29, 206–214. [Google Scholar] [CrossRef] [PubMed]
- Varshney, R.K.; Chabane, K.; Hendre, P.S.; Aggarwal, R.K.; Graner, A. Comparative assessment of EST-SSR, EST-SNP and AFLP markers for evaluation of genetic diversity and conservation of genetic resources using wild, cultivated and elite barleys. Plant Sci. 2007, 173, 638–649. [Google Scholar] [CrossRef]
- Gao, Y.; Zhang, R.; Hu, X.; Li, N. Application of genomic technologies to the improvement of meat quality of farm animals. Meat Sci. 2007, 77, 36–45. [Google Scholar] [CrossRef] [PubMed]
- Vignal, A.; Milan, D.; SanCristobal, M.; Eggen, A. A review on SNP and other types of molecular markers and their use in animal genetics. Genet. Sel. Evol. 2002, 34, 275–306. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Misra, A. SNP genotyping: Technologies and biomedical applications. Ann. Rev. Biomed. Eng. 2007, 9, 289–320. [Google Scholar] [CrossRef] [PubMed]
- Landegren, U.; Kaiser, R.; Sanders, J.; Hood, L. A ligase-mediated gene detection technique. Science 1988, 241, 1077–1080. [Google Scholar] [CrossRef] [PubMed]
- Tong, A.K.; Li, Z.; Jones, G.S.; Russo, J.J.; Ju, J. Combinatorial fluorescence energy transfer tags for multiplex biological assays. Nat. Biotechnol. 2001, 19, 756–759. [Google Scholar] [CrossRef] [PubMed]
- Botstein, D.; White, R.L.; Skolnick, M.; Davis, R.W. Construction of a genetic-linkage map in man using restriction fragment length polymorphisms. Am. J. Hum. Genet. 1980, 32, 314–331. [Google Scholar] [PubMed]
- Lyamichev, V.; Mast, A.L.; Hall, J.G.; Prudent, J.R.; Kaiser, M.W.; Takova, T.; Kwiatkowski, R.W.; Sander, T.J.; de Arruda, M.; et al. Polymorphism identification and quantitative detection of genomic DNA by invasive cleavage of oligonucleotide probes. Nat. Biotechnol. 1999, 17, 292–296. [Google Scholar] [PubMed]
- Sokolov, B.P. Primer extension technique for the detection of single nucleotide in genomic DNA. Nucleic Acids Res. 1990, 18. [Google Scholar] [CrossRef]
- Takatsu, K.; Yokomaku, T.; Kurata, S.; Kanagawa, T. A FRET-based analysis of SNPs without fluorescent probes. Nucleic Acids Res. 2004, 32. [Google Scholar] [CrossRef] [PubMed]
- Kolpashchikov, D.M. Split DNA enzyme for visual single nucleotide polymorphism typing. J. Am. Chem. Soc. 2008, 130, 2934–2935. [Google Scholar] [CrossRef] [PubMed]
- Neo, J.L.; Aw, K.D.; Uttamchandani, M. Visual SNP genotyping using asymmetric PCR and split DNA enzymes. Analyst 2011, 136, 1569–1572. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Liu, Z.; Monroe, H.; Culiat, C.T. Integrated platform for detection of DNA sequence variants using capillary array electrophoresis. Electrophoresis 2002, 23, 1499–1511. [Google Scholar] [CrossRef]
- Ronaghi, M.; Uhlen, M.; Nyren, P. A sequencing method based on real-time pyrophosphate. Science 1998, 281, 363–365. [Google Scholar] [CrossRef] [PubMed]
- Tost, J.; Gut, I.G. Genotyping single nucleotide polymorphisms by MALDI mass spectrometry in clinical applications. Clin. Biochem. 2005, 38, 335–350. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.-T.; Lin, C.Y.; Liou, J.S.; Fan, Y.H.; Chiou, S.H.; Huang, C.W.; Wu, W.-P.; Lin, E.-C.; Chen, C.-F.; Lee, Y.-P.; et al. Differentially expressed transcripts in shell glands from low and high egg production strains of chickens using cDNA microarrays. Anim. Reprod. Sci. 2007, 101, 113–124. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, J.; Kagiala, G.V.; Pushpakom, S.; Lauzon, J.; Makin, A.; Atrazhev, A.; Stickel, A.; Newman, W.G.; Backhouse, C.J.; Pilarski, L.M. Microfluidic platform for single nucleotide polymorphism genotyping of the thiopurine S-methyltransferase gene to evaluate risk for adverse drug events. J. Mol. Diagn. 2007, 9, 521–529. [Google Scholar] [CrossRef] [PubMed]
- Jung, Y.K.; Kim, J.; Mathies, R.A. Microfluidic Linear Hydrogel Array for Multiplexed Single Nucleotide Polymorphism (SNP) Detection. Anal. Chem. 2015, 87, 3165–3170. [Google Scholar] [CrossRef] [PubMed]
- Schmalzing, D.; Belenky, A.; Novotny, M.A.; Koutny, L.; Salas-Solano, O.; El-Difrawy, S.; Adourian, A.; Matsudaira, P.; Ehrlich, D. Microchip electrophoresis: a method for high-speed SNP detection. Nucleic Acids Res. 2000, 28. [Google Scholar] [CrossRef]
- Rege, J. Biotechnology options for improving livestock production in developing countries, with special reference to sub-Saharan Africa. In Proceedings of the Third Biennial Conference of the African Small Ruminant Research Network, UICC, Kampala, Uganda, 5–9 December 1994.
