SNP Array in Hematopoietic Neoplasms: A Review
Abstract
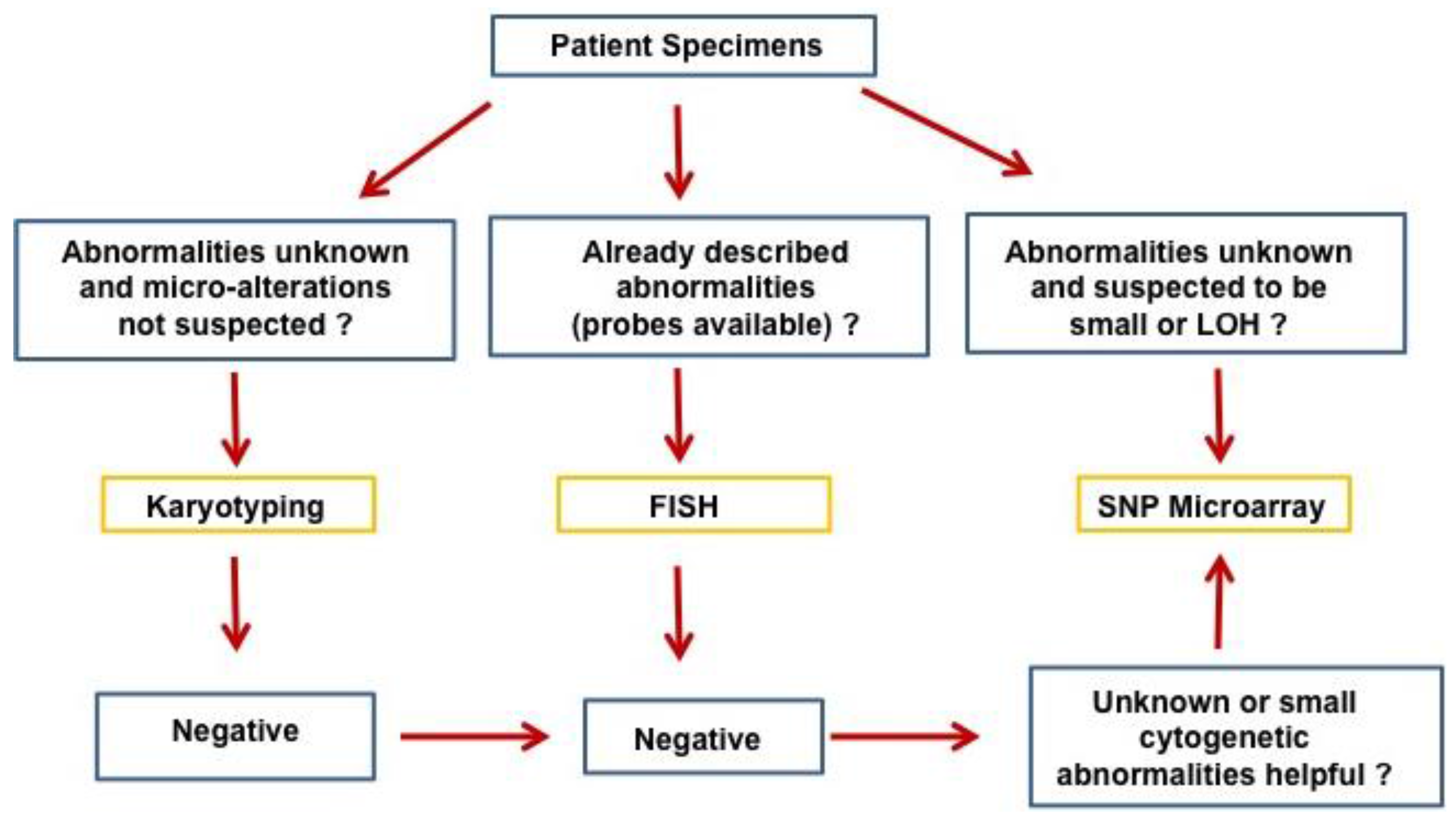
:1. Introduction
2. Acute Lymphoblastic Leukemia/Lymphoma
3. Acute Myeloid Leukemia
4. Myelodysplastic Syndrome
5. Chronic Myelogenous Leukemia
6. Polycythemia Vera, Essential Thrombocythemia and Primary Myelofibrosis
7. Myelodysplastic/Myeloproliferative Neoplasms
8. Classical Hodgkin Lymphoma
9. Mature B-Cell Lymphoproliferative Disorder
10. Mature T/NK-Cell Lymphoproliferative Disorders
11. Conclusions
| Disease | CNVs/CNAs and/or Associated Genes | LOH/UPD and/or Associated Genes | Prognostic Association | Ref. |
|---|---|---|---|---|
| B-ALL | Deletion of PAX5, EBF1, TCF3, LEF1, IKZF1 (IKAROS), IKZF3 (AIOLOS), ETV6, and CDKN2A/p16INK4A | 9p (CDKN2A/B) | - | [26,27,28,29,35] |
| T-ALL | Deletion of TAL1, RB1, PTEN, CDKN2A, CDKN2B, LEF1, and STIL; Gains of MYB | 9p (CDKN2A) | - | [32] |
| AML | Deletion of 3p14.1–p13 (FOXP1, RYBP, FHIT), 6q27 (RPS6KA2), 8q23.3 (TRPS1), 10q11.21 (HNRPF), 11q25, 12p13.2 (ETV6), 15q21.3 (RFXDC2), 5q31.1 (CTNNA1), 16q22.1 (CBFB), 17p13.1 (TP53), 17q11.2 (NF1) | 13q (FLT3), 11p (WT1, PU1) and 11q (MLL), 19q (CEBPA), 6p and 21q (RUNX1) | Worse prognosis: ≥2 genomic lesions detected by SNP array | [42,43,44,45,47,48,49] |
| Amplifications of 8q23.2 (MYC), 11q23.3 (MLL), and 21q22.2 (ETS2) | ||||
| MDS | Deletion and aUPD of chromosomes 1, 5q, 7, 11, 17, and 21 | Worse prognosis: UPDs of 7q; New genetic lesions detected by SNP array; EZH2 mutations | [57,58,60,61,62,63,64] | |
| Deletion of EZH2 and TET2 | ||||
| UPD 20p (BMP2 and TRIB3) | ||||
| CML | Frequent amplifications in chronic phase | 1, 8, 9, 17, 19, and 22 in TKI-resistant CML | - | [66,67] |
| Deletion of IKFZ1 in lymphoid blast phase | ||||
| MPN | Rare in ET and PV | 9p (JAK2) | Worse prognosis: CNN-LOH on 7q or 9p (JAK2 V617F); Genetic aberrations of chromosome 5, 7, or 17p | [68,69,70,73,74] |
| Deletion of 13q14 (RB) or 17q11 (NF1) in PMF | ||||
| CMML | Frequent microdeletions | 7q (EZH2), 11q (CBL), and 4q (TET2) | Worse prognosis: Multiple chromosomal defects detected by SNP array | [75,76] |
| cHL | Gain of MAP3K14 | 14q (TRAF3) | - | [79] |
| DLBCL | Frequent gains and deletions; gains HDAC7A on chromosome 12 predominantly in GCB-DLBCL, losses of BACH2 and CASP8AP2 on chromosome 6 predominantly in ABC-DLBCL; Potential tumor suppressor genes: CASP3, IL5RA ARID1B, ROBO2 and MRS1; Potential oncogenes: KLHL6, IL31 and LRP1 | 11p11.2 (PTPRJ), | Worse prognosis: Loss of 8p23.1 | [81,82,83,84] |
| FL | CDKN2A, CDKN2B, FHIT, KIT, PEX14, and PTPRD | 1p36 (TNFRSF14), 6p, 6q, 9p (CDKN2A), 10q, 12q, 16p, and 17p (TP53) | Worse prognosis: >3 SNP abnormalities; aUPD and deletion of 1p36, aUPD of 16p | [85,86] |
| CLL | Deletion of 17p13 (TP53), 11q22 (ATM) and 13q14 (DLEU1 and DLEU2), 2p16.1–2p15, 8q24.21, 6q21 (AIM1) | 13q, 13 (miR-15a/miR-16-1), 17p, and 11q | Worse prognosis: Genomic complexity; large genomic aberrations; large (type II) 13q14 deletions | [91,92,93,101] |
| Gain of 12, 2p16 (REL, BCL11A) | ||||
| MCL | Deletion of INK4A/ARF, ATM, TP53, 1p, 6q, CDKN2C, BCL2L11, CDKN2A, and RB1, FAF1, MAP2, SP100, MOBKL2B, ZNF280A, and PRAME. | 9p, 9, 17p (TP53) | - | [104,105] |
| Amplification of MYC, 11q13(cyclin D1), 13q (miR17-92, C13 or f25), dup(3q), 18q (BCL2) | ||||
| MZL | Deletion of 6q23 (TNFAIP3, A20), 9p | 6q (A20), 3q | - | [106,107,108] |
| Gain of 3, 18, 6p and 21q | ||||
| Gains of REL, BCL11A, ETS1, PTPN1, PTEN and KRAS in transformation to DLBCL | ||||
| BL | Losses of 6q14.1–q22.33, 9p21.3 (CDKN2A), and 13q14.2–q14.3 | 6p12.2-pter, 9p23-pter, and 17p11.2-pter (TP53). | - | [109] |
| Gains of 1q23.3–q31.3, 7, 13q31.3, | ||||
| MM | Genomic alterations at 1p, 1q, 6q, 8p, 13, and 16q | 1q, 16q (CYLD), and X | Worse prognosis: Amplifications in 1q and deletions in 1p, 12p, 14q, 16q, and 22q | [111,112,115,116,117] |
| PTCL, NOS | Losses of 1p35-36, 3q, 5q33, 6p22, 6q16, 6q21–22, 8p21–23, 9p21, 10p11–12, 10q11-22, 10q25–26, 13q14, 15q24, 16q22, 16q24, 17p11, 17p13 and Xp22 | 2q32.3 | - | [120,121] |
| Gains of 1q32–43, 2p15–16 (REL), 7, 8q24, 11q14–25, 17q11–21 and 21q11–21, 9p and 19q | ||||
| AILT | Loss of 3q and 9p | 2q32.3 | Worse prognosis: The presence of CNAs; overexpression of CARMA1 at 7p22 and MYCBP2 at 13q22 | [121] |
| Gains of 8q, 9p and 19q | ||||
| ATLL | Deletion of 10p11.2 ( TCF8) | - | - | [122] |
| T-PLL | Loss in 6q, 8p, 10p, 11q (microRNA 34b/c, ETS1 and FLI1), and 18p and Gains of 6p, 8q | 3q, 17q | - | [124,125] |
| Aberrations in 5p, 12p, 13q, 17 and 22 (DNAH5, ETV6, miR-15a and miR-16-1, p53, BIRC5, and SOCS3) | ||||
| SS | Loss at 4q35 (FAT), 4q34 (VEGFC), 12 (NFIB), and 17p11.2 (TRIM16) | 9q31q34, 10p11q26, and 13q11q12 | - | [126] |
Author Contributions
Conflicts of Interest
References
- Hahn, W.C.; Weinberg, R.A. Rules for making human tumor cells. N. Engl. J. Med. 2002, 347, 1593–1603. [Google Scholar] [CrossRef] [PubMed]
- Bayani, J.S.J. Traditional banding of chromosomes for cytogenetic analysis. Curr. Protoc. Cell Biol. 2004, S23. [Google Scholar] [CrossRef]
- Levsky, J.M.; Singer, R.H. Fluorescence in situ hybridization: Past, present and future. J. Cell Sci. 2003, 116, 2833–2838. [Google Scholar] [CrossRef] [PubMed]
- Kallioniemi, A.; Kallioniemi, O.P.; Sudar, D.; Rutovitz, D.; Gray, J.W.; Waldman, F.; Pinkel, D. Comparative genomic hybridization for molecular cytogenetic analysis of solid tumors. Science 1992, 258, 818–821. [Google Scholar] [CrossRef] [PubMed]
- Du Manoir, S.; Speicher, M.R.; Joos, S.; Schrock, E.; Popp, S.; Dohner, H.; Kovacs, G.; Robert-Nicoud, M.; Lichter, P.; Cremer, T. Detection of complete and partial chromosome gains and losses by comparative genomic in situ hybridization. Hum. Genet. 1993, 90, 590–610. [Google Scholar] [CrossRef] [PubMed]
- Pinkel, D.; Albertson, D.G. Comparative genomic hybridization. Annu Rev. Genomics Hum. Genet. 2005, 6, 331–354. [Google Scholar] [CrossRef] [PubMed]
- De Ravel, T.J.; Devriendt, K.; Fryns, J.P.; Vermeesch, J.R. What’s new in karyotyping? The move towards array comparative genomic hybridisation (CGH). Eur. J. Pediatr. 2007, 166, 637–643. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, G.C.; Matsuzaki, H.; Dong, S.; Liu, W.M.; Huang, J.; Liu, G.; Su, X.; Cao, M.; Chen, W.; Zhang, J.; et al. Large-scale genotyping of complex DNA. Nat. Biotechnol. 2003, 21, 1233–1237. [Google Scholar] [CrossRef] [PubMed]
- Lindblad-Toh, K.; Tanenbaum, D.M.; Daly, M.J.; Winchester, E.; Lui, W.O.; Villapakkam, A.; Stanton, S.E.; Larsson, C.; Hudson, T.J.; Johnson, B.E.; et al. Loss-of-heterozygosity analysis of small-cell lung carcinomas using single-nucleotide polymorphism arrays. Nat. Biotechnol. 2000, 18, 1001–1005. [Google Scholar] [PubMed]
- Wong, K.K.; Tsang, Y.T.; Shen, J.; Cheng, R.S.; Chang, Y.M.; Man, T.K.; Lau, C.C. Allelic imbalance analysis by high-density single-nucleotide polymorphic allele (SNP) array with whole genome amplified DNA. Nucleic Acids Res. 2004, 32. [Google Scholar] [CrossRef] [PubMed]
- La Framboise, T. Single nucleotide polymorphism arrays: A decade of biological, computational and technological advances. Nucleic Acids Res. 2009, 37, 4181–4193. [Google Scholar] [CrossRef] [PubMed]
- Mardis, E.R.; Ding, L.; Dooling, D.J.; Larson, D.E.; McLellan, M.D.; Chen, K.; Koboldt, D.C.; Fulton, R.S.; Delehaunty, K.D.; McGrath, S.D.; et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N. Engl. J. Med. 2009, 361, 1058–1066. [Google Scholar] [CrossRef] [PubMed]
- Ley, T.J.; Mardis, E.R.; Ding, L.; Fulton, B.; McLellan, M.D.; Chen, K.; Dooling, D.; Dunford-Shore, B.H.; McGrath, S.; Hickenbotham, M.; et al. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature 2008, 456, 66–72. [Google Scholar] [CrossRef] [PubMed]
- Stiller, C.A.; Parkin, D.M. Geographic and ethnic variations in the incidence of childhood cancer. Br. Med. Bull. 1996, 52, 682–703. [Google Scholar] [CrossRef] [PubMed]