- Teale, A.; Tan, S.; Tan, J.-H. Applications of molecular genetic and reproductive technologies in the conservation of domestic animal diversity. In Proceedings of the 5th World Congress on Genetics Applied to Livestock Production, Guelph, ON, Canada, 7–12 August 1994.
- Girkin, J.M.; Mohammed, M.-I.; Ellis, E.M. A miniaturised integrated biophotonic point-of-care genotyping system. Faraday Discuss. 2011, 149, 115–123. [Google Scholar] [CrossRef] [PubMed]
- Horejsh, D.; Martini, F.; Poccia, F.; Ippolito, G.; Di Caro, A.; Capobianchi, M.R. A molecular beacon, bead-based assay for the detection of nucleic acids by flow cytometry. Nucleic Acids Res. 2005, 33. [Google Scholar] [CrossRef] [PubMed]
- Ng, J.; Liu, W.-T. Miniaturized platforms for the detection of single-nucleotide polymorphisms. Anal. Bioanal. Chem. 2006, 386, 427–434. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Park, S.; Liu, K.; Tsuan, J.; Yang, S.; Wang, T.-H. A surface topography assisted droplet manipulation platform for biomarker detection and pathogen identification. Lab Chip 2011, 11, 398–406. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, J.; Evander, M.; Hammarström, B.; Laurell, T. Review of cell and particle trapping in microfluidic systems. Anal. Chim. Acta 2009, 649, 141–157. [Google Scholar] [CrossRef] [PubMed]
- Riahi, R.; Mach, K.E.; Mohan, R.; Liao, J.C.; Wong, P.K. Molecular Detection of Bacterial Pathogens Using Microparticle Enhanced Double-Stranded DNA Probes. Anal. Chem. 2011, 83, 6349–6354. [Google Scholar] [CrossRef] [PubMed]
- Ramji, R.; Wang, M.; Bhagat, A.A.S.; Tan Shao Weng, D.; Thakor, N.V.; Teck Lim, C.; Chen, C.-H. Single cell kinase signaling assay using pinched flow coupled droplet microfluidics. Biomicrofluidics 2014, 8. [Google Scholar] [CrossRef] [PubMed]
- Kao, P.-C.; Ding, S.-T.; Lin, E.-C.; Li, K.-C.; Wang, L.; Lu, Y.-W. A bead-based single nucleotide polymorphism (SNP) detection using melting temperature on a microchip. Microfluid. Nanofluidics 2014, 17, 477–488. [Google Scholar] [CrossRef]
- Seidel, G.E. Brief introduction to whole-genome selection in cattle using single nucleotide polymorphisms. Reprod. Fertil. Dev. 2010, 22, 138–144. [Google Scholar] [CrossRef] [PubMed]
- Ai, H; Huang, L.; Ren, J. Genetic diversity, linkage disequilibrium and selection signatures in chinese and Western pigs revealed by genome-wide SNP markers. PLoS ONE 2013, 8, e56001. [Google Scholar]
- Edea, Z.; Bhuiyan, M.S.; Dessie, T.; Rothschild, M.F.; Dadi, H.; Kim, K.S. Genome-wide genetic diversity, population structure and admixture analysis in African and Asian cattle breeds. Animal 2015, 9, 218–226. [Google Scholar] [CrossRef] [PubMed]
- Muir, W.M.; Wong, G.K.S.; Zhang, Y.; Wang, J.; Groenen, M.A.; Crooijmans, R.P. Genome-wide assessment of worldwide chicken SNP genetic diversity indicates significant absence of rare alleles in commercial breeds. Proc. Natl. Acad. Sci. 2008, 105, 17312–17317. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.; Shin, D.H.; Cho, S.; Kang, H.S.; Kim, S.H.; Lee, H.K.; Kim, H.; Seo, K.S. Genome-wide Association Study of Integrated Meat Quality-related Traits of the Duroc Pig Breed. Asian Australas. J. Anim. Sci. 2014, 27, 303–309. [Google Scholar] [CrossRef] [PubMed]
- Tu, P.A.; Lo, L.L.; Chen, Y.C.; Hsu, C.C.; Shiau, J.W.; Lin, E.C.; Lin, R.S.; Wang, P.H. Polymorphisms in the promoter region of myostatin gene are associated with carcass traits in pigs. J. Anim. Breed. Genet. 2014, 131, 116–122. [Google Scholar] [CrossRef] [PubMed]
- Uimari, P.; Tapio, M. Extent of linkage disequilibrium and effective population size in Finnish Landrace and Finnish Yorkshire pig breeds. J. Anim. Sci. 2011, 89, 609–614. [Google Scholar] [CrossRef] [PubMed]