- Swerdlow, S.H.; Campo, E.; Harris, N.L.; Jaffe, E.S.; Pileri, S.A.; Stein, H.; Thiele, J.; Vardiman, J.W. Who Classification of Tumours of Haematopoietic and Lymphoid Tissues; International Agency for Research on Cancer: Lyon, France, 2008; pp. 167–178. [Google Scholar]
- Schrappe, M.; Reiter, A.; Ludwig, W.D.; Harbott, J.; Zimmermann, M.; Hiddemann, W.; Niemeyer, C.; Henze, G.; Feldges, A.; Zintl, F.; et al. Improved outcome in childhood acute lymphoblastic leukemia despite reduced use of anthracyclines and cranial radiotherapy: Results of trial ALL-BFM 90. German-Austrian-Swiss ALL-BFM study group. Blood 2000, 95, 3310–3322. [Google Scholar] [PubMed]
- Gaynon, P.S.; Trigg, M.E.; Heerema, N.A.; Sensel, M.G.; Sather, H.N.; Hammond, G.D.; Bleyer, W.A. Children’s cancer group trials in childhood acute lymphoblastic leukemia: 1983–1995. Leukemia 2000, 14, 2223–2233. [Google Scholar] [CrossRef] [PubMed]
- Harms, D.O.; Janka-Schaub, G.E. Co-operative study group for childhood acute lymphoblastic leukemia (COALL): Long-term follow-up of trials 82, 85, 89 and 92. Leukemia 2000, 14, 2234–2239. [Google Scholar] [CrossRef] [PubMed]
- Silverman, L.B.; Gelber, R.D.; Dalton, V.K.; Asselin, B.L.; Barr, R.D.; Clavell, L.A.; Hurwitz, C.A.; Moghrabi, A.; Samson, Y.; Schorin, M.A.; et al. Improved outcome for children with acute lymphoblastic leukemia: Results of dana-farber consortium protocol 91–01. Blood 2001, 97, 1211–1218. [Google Scholar] [CrossRef] [PubMed]
- Gustafsson, G.; Schmiegelow, K.; Forestier, E.; Clausen, N.; Glomstein, A.; Jonmundsson, G.; Mellander, L.; Makipernaa, A.; Nygaard, R.; Saarinen-Pihkala, U.M. Improving outcome through two decades in childhood all in the nordic countries: The impact of high-dose methotrexate in the reduction of CNS irradiation. Nordic society of pediatric haematology and oncology (NOPHO). Leukemia 2000, 14, 2267–2275. [Google Scholar] [CrossRef] [PubMed]
- Pui, C.H.; Sandlund, J.T.; Pei, D.; Rivera, G.K.; Howard, S.C.; Ribeiro, R.C.; Rubnitz, J.E.; Razzouk, B.I.; Hudson, M.M.; Cheng, C.; et al. Results of therapy for acute lymphoblastic leukemia in black and white children. JAMA 2003, 290, 2001–2007. [Google Scholar] [CrossRef] [PubMed]
- Gokbuget, N.; Hoelzer, D. Recent approaches in acute lymphoblastic leukemia in adults. Rev. Clin. Exp. Hematol. 2002, 6, 114–141. [Google Scholar] [CrossRef] [PubMed]
- Kantarjian, H.M.; O’Brien, S.; Smith, T.L.; Cortes, J.; Giles, F.J.; Beran, M.; Pierce, S.; Huh, Y.; Andreeff, M.; Koller, C.; et al. Results of treatment with hyper-CVAD, a dose-intensive regimen, in adult acute lymphocytic leukemia. J. Clin. Oncol. 2000, 18, 547–561. [Google Scholar] [PubMed]
- Linker, C.; Damon, L.; Ries, C.; Navarro, W. Intensified and shortened cyclical chemotherapy for adult acute lymphoblastic leukemia. J. Clin. Oncol. 2002, 20, 2464–2471. [Google Scholar] [CrossRef] [PubMed]
- Greaves, M.F.; Wiemels, J. Origins of chromosome translocations in childhood leukaemia. Nat. Rev. Cancer 2003, 3, 639–649. [Google Scholar] [CrossRef] [PubMed]
- Irving, J.A.; Bloodworth, L.; Bown, N.P.; Case, M.C.; Hogarth, L.A.; Hall, A.G. Loss of heterozygosity in childhood acute lymphoblastic leukemia detected by genome-wide microarray single nucleotide polymorphism analysis. Cancer Res. 2005, 65, 3053–3058. [Google Scholar] [PubMed]
- Mullighan, C.G.; Goorha, S.; Radtke, I.; Miller, C.B.; Coustan-Smith, E.; Dalton, J.D.; Girtman, K.; Mathew, S.; Ma, J.; Pounds, S.B.; et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature 2007, 446, 758–764. [Google Scholar] [CrossRef] [PubMed]
- Kawamata, N.; Ogawa, S.; Zimmermann, M.; Kato, M.; Sanada, M.; Hemminki, K.; Yamatomo, G.; Nannya, Y.; Koehler, R.; Flohr, T.; et al. Molecular allelokaryotyping of pediatric acute lymphoblastic leukemias by high-resolution single nucleotide polymorphism oligonucleotide genomic microarray. Blood 2008, 111, 776–784. [Google Scholar] [CrossRef] [PubMed]
- Bungaro, S.; Dell’Orto, M.C.; Zangrando, A.; Basso, D.; Gorletta, T.; Lo Nigro, L.; Leszl, A.; Young, B.D.; Basso, G.; Bicciato, S.; et al. Integration of genomic and gene expression data of childhood all without known aberrations identifies subgroups with specific genetic hallmarks. Genes Chromosomes Cancer 2009, 48, 22–38. [Google Scholar] [CrossRef] [PubMed]
- Harrison, C.J.; Foroni, L. Cytogenetics and molecular genetics of acute lymphoblastic leukemia. Rev. Clin. Exp. Hematol. 2002, 6, 91–113. [Google Scholar] [CrossRef] [PubMed]
- Graux, C.; Cools, J.; Michaux, L.; Vandenberghe, P.; Hagemeijer, A. Cytogenetics and molecular genetics of T-cell acute lymphoblastic leukemia: From thymocyte to lymphoblast. Leukemia 2006, 20, 1496–1510. [Google Scholar] [CrossRef] [PubMed]
- Mullighan, C.G.; Williams, R.T.; Downing, J.R.; Sherr, C.J. Failure of CDKN2A/b (INK4A/b-ARF)-mediated tumor suppression and resistance to targeted therapy in acute lymphoblastic leukemia induced by BCR-ABL. Genes Dev. 2008, 22, 1411–1415. [Google Scholar] [CrossRef] [PubMed]
- Karrman, K.; Castor, A.; Behrendtz, M.; Forestier, E.; Olsson, L.; Ehinger, M.; Biloglav, A.; Fioretos, T.; Paulsson, K.; Johansson, B. Deep sequencing and SNP array analyses of pediatric T-cell acute lymphoblastic leukemia reveal NOTCH1 mutations in minor subclones and a high incidence of uniparental isodisomies affecting CDKN2A. J. Hematol. Oncol. 2015, 8. [Google Scholar] [CrossRef] [PubMed]
- Okamoto, R.; Ogawa, S.; Nowak, D.; Kawamata, N.; Akagi, T.; Kato, M.; Sanada, M.; Weiss, T.; Haferlach, C.; Dugas, M.; et al. Genomic profiling of adult acute lymphoblastic leukemia by single nucleotide polymorphism oligonucleotide microarray and comparison to pediatric acute lymphoblastic leukemia. Haematologica 2010, 95, 1481–1488. [Google Scholar] [CrossRef] [PubMed]
- Safavi, S.; Hansson, M.; Karlsson, K.; Biloglav, A.; Johansson, B.; Paulsson, K. Novel gene targets detected by genomic profiling in a consecutive series of 126 adults with acute lymphoblastic leukemia. Haematologica 2015, 100, 55–61. [Google Scholar] [CrossRef] [PubMed]
- Trevino, L.R.; Yang, W.; French, D.; Hunger, S.P.; Carroll, W.L.; Devidas, M.; Willman, C.; Neale, G.; Downing, J.; Raimondi, S.C.; et al. Germline genomic variants associated with childhood acute lymphoblastic leukemia. Nat. Genet. 2009, 41, 1001–1005. [Google Scholar] [CrossRef] [PubMed]
- Papaemmanuil, E.; Hosking, F.J.; Vijayakrishnan, J.; Price, A.; Olver, B.; Sheridan, E.; Kinsey, S.E.; Lightfoot, T.; Roman, E.; Irving, J.A.; et al. Loci on 7p12.2, 10q21.2 and 14q11.2 are associated with risk of childhood acute lymphoblastic leukemia. Nat. Genet. 2009, 41, 1006–1010. [Google Scholar] [CrossRef] [PubMed]
- Prasad, R.B.; Hosking, F.J.; Vijayakrishnan, J.; Papaemmanuil, E.; Koehler, R.; Greaves, M.; Sheridan, E.; Gast, A.; Kinsey, S.E.; Lightfoot, T.; et al. Verification of the susceptibility loci on 7p12.2, 10q21.2, and 14q11.2 in precursor B-cell acute lymphoblastic leukemia of childhood. Blood 2010, 115, 1765–1767. [Google Scholar] [CrossRef] [PubMed]
- Thiede, C.; Steudel, C.; Mohr, B.; Schaich, M.; Schakel, U.; Platzbecker, U.; Wermke, M.; Bornhauser, M.; Ritter, M.; Neubauer, A.; et al. Analysis of FLT3-activating mutations in 979 patients with acute myelogenous leukemia: Association with FAB subtypes and identification of subgroups with poor prognosis. Blood 2002, 99, 4326–4335. [Google Scholar] [CrossRef] [PubMed]
- Frohling, S.; Schlenk, R.F.; Stolze, I.; Bihlmayr, J.; Benner, A.; Kreitmeier, S.; Tobis, K.; Dohner, H.; Dohner, K. CEBPA mutations in younger adults with acute myeloid leukemia and normal cytogenetics: Prognostic relevance and analysis of cooperating mutations. J. Clin. Oncol. 2004, 22, 624–633. [Google Scholar] [CrossRef] [PubMed]
- Raghavan, M.; Lillington, D.M.; Skoulakis, S.; Debernardi, S.; Chaplin, T.; Foot, N.J.; Lister, T.A.; Young, B.D. Genome-wide single nucleotide polymorphism analysis reveals frequent partial uniparental disomy due to somatic recombination in acute myeloid leukemias. Cancer Res. 2005, 65, 375–378. [Google Scholar] [PubMed]