- Badke, Y.M.; Bates, R.O.; Ernst, C.W.; Schwab, C.; Steibel, J.P. Estimation of linkage disequilibrium in four US pig breeds. BMC Genomics 2012, 13. [Google Scholar] [CrossRef] [PubMed]
- Van Heirstraeten, L.; Spang, P.; Schwind, C.; Drese, K.S.; Ritzi-Lehnert, M.; Nieto, B.; Camps, M.; Landgraf, B.; Guasch, F.; Corbera, A.H.; et al. Integrated DNA and RNA extraction and purification on an automated microfluidic cassette from bacterial and viral pathogens causing community-acquired lower respiratory tract infections. Lab Chip 2014, 14, 1519–1526. [Google Scholar] [CrossRef] [PubMed]
- Beaulieu, M.; Larson, G.P.; Geller, L.; Flanagan, S.D.; Krontiris, T.G. PCR candidate region mismatch scanning: Adaptation to quantitative, high-throughput genotyping. Nucleic Acids Res. 2001, 29, 1114–1124. [Google Scholar] [CrossRef] [PubMed]
- Haliassos, A.; Chomel, J.C.; Grandjouan, S.; Kruh, J.; Kaplan, J.C.; Kitzis, A. Detection of minority point mutations by modified PCR technique: A new approach for a sensitive diagnosis of tumor-progression markers. Nucleic Acids Res. 1989, 17, 8093–8099. [Google Scholar] [CrossRef] [PubMed]
- Haliassos, A.; Chomel, J.C.; Tesson, L.; Baudis, M.; Kruh, J.; Kaplan, J.C.; Kitzis, A. Modification of enzymatically amplified DNA for the detection of point mutations. Nucleic Acids Res. 1989, 17, 3606. [Google Scholar] [CrossRef] [PubMed]
- Comai, L.; Henikoff, S. TILLING: Practical single-nucleotide mutation discovery. Plant J. 2006, 45, 684–694. [Google Scholar] [CrossRef] [PubMed]
- Pareek, C.S.; Smoczynski, R.; Tretyn, A. Sequencing technologies and genome sequencing. J. Appl. Genet. 2011, 52, 413–435. [Google Scholar] [CrossRef] [PubMed]
- Milne, I.; Bayer, M.; Cardle, L.; Shaw, P.; Stephen, G.; Wright, F.; Marshall, D. Tablet—Next generation sequence assembly visualization. Bioinformatics 2010, 26, 401–402. [Google Scholar] [CrossRef] [PubMed]
- Quinlan, A.R.; Stewart, D.A.; Stromberg, M.P.; Marth, G.T. Pyrobayes: An improved base caller for SNP discovery in pyrosequences. Nat. Methods 2008, 5, 179–181. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Li, Y.; Fang, X.; Yang, H.; Wang, J.; Kristiansen, K.; Wang, J. SNP detection for massively parallel whole-genome resequencing. Genome Res. 2009, 19, 1124–1132. [Google Scholar] [CrossRef] [PubMed]
- Koboldt, D.C.; Chen, K.; Wylie, T.; Larson, D.E.; McLellan, M.D.; Mardis, E.R.; Weinstock, G.M.; Wilson, R.K.; Ding., L. VarScan: Variant detection in massively parallel sequencing of individual and pooled samples. Bioinformatics 2009, 25, 2283–2285. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Ruan, J.; Durbin, R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008, 18, 1851–1858. [Google Scholar] [CrossRef] [PubMed]
- Hou, H.; Zhao, F.; Zhou, L.; Zhu, E.; Teng, H.; Li, X.; Bao, Q.; Wu, J.; Sun, Z. MagicViewer: Integrated solution for next-generation sequencing data visualization and genetic variation detection and annotation. Nucleic Acids Res. 2010, 38, W732–W736. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Wan, Z.; Coarfa, C.; Drabek, R.; Chen, L.; Ostrowski, E.A.; Liu, Y.; Weinstock, G.M.; Wheeler, D.A.; Gibbs, R.A.; Yu, F. A SNP discovery method to assess variant allele probability from next-generation resequencing data. Genome Res. 2010, 20, 273–280. [Google Scholar] [CrossRef] [PubMed]
- Tsuchihashi, Z.; Dracopoli, N. Progress in high throughput SNP genotyping methods. Pharmacogenomics J. 2002, 2, 103–110. [Google Scholar] [CrossRef] [PubMed]
- Sobrino, B.; Brion, M.; Carracedo, A. SNPs in forensic genetics: A review on SNP typing methodologies. Forensic Sci. Int. 2005, 154, 181–194. [Google Scholar] [CrossRef] [PubMed]