- Fitzgibbon, J.; Smith, L.L.; Raghavan, M.; Smith, M.L.; Debernardi, S.; Skoulakis, S.; Lillington, D.; Lister, T.A.; Young, B.D. Association between acquired uniparental disomy and homozygous gene mutation in acute myeloid leukemias. Cancer Res. 2005, 65, 9152–9154. [Google Scholar] [CrossRef] [PubMed]
- Gupta, M.; Raghavan, M.; Gale, R.E.; Chelala, C.; Allen, C.; Molloy, G.; Chaplin, T.; Linch, D.C.; Cazier, J.B.; Young, B.D. Novel regions of acquired uniparental disomy discovered in acute myeloid leukemia. Genes Chromosomes Cancer 2008, 47, 729–739. [Google Scholar] [CrossRef] [PubMed]
- Bullinger, L.; Kronke, J.; Schon, C.; Radtke, I.; Urlbauer, K.; Botzenhardt, U.; Gaidzik, V.; Cario, A.; Senger, C.; Schlenk, R.F.; et al. Identification of acquired copy number alterations and uniparental disomies in cytogenetically normal acute myeloid leukemia using high-resolution single-nucleotide polymorphism analysis. Leukemia 2009, 24, 438–449. [Google Scholar] [CrossRef] [PubMed]
- Walter, M.J.; Payton, J.E.; Ries, R.E.; Shannon, W.D.; Deshmukh, H.; Zhao, Y.; Baty, J.; Heath, S.; Westervelt, P.; Watson, M.A.; et al. Acquired copy number alterations in adult acute myeloid leukemia genomes. Proc. Natl. Acad. Sci. USA 2009, 106, 12950–12955. [Google Scholar] [CrossRef] [PubMed]
- Radtke, I.; Mullighan, C.G.; Ishii, M.; Su, X.; Cheng, J.; Ma, J.; Ganti, R.; Cai, Z.; Goorha, S.; Pounds, S.B.; et al. Genomic analysis reveals few genetic alterations in pediatric acute myeloid leukemia. Proc. Natl. Acad. Sci. USA 2009, 106, 12944–12949. [Google Scholar] [CrossRef] [PubMed]
- Parkin, B.; Erba, H.; Ouillette, P.; Roulston, D.; Purkayastha, A.; Karp, J.; Talpaz, M.; Kujawski, L.; Shakhan, S.; Li, C.; et al. Acquired genomic copy number aberrations and survival in adult acute myelogenous leukemia. Blood 2010, 116, 4958–4967. [Google Scholar] [CrossRef] [PubMed]
- Tiu, R.V.; Gondek, L.P.; O’Keefe, C.L.; Huh, J.; Sekeres, M.A.; Elson, P.; McDevitt, M.A.; Wang, X.F.; Levis, M.J.; Karp, J.E.; et al. New lesions detected by single nucleotide polymorphism array-based chromosomal analysis have important clinical impact in acute myeloid leukemia. J. Clin. Oncol. 2009, 27, 5219–5226. [Google Scholar] [CrossRef] [PubMed]
- Yi, J.H.; Huh, J.; Kim, H.J.; Kim, S.H.; Kim, Y.K.; Sohn, S.K.; Moon, J.H.; Kim, K.H.; Won, J.H.; Mun, Y.C.; et al. Adverse prognostic impact of abnormal lesions detected by genome-wide single nucleotide polymorphism array-based karyotyping analysis in acute myeloid leukemia with normal karyotype. J. Clin. Oncol. 2011, 29, 4702–4708. [Google Scholar] [CrossRef] [PubMed]
- Tefferi, A.; Vardiman, J.W. Myelodysplastic syndromes. N. Engl. J. Med. 2009, 361, 1872–1885. [Google Scholar] [CrossRef] [PubMed]
- Greenberg, P.; Cox, C.; LeBeau, M.M.; Fenaux, P.; Morel, P.; Sanz, G.; Sanz, M.; Vallespi, T.; Hamblin, T.; Oscier, D.; et al. International scoring system for evaluating prognosis in myelodysplastic syndromes. Blood 1997, 89, 2079–2088. [Google Scholar] [PubMed]
- Greenberg, P.L.; Tuechler, H.; Schanz, J.; Sanz, G.; Garcia-Manero, G.; Sole, F.; Bennett, J.M.; Bowen, D.; Fenaux, P.; Dreyfus, F.; et al. Revised international prognostic scoring system for myelodysplastic syndromes. Blood 2012, 120, 2454–2465. [Google Scholar] [CrossRef] [PubMed]
- Gondek, L.P.; Tiu, R.; Haddad, A.S.; O’Keefe, C.L.; Sekeres, M.A.; Theil, K.S.; Maciejewski, J.P. Single nucleotide polymorphism arrays complement metaphase cytogenetics in detection of new chromosomal lesions in mds. Leukemia 2007, 21, 2058–2061. [Google Scholar] [CrossRef] [PubMed]
- Gondek, L.P.; Haddad, A.S.; O’Keefe, C.L.; Tiu, R.; Wlodarski, M.W.; Sekeres, M.A.; Theil, K.S.; Maciejewski, J.P. Detection of cryptic chromosomal lesions including acquired segmental uniparental disomy in advanced and low-risk myelodysplastic syndromes. Exp. Hematol. 2007, 35, 1728–1738. [Google Scholar] [CrossRef] [PubMed]
- Mohamedali, A.; Gaken, J.; Twine, N.A.; Ingram, W.; Westwood, N.; Lea, N.C.; Hayden, J.; Donaldson, N.; Aul, C.; Gattermann, N.; et al. Prevalence and prognostic significance of allelic imbalance by single-nucleotide polymorphism analysis in low-risk myelodysplastic syndromes. Blood 2007, 110, 3365–3373. [Google Scholar] [CrossRef] [PubMed]
- Nowak, D.; Nolte, F.; Mossner, M.; Nowak, V.; Baldus, C.D.; Hopfer, O.; Noll, S.; Thiel, E.; Wagner, F.; Hofmann, W.K. Genome-wide DNA-mapping of CD34+ cells from patients with myelodysplastic syndrome using 500k SNP arrays identifies significant regions of deletion and uniparental disomy. Exp. Hematol. 2009, 37, 215–224. [Google Scholar] [CrossRef] [PubMed]
- Heinrichs, S.; Kulkarni, R.V.; Bueso-Ramos, C.E.; Levine, R.L.; Loh, M.L.; Li, C.; Neuberg, D.; Kornblau, S.M.; Issa, J.P.; Gilliland, D.G.; et al. Accurate detection of uniparental disomy and microdeletions by SNP array analysis in myelodysplastic syndromes with normal cytogenetics. Leukemia 2009, 23, 1605–1613. [Google Scholar] [CrossRef] [PubMed]
- Tiu, R.V.; Gondek, L.P.; O’Keefe, C.L.; Elson, P.; Huh, J.; Mohamedali, A.; Kulasekararaj, A.; Advani, A.S.; Paquette, R.; List, A.F.; et al. Prognostic impact of SNP array karyotyping in myelodysplastic syndromes and related myeloid malignancies. Blood 2011, 117, 4552–4560. [Google Scholar] [CrossRef] [PubMed]
- Afable, M.G., II; Wlodarski, M.; Makishima, H.; Shaik, M.; Sekeres, M.A.; Tiu, R.V.; Kalaycio, M.; O’Keefe, C.L.; Maciejewski, J.P. SNP array-based karyotyping: Differences and similarities between aplastic anemia and hypocellular myelodysplastic syndromes. Blood 2011, 117, 6876–6884. [Google Scholar] [CrossRef] [PubMed]
- Langemeijer, S.M.; Kuiper, R.P.; Berends, M.; Knops, R.; Aslanyan, M.G.; Massop, M.; Stevens-Linders, E.; van Hoogen, P.; van Kessel, A.G.; Raymakers, R.A.; et al. Acquired mutations in TET2 are common in myelodysplastic syndromes. Nat. Genet. 2009, 41, 838–842. [Google Scholar] [CrossRef] [PubMed]
- Ernst, T.; Chase, A.J.; Score, J.; Hidalgo-Curtis, C.E.; Bryant, C.; Jones, A.V.; Waghorn, K.; Zoi, K.; Ross, F.M.; Reiter, A.; et al. Inactivating mutations of the histone methyltransferase gene EZH2 in myeloid disorders. Nat. Genet. 2010, 42, 722–726. [Google Scholar] [CrossRef] [PubMed]
- Nikoloski, G.; Langemeijer, S.M.; Kuiper, R.P.; Knops, R.; Massop, M.; Tonnissen, E.R.; van der Heijden, A.; Scheele, T.N.; Vandenberghe, P.; de Witte, T.; et al. Somatic mutations of the histone methyltransferase gene EZH2 in myelodysplastic syndromes. Nat. Genet. 2010, 42, 665–667. [Google Scholar] [CrossRef] [PubMed]
- Bejar, R.; Lord, A.; Stevenson, K.; Bar-Natan, M.; Perez-Ladaga, A.; Zaneveld, J.; Wang, H.; Caughey, B.; Stojanov, P.; Getz, G.; et al. TET2 mutations predict response to hypomethylating agents in myelodysplastic syndrome patients. Blood 2014, 124, 2705–2712. [Google Scholar] [CrossRef] [PubMed]
- Merkerova, M.D.; Bystricka, D.; Belickova, M.; Krejcik, Z.; Zemanova, Z.; Polak, J.; Hajkova, H.; Brezinova, J.; Michalova, K.; Cermak, J. From cryptic chromosomal lesions to pathologically relevant genes: Integration of SNP-array with gene expression profiling in myelodysplastic syndrome with normal karyotype. Genes Chromosomes Cancer 2012, 51, 419–428. [Google Scholar] [CrossRef] [PubMed]
- Mullighan, C.G.; Miller, C.B.; Radtke, I.; Phillips, L.A.; Dalton, J.; Ma, J.; White, D.; Hughes, T.P.; le Beau, M.M.; Pui, C.H.; et al. BCR-ABl1 lymphoblastic leukaemia is characterized by the deletion of Ikaros. Nature 2008, 453, 110–114. [Google Scholar] [CrossRef] [PubMed]
- Khorashad, J.S.; de Melo, V.A.; Fiegler, H.; Gerrard, G.; Marin, D.; Apperley, J.F.; Goldman, J.M.; Foroni, L.; Reid, A.G. Multiple sub-microscopic genomic lesions are a universal feature of chronic myeloid leukaemia at diagnosis. Leukemia 2008, 22, 1806–1807. [Google Scholar] [CrossRef] [PubMed]