- Giancola, S.; McKhann, H.I.; Bérard, A.; Camilleri, C.; Durand, S.; Libeau, P.; Roux, F.; Reboud, X.; Gut, I.G.; Brunel, D. Utilization of the three high-throughput SNP genotyping methods, the GOOD assay, Amplifluor and TaqMan, in diploid and polyploid plants. Theor. Appl. Genet. 2006, 112, 1115–1124. [Google Scholar] [CrossRef] [PubMed]
- Gupta, P.K.; Rustgi, S.; Mir, R.R. Array-based high-throughput DNA markers for crop improvement. Heredity 2008, 101, 5–18. [Google Scholar] [CrossRef] [PubMed]
- Ragoussis, J. Genotyping technologies for genetic research. Ann. Rev. Genomics Hum. Genet. 2009, 10, 117–133. [Google Scholar] [CrossRef] [PubMed]
- Garvin, M.; Saitoh, K.; Gharrett, A. Application of single nucleotide polymorphisms to non-model species: A technical review. Mol. Ecol. Res. 2010, 10, 915–934. [Google Scholar] [CrossRef] [PubMed]
- Bagge, M.; Xia, X.; Lubberstedt, T. Functional markers in wheat. Curr. Opin. Plant Biol. 2007, 10, 211–216. [Google Scholar] [CrossRef] [PubMed]
- Syvänen, A.-C. Accessing genetic variation: Genotyping single nucleotide polymorphisms. Nat. Rev. Genet. 2001, 2, 930–942. [Google Scholar] [CrossRef] [PubMed]
- Oraguzie, N.C.; Rikkerink, E.H.A.; Gardiner, S.E.; de Silva, H.N. Association Mapping in Plants; Springer: Berlin, Germany, 2007. [Google Scholar]
- Gunderson, K.L.; Steemers, F.J.; Lee, G.; Mendoza, L.G.; Chee, M.S. A genome-wide scalable SNP genotyping assay using microarray technology. Nat. Genet. 2005, 37, 549–554. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.B.; Chee, M.S.; Gunderson, K.L. Highly parallel genomic assays. Nat. Rev. Genet. 2006, 7, 632–644. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, A.I.; Pérez-Montarelo, D.; Barragán, C.; Ramayo-Caldas, Y.; Ibáñez-Escriche, N.; Castelló, A.; Noguera, J.L.; Silió, L.; Folch, J.M.; Rodríguez, M.C. Genome-wide linkage analysis of QTL for growth and body composition employing the PorcineSNP60 BeadChip. BMC Genet. 2012, 13. [Google Scholar] [CrossRef] [PubMed]
- Baird, N.A.; Etter, P.D.; Atwood, T.S.; Currey, M.C.; Shiver, A.L.; Lewis, Z.A.; Selker, E.U.; Cresko, W.A.; Johnson, E.C. Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS ONE 2008, 3, e3376. [Google Scholar] [CrossRef] [PubMed]
- Ahmadian, A.; Gharizadeh, B.; Gustafsson, A.C.; Sterky, F.; Nyren, P.; Uhlen, M.; Lundeberg, J. Single-nucleotide polymorphism analysis by pyrosequencing. Anal. Biochem. 2000, 280, 103–110. [Google Scholar] [CrossRef] [PubMed]
- Gabriel, S.; Ziaugra, L.; Tabbaa, D. SNP genotyping using the Sequenom MassARRAY iPLEX platform. Curr. Protoc. Hum. Genet. 2009, 2. [Google Scholar] [CrossRef]
- Bell, P.A.; Chaturvedi, S.; Gelfand, C.A.; Huang, C.Y.; Kochersperger, M.; Kopla, R.; Modica, F.; Pohl, M.; Varde, S.; Zhao, R.; et al. SNPstream UHT: Ultra-high throughput SNP genotyping for pharmacogenomics and drug discovery. Biotechniques 2002, 74, 76–77. [Google Scholar]
- Bouakaze, C.; Keyser, C.; de Martino, S.J.; Sougakoff, W.; Veziris, N.; Dabernat, H.; Ludes, B. Identification and genotyping of mycobacterium tuberculosis complex species by use of a SNaPshot Minisequencing-based assay. J. Clin. Microbiol. 2010, 48, 1758–1766. [Google Scholar] [CrossRef] [PubMed]
- Kamińiski, S.; Brym, P.; Ruść, A.; Wojcik, E. Microarray of SNPs for diverse applications in commercial pig breeding. Pol. J. Vet. Sci. 2008, 12, 69–74. [Google Scholar]
- Kurg, A.; Tõnisson, N.; Georgiou, I.; Shumaker, J.; Tollett, J.; Metspalu, A. Arrayed primer extension: Solid-phase four-color DNA resequencing and mutation detection technology. Genet. Test. 2000, 4, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Bruse, S.E.; Moreau, M.P.; Azaro, M.A.; Zimmerman, R.; Brzustowicz, L.M. Improvements to bead-based oligonucleotide ligation SNP genotyping assays. Biotechniques 2008, 45, 559–571. [Google Scholar] [CrossRef] [PubMed]
- Prince, J.A.; Feuk, L.; Howell, W.M.; Jobs, M.; Emahazion, T.; Blennow, K.; Brookes, A.J. Robust and accurate single nucleotide polymorphism genotyping by dynamic allele-specific hybridization (DASH): Design criteria and assay validation. Genome Res. 2001, 11, 152–162. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Liu, J.; Sun, D.; Ma, P.; Ding, X.; Yu, Y.; Zhang, Q. Genome wide association studies for milk production traits in Chinese Holstein population. PLoS ONE 2010, 5, e13661. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Yang, J.; Zhou, L.; Ren, J.; Liu, X.; Zhang, H.; Yang, B.; Zhang, Z.; Ma, H.; Xie, X. A splice mutation in the PHKG1 gene causes high glycogen content and low meat quality in pig skeletal muscle. PLoS Genet. 2014, 10, e1004710. [Google Scholar] [CrossRef] [PubMed]
- Houston, R.D.; Davey, J.W.; Bishop, S.C.; Lowe, N.R.; Mota-Velasco, J.C.; Hamilton, A.; Guy, D.R.; Tinch, A.E.; Thomson, M.L.; Blaxter, M.L.; et al. Characterisation of QTL-linked and genome-wide restriction site-associated DNA (RAD) markers in farmed Atlantic salmon. BMC Genomics 2012, 13. [Google Scholar] [CrossRef] [PubMed]
- Lo, R.C.; Ugaz, V.M. Microchip DNA electrophoresis with automated whole-gel scanning detection. Lab Chip 2008, 8, 2135–2145. [Google Scholar] [CrossRef] [PubMed]
- Julich, S.; Riedel, M.; Kielpinski, M.; Urban, M.; Kretschmer, R.; Wagner, S.; Fritzsche, W.; Henkel, T.; Möller, R.; Werres, S. Development of a Lab-on-a-Chip device for diagnosis of plant pathogens. Biosens. Bioelectron. 2011, 26, 4070–4075. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.G.; Cheong, K.H.; Huh, N.; Kim, S.; Choi, J.W.; Ko, C. Microchip-based one step DNA extraction and real-time PCR in one chamber for rapid pathogen identification. Lab Chip 2006, 6, 886–895. [Google Scholar] [CrossRef] [PubMed]
- Marasso, S.L.; Mombello, D.; Cocuzza, M.; Casalena, D.; Ferrante, I.; Nesca, A.; Poiklik, P.; Rekker, K.; Aaspollu, A.; Ferrero, S. A polymer Lab-on-a-Chip for genetic analysis using the arrayed primer extension on microarray chips. Biomed. Microdevices 2014, 16, 661–670. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.H.; Su, T.Y.; Liu, Y.J.; Chang, H.Y.; Yao, D.J. Single-Nucleotide Polymorphism Detection Based on a Temperature-Controllable Electrowetting on Dielectrics Digital Microfluidic System. Sens. Mater. 2013, 25, 643–651. [Google Scholar]
- Kolchinsky, A.; Mirzabekov, A. Analysis of SNPs and other genomic variations using gel-based chips. Hum. Mutat. 2002, 19, 343–360. [Google Scholar] [CrossRef] [PubMed]
- Dubiley, S.; Kirillov, E.; Mirzabekov, A. Polymorphism analysis and gene detection by minisequencing on an array of gel-immobilized primers. Nucleic Acids Res. 1999, 27. [Google Scholar] [CrossRef]
- Russom, A.; Haasl, S.; Brookes, A.J.; Andersson, H.; Stemme, G. Rapid melting curve analysis on monolayered beads for high-throughput genotyping of single-nucleotide polymorphisms. Anal. Chemis. 2006, 78, 2220–2225. [Google Scholar] [CrossRef] [PubMed]
- Shin, D.; Zhang, Y.; Wang, T.-H. A droplet microfluidic approach to single-stream nucleic acid isolation and mutation detection. Microfluid. Nanofluidics 2014, 17, 425–430. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Shin, D.J.; Wang, T.H. Detecting genetic variations in a droplet. In Proceedings of the 15th International Conference on Miniaturized Chemical and Biochemical Analysis Systems (Micro-TAS 2011), Seattle, WA, USA, 2–6 October 2011; Volume 978, pp. 4–5.