- Nowak, D.; Ogawa, S.; Muschen, M.; Kato, M.; Kawamata, N.; Meixel, A.; Nowak, V.; Kim, H.S.; Kang, S.; Paquette, R.; et al. SNP array analysis of tyrosine kinase inhibitor-resistant chronic myeloid leukemia identifies heterogeneous secondary genomic alterations. Blood 2010, 115, 1049–1053. [Google Scholar] [CrossRef] [PubMed]
- Stegelmann, F.; Bullinger, L.; Griesshammer, M.; Holzmann, K.; Habdank, M.; Kuhn, S.; Maile, C.; Schauer, S.; Dohner, H.; Dohner, K. High-resolution single-nucleotide polymorphism array-profiling in myeloproliferative neoplasms identifies novel genomic aberrations. Haematologica 2010, 95, 666–669. [Google Scholar] [CrossRef] [PubMed]
- Rice, K.L.; Lin, X.; Wolniak, K.; Ebert, B.L.; Berkofsky-Fessler, W.; Buzzai, M.; Sun, Y.; Xi, C.; Elkin, P.; Levine, R.; et al. Analysis of genomic aberrations and gene expression profiling identifies novel lesions and pathways in myeloproliferative neoplasms. Blood Cancer J. 2011, 1. [Google Scholar] [CrossRef] [PubMed]
- Kawamata, N.; Ogawa, S.; Yamamoto, G.; Lehmann, S.; Levine, R.L.; Pikman, Y.; Nannya, Y.; Sanada, M.; Miller, C.W.; Gilliland, D.G.; et al. Genetic profiling of myeloproliferative disorders by single-nucleotide polymorphism oligonucleotide microarray. Exp. Hematol. 2008, 36, 1471–1479. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Swierczek, S.I.; Lanikova, L.; Kim, S.J.; Hickman, K.; Walker, K.; Wang, K.; Drummond, J.; Doddapaneni, H.; Reid, J.G.; et al. The relationship of JAK2(V617F) and acquired upd at chromosome 9p in polycythemia vera. Leukemia 2014, 28, 938–941. [Google Scholar] [CrossRef] [PubMed]
- Kilpivaara, O.; Mukherjee, S.; Schram, A.M.; Wadleigh, M.; Mullally, A.; Ebert, B.L.; Bass, A.; Marubayashi, S.; Heguy, A.; Garcia-Manero, G.; et al. A germline JAK2 SNP is associated with predisposition to the development of JAK2(V617F)-positive myeloproliferative neoplasms. Nat. Genet. 2009, 41, 455–459. [Google Scholar] [CrossRef] [PubMed]
- Thoennissen, N.H.; Krug, U.O.; Lee, D.H.; Kawamata, N.; Iwanski, G.B.; Lasho, T.; Weiss, T.; Nowak, D.; Koren-Michowitz, M.; Kato, M.; et al. Prevalence and prognostic impact of allelic imbalances associated with leukemic transformation of philadelphia chromosome-negative myeloproliferative neoplasms. Blood 2010, 115, 2882–2890. [Google Scholar] [CrossRef] [PubMed]
- Rumi, E.; Harutyunyan, A.; Elena, C.; Pietra, D.; Klampfl, T.; Bagienski, K.; Berg, T.; Casetti, I.; Pascutto, C.; Passamonti, F.; et al. Identification of genomic aberrations associated with disease transformation by means of high-resolution SNP array analysis in patients with myeloproliferative neoplasm. Am. J. Hematol. 2011, 86, 974–979. [Google Scholar] [CrossRef] [PubMed]
- Jankowska, A.M.; Makishima, H.; Tiu, R.V.; Szpurka, H.; Huang, Y.; Traina, F.; Visconte, V.; Sugimoto, Y.; Prince, C.; O’Keefe, C.; et al. Mutational spectrum analysis of chronic myelomonocytic leukemia includes genes associated with epigenetic regulation: UTX, EZH2, AND DNMT3A. Blood 2011, 118, 3932–3941. [Google Scholar] [CrossRef] [PubMed]
- Dunbar, A.J.; Gondek, L.P.; O’Keefe, C.L.; Makishima, H.; Rataul, M.S.; Szpurka, H.; Sekeres, M.A.; Wang, X.F.; McDevitt, M.A.; Maciejewski, J.P. 250k single nucleotide polymorphism array karyotyping identifies acquired uniparental disomy and homozygous mutations, including novel missense substitutions of c-Cbl, in myeloid malignancies. Cancer Res. 2008, 68, 10349–10357. [Google Scholar] [CrossRef] [PubMed]
- Yi, J.H.; Huh, J.; Kim, H.J.; Kim, S.H.; Kim, K.H.; Do, Y.R.; Mun, Y.C.; Kim, H.; Kim, M.K.; Kim, T.; et al. Genome-wide single-nucleotide polymorphism array-based karyotyping in myelodysplastic syndrome and chronic myelomonocytic leukemia and its impact on treatment outcomes following decitabine treatment. Ann. Hematol. 2013, 92, 459–469. [Google Scholar] [CrossRef] [PubMed]