- Li, K.C.; Ding, S.T.; Lin, E.C.; Wang, L.A.; Lu, Y.W. Melting analysis on microbeads in rapid temperature-gradient inside microchannels for single nucleotide polymorphisms detection. Biomicrofluidics 2014, 8. [Google Scholar] [CrossRef] [PubMed]
- Sochol, R.D.; Casavant, B.P.; Dueck, M.E.; Lee, L.P.; Lin, L. A dynamic bead-based microarray for parallel DNA detection. J. Micromech. Microeng. 2011, 21, 054019. [Google Scholar] [CrossRef]
- Koopaee, H.K.; Koshkoiyeh, A.E. SNPs genotyping technologies and their applications in farm animals breeding programs: Review. Braz. Arch. Biol. Technol. 2014, 57, 87–95. [Google Scholar] [CrossRef]
- Schroyen, M.; Stinckens, A.; Verhelst, R.; Niewold, T.; Buys, N. The search for the gene mutations underlying enterotoxigenic Escherichia coli F4ab/ac susceptibility in pigs: A review. Vet. Res. 2012, 43. [Google Scholar] [CrossRef] [PubMed]
- Rothschild, M.F. Genetics and reproduction in the pig. Anim. Reprod. Sci. 1996, 42, 143–151. [Google Scholar] [CrossRef]
- Onteru, S.; Fan, B.; Du, Z.Q.; Garrick, D.; Stalder, K.; Rothschild, M. A whole-genome association study for pig reproductive traits. Anim. Genet. 2012, 43, 18–26. [Google Scholar] [CrossRef] [PubMed]
- Cao, S.; Han, J.; Wu, J.; Li, Q.; Liu, S.; Zhang, W.; Pei, Y.; Ruan, X.; Liu, Z.; Wang, X.; et al. Specific gene-regulation networks during the pre-implantation development of the pig embryo as revealed by deep sequencing. BMC Genomics 2014, 15. [Google Scholar] [CrossRef] [PubMed]
- Wilson, M.E.; Biensen, N.J.; Ford, S.P. Novel insight into the control of litter size in pigs, using placental efficiency as a selection tool. J. Anim. Sci. 1999, 77, 1654–1658. [Google Scholar] [PubMed]
- Chang, P. Using High-Throughput Single Nucleotide Polymorphism Genotyping to Reveal Litter Size-Related Molecular Markers in Landrace Sows. In Animal Science; National Taiwan University: Taipei, Taiwan, 2010. [Google Scholar]
- Rothschild, M.; Jacobson, C.; Vaske, D.; Tuggle, C.; Wang, L.; Short, T.; Eckardt, G.; Sasaki, S.; Vincent, A.; McLaren, D.; et al. The estrogen receptor locus is associated with a major gene influencing litter size in pigs. Proc. Natl. Acad. Sci. USA 1996, 93, 201–205. [Google Scholar] [CrossRef] [PubMed]
- Munoz, G.; Ovilo, C.; Amills, M.; Rodriguez, C. Mapping of the porcine oestrogen receptor 2 gene and association study with litter size in Iberian pigs. Anim. Genet. 2004, 35, 242–244. [Google Scholar] [CrossRef] [PubMed]
- Terman, A.; Kumalska, M. The effect of a SNP in ESR gene on the reproductive performance traits in Polish sows. Russ. J. Genet. 2012, 48, 1260–1263. [Google Scholar] [CrossRef]
- Judyma, D. Polymorphism in the PRLR/AluI gene and its effect on litter size in Large White sows. Anim. Sci. Pap. Rep. 2004, 22, 523–527. [Google Scholar]
- Hirose, K.; Ito, T.; Fukawa, K.; Arakawa, A.; Mikawa, S.; Hayashi, Y.; Tanaka, K. Evaluation of effects of multiple candidate genes (LEP, LEPR, MC4R, PIK3C3, and VRTN) on production traits in Duroc pigs. Anim. Sci. J. 2014, 85, 198–206. [Google Scholar] [CrossRef] [PubMed]
- Muñoz, G.; Alcázar, E.; Fernández, A.; Barragán, C.; Carrasco, A.; de Pedro, E.; Silió, L.; Sánchez, J.L.; Rodríguez, M.C. Effects of porcine MC4R and LEPR polymorphisms, gender and Duroc sire line on economic traits in Duroc × Iberian crossbred pigs. Meat Sci. 2011, 88, 169–173. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Montarelo, D.; Rodríguez, M.C.; Fernández, A.; Benítez, R.; García, F.; Silió, L.; Fernández, A. Haplotypic diversity of porcine LEP and LEPR genes involved in growth and fatness regulation. J. Appl. Genet. 2015, 56, 525–533. [Google Scholar] [CrossRef] [PubMed]
- Balcells, I.; Castelló, A.; Noguera, J.L.; Fernández-Rodríguez, A.; Sánchez, A.; Tomás, A. Sequencing and gene expression of the porcine ITIH SSC13 cluster and its effect on litter size in an Iberian × Meishan F2 population. Anim. Reprod. Sci. 2011, 128, 85–92. [Google Scholar] [CrossRef] [PubMed]
- Rothschild, M.F. Porcine genomics delivers new tools and results: This little piggy did more than just go to market. Genet. Res. 2004, 83, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Bongiorni, S.; Tilesi, F.; Bicorgna, S.; Iacoponi, F.; Willems, D.; Gargani, M.; D'Andrea, M.; Pilla, F.; Valentini, A. Promoter polymorphisms in genes involved in porcine myogenesis influence their transcriptional activity. BMC Genet. 2014, 15. [Google Scholar] [CrossRef] [PubMed]
- Van Laere, A.-S.; Nguyen, M.; Braunschweig, M.; Nezer, C.; Collette, C.; Moreau, L.; Archibald, A.L.; Haley, C.S.; Buys, N.; Tally, M. A regulatory mutation in IGF2 causes a major QTL effect on muscle growth in the pig. Nature 2003, 425, 832–836. [Google Scholar] [CrossRef] [PubMed]
- Vykoukalova, Z.; Knoll, A.; Dvořák, J.; Čepica, S. New SNPs in the IGF2 gene and association between this gene and backfat thickness and lean meat content in Large White pigs. J. Anim. Breed. Genet. 2006, 123, 204–207. [Google Scholar] [CrossRef] [PubMed]
- Jeon, J.-T.; Carlborg, O.; Törnsten, A.; Giuffra, E.; Amarger, V.; Chardon, P.; Andersson-Eklund, L.; Andersson, K.; Hansson, I.; Lundström, K.; et al. A paternally expressed QTL affecting skeletal and cardiac muscle mass in pigs maps to the IGF2 locus. Nat. Genet. 1999, 21, 157–158. [Google Scholar] [PubMed]
- Tuggle, C.; Yu, T.P.; Helm, J.; Rothschild, M. Cloning and restriction fragment length polymorphism analysis of a cDNA for swine PIT-1, a gene controlling growth hormone expression. Anim. Genet. 1993, 24, 17–21. [Google Scholar] [CrossRef] [PubMed]
- Klont, R.E.; Lambooy, E.; van Logtestijn, J.G. Effect of dantrolene treatment on muscle metabolism and meat quality of anesthetized pigs of different halothane genotypes. J. Anim. Sci. 1994, 72, 2008–2016. [Google Scholar] [PubMed]
- Liu, M.; Peng, J.; Xu, D.Q.; Zheng, R.; Li, F.G.; Li, J.L.; Zuo, B.; Lei, M.G.; Xiong, Y.Z.; Deng, C.Y.; et al. Associations of MYF5 gene polymorphisms with meat quality traits in different domestic pig (Sus scrofa) populations. Genetics Mol. Biol. 2007, 30, 370–374. [Google Scholar]
- Stupka, R.; Citek, J.; Sprysl, M.; Okrouhla, M.; Brzobohaty, L. The impact of MYOG, MYF6 and MYOD1 genes on meat quality traits in crossbred pigs. Afr. J. Biotechnol. 2012, 11, 15405–15409. [Google Scholar]
- Wright, D.; Kerje, S.; Lundström, K.; Babol, J.; Schütz, K.; Jensen, P.; Andersson, L. Quantitative trait loci analysis of egg and meat production traits in a red junglefowl × White Leghorn cross. Anim. Genet. 2006, 37, 529–534. [Google Scholar] [CrossRef] [PubMed]
- Rubin, C.-J.; Zody, M.C.; Eriksson, J.; Meadows, J.R.S.; Sherwood, E.; Webster, W.T.; Jiang, L.; Ingman, M.; Sharpe, T.; Ka, S.; et al. Whole-genome resequencing reveals loci under selection during chicken domestication. Nature 2010, 464, 587–591. [Google Scholar] [CrossRef] [PubMed]
- Kranis, A.; Gheyas, A.A.; Boschiero, C.; Turner, F.; Yu, L.; Smith, S.; Talbot, R.; Pirani, A.; Brew, F.; Kaiser, P.; et al. Development of a high density 600K SNP genotyping array for chicken. BMC Genomics 2013, 14. [Google Scholar] [CrossRef] [PubMed]
- Yao, J.; Chen, Z.; Xu, G.; Wang, X.; Ning, Z.; Zheng, J.; Qu, L.; Yang, N. Low-density lipoprotein receptor-related protein 8 gene association with egg traits in dwarf chickens. Poult. Sci. 2010, 89, 883–886. [Google Scholar] [CrossRef] [PubMed]
- Sundaresan, N.; Marcus Leo, M.D.; Subramani, J.; Anish, D.; Sudhagar, M.; Ahmed, K.A.; Saxena, M.; Tyagi, J.S.; Sastry, K.V.; Saxena, V.K. Expression analysis of melatonin receptor subtypes in the ovary of domestic chicken. Vet. Res. Commun. 2009, 33, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Zhang, L.; Smith, D.; Xu, H.; Liu, Y.; Zhao, X.; Wang, Y.; Zhu, Q. Genetic effects of melatonin receptor genes on chicken reproductive traits. Czech J. Anim. Sci. 2013, 58, 58–64. [Google Scholar]
- Yu, S.; Chu, W.; Zhang, L.; Han, H.; Zhao, R.; Wu, W.; Zhu, J.; Dodson, M.V.; Wei, W.; Liu, H.; Chen, J. Identification of Laying-Related SNP Markers in Geese Using RAD Sequencing. PLoS ONE 2015, 10, e0131572. [Google Scholar] [CrossRef] [PubMed]
- Jiang, R.; Li, J.; Qu, L.; Li, H.; Yang, N. A new single nucleotide polymorphism in the chicken pituitary-specific transcription factor (POU1F1) gene associated with growth rate. Anim. Genet. 2004, 35, 344–346. [Google Scholar] [CrossRef] [PubMed]
- Nie, Q.; Fang, M.; Xie, L.; Zhou, M.; Liang, Z.; Luo, Z.; Wang, G.; Bi, W.; Liang, C.; Zhang, W. The PIT1 gene polymorphisms were associated with chicken growth traits. BMC Genet. 2008, 9. [Google Scholar] [CrossRef] [PubMed]
- Qiu, X.; Li, N.; Deng, X.; Zhao, X.; Meng, Q.; Wang, X. The single nucleotide polymorphisms of chicken melanocortin-4 receptor (MC4R) gene and their association analysis with carcass traits. Sci. China C Life Sci. 2006, 49, 560–566. [Google Scholar] [CrossRef] [PubMed]
- Ansorge, W.J. Next-generation DNA sequencing techniques. New Biotechnol. 2009, 25, 195–203. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, C.-W.; Lin, Y.-T.; Ding, S.-T.; Lo, L.-L.; Wang, P.-H.; Lin, E.-C.; Liu, F.-W.; Lu, Y.-W. Efficient SNP Discovery by Combining Microarray and Lab-on-a-Chip Data for Animal Breeding and Selection. Microarrays 2015, 4, 570-595. https://doi.org/10.3390/microarrays4040570
Huang C-W, Lin Y-T, Ding S-T, Lo L-L, Wang P-H, Lin E-C, Liu F-W, Lu Y-W. Efficient SNP Discovery by Combining Microarray and Lab-on-a-Chip Data for Animal Breeding and Selection. Microarrays. 2015; 4(4):570-595. https://doi.org/10.3390/microarrays4040570
Chicago/Turabian StyleHuang, Chao-Wei, Yu-Tsung Lin, Shih-Torng Ding, Ling-Ling Lo, Pei-Hwa Wang, En-Chung Lin, Fang-Wei Liu, and Yen-Wen Lu. 2015. "Efficient SNP Discovery by Combining Microarray and Lab-on-a-Chip Data for Animal Breeding and Selection" Microarrays 4, no. 4: 570-595. https://doi.org/10.3390/microarrays4040570
APA StyleHuang, C.-W., Lin, Y.-T., Ding, S.-T., Lo, L.-L., Wang, P.-H., Lin, E.-C., Liu, F.-W., & Lu, Y.-W. (2015). Efficient SNP Discovery by Combining Microarray and Lab-on-a-Chip Data for Animal Breeding and Selection. Microarrays, 4(4), 570-595. https://doi.org/10.3390/microarrays4040570