- Flotho, C.; Steinemann, D.; Mullighan, C.G.; Neale, G.; Mayer, K.; Kratz, C.P.; Schlegelberger, B.; Downing, J.R.; Niemeyer, C.M. Genome-wide single-nucleotide polymorphism analysis in juvenile myelomonocytic leukemia identifies uniparental disomy surrounding the NF1 locus in cases associated with neurofibromatosis but not in cases with mutant RAS or PTPN11. Oncogene 2007, 26, 5816–5821. [Google Scholar] [CrossRef] [PubMed]
- Otto, C.; Giefing, M.; Massow, A.; Vater, I.; Gesk, S.; Schlesner, M.; Richter, J.; Klapper, W.; Hansmann, M.L.; Siebert, R.; et al. Genetic lesions of the TRAF3 and MAP3K14 genes in classical hodgkin lymphoma. Br. J. Haematol. 2012, 157, 702–708. [Google Scholar] [CrossRef] [PubMed]
- Cozen, W.; Li, D.; Best, T.; Van Den Berg, D.J.; Gourraud, P.A.; Cortessis, V.K.; Skol, A.D.; Mack, T.M.; Glaser, S.L.; Weiss, L.M.; et al. A genome-wide meta-analysis of nodular sclerosing hodgkin lymphoma identifies risk loci at 6p21.32. Blood 2012, 119, 469–475. [Google Scholar] [CrossRef] [PubMed]
- Scholtysik, R.; Kreuz, M.; Hummel, M.; Rosolowski, M.; Szczepanowski, M.; Klapper, W.; Loeffler, M.; Trumper, L.; Siebert, R.; Kuppers, R. Characterization of genomic imbalances in diffuse large B-cell lymphoma by detailed SNP-chip analysis. Int. J. Cancer 2015, 136, 1033–1042. [Google Scholar] [CrossRef] [PubMed]
- Aya-Bonilla, C.; Green, M.R.; Camilleri, E.; Benton, M.; Keane, C.; Marlton, P.; Lea, R.; Gandhi, M.K.; Griffiths, L.R. High-resolution loss of heterozygosity screening implicates PTPRJ as a potential tumor suppressor gene that affects susceptibility to non-hodgkin’s lymphoma. Genes Chromosomes Cancer 2013, 52, 467–479. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trifonov, V.; Pasqualucci, L.; Dalla Favera, R.; Rabadan, R. Mutcomfocal: An integrative approach to identifying recurrent and focal genomic alterations in tumor samples. BMC Syst. Biol. 2013, 7. [Google Scholar] [CrossRef] [PubMed]
- Scandurra, M.; Mian, M.; Greiner, T.C.; Rancoita, P.M.; De Campos, C.P.; Chan, W.C.; Vose, J.M.; Chigrinova, E.; Inghirami, G.; Chiappella, A.; et al. Genomic lesions associated with a different clinical outcome in diffuse large B-cell lymphoma treated with R-CHOP-21. Br. J. Haematol. 2010, 151, 221–231. [Google Scholar] [CrossRef] [PubMed]
- Fitzgibbon, J.; Iqbal, S.; Davies, A.; O’Shea, D.; Carlotti, E.; Chaplin, T.; Matthews, J.; Raghavan, M.; Norton, A.; Lister, T.A.; et al. Genome-wide detection of recurring sites of uniparental disomy in follicular and transformed follicular lymphoma. Leukemia 2007, 21, 1514–1520. [Google Scholar] [CrossRef] [PubMed]
- O’Shea, D.; O’Riain, C.; Gupta, M.; Waters, R.; Yang, Y.; Wrench, D.; Gribben, J.; Rosenwald, A.; Ott, G.; Rimsza, L.M.; et al. Regions of acquired uniparental disomy at diagnosis of follicular lymphoma are associated with both overall survival and risk of transformation. Blood 2009, 113, 2298–2301. [Google Scholar] [CrossRef] [PubMed]
- Cheung, K.J.; Delaney, A.; Ben-Neriah, S.; Schein, J.; Lee, T.; Shah, S.P.; Cheung, D.; Johnson, N.A.; Mungall, A.J.; Telenius, A.; et al. High resolution analysis of follicular lymphoma genomes reveals somatic recurrent sites of copy-neutral loss of heterozygosity and copy number alterations that target single genes. Genes Chromosomes Cancer 2010, 49, 669–681. [Google Scholar] [CrossRef] [PubMed]
- Cheung, K.J.; Rogic, S.; Ben-Neriah, S.; Boyle, M.; Connors, J.M.; Gascoyne, R.D.; Horsman, D.E. SNP analysis of minimally evolved t(14;18)(q32;q21)-positive follicular lymphomas reveals a common copy-neutral loss of heterozygosity pattern. Cytogenet. Genome Res. 2012, 136, 38–43. [Google Scholar] [CrossRef] [PubMed]
- Cheung, K.J.; Johnson, N.A.; Affleck, J.G.; Severson, T.; Steidl, C.; Ben-Neriah, S.; Schein, J.; Morin, R.D.; Moore, R.; Shah, S.P.; et al. Acquired TNFRSF14 mutations in follicular lymphoma are associated with worse prognosis. Cancer Res. 2010, 70, 9166–9174. [Google Scholar] [CrossRef] [PubMed]
- Leich, E.; Salaverria, I.; Bea, S.; Zettl, A.; Wright, G.; Moreno, V.; Gascoyne, R.D.; Chan, W.C.; Braziel, R.M.; Rimsza, L.M.; et al. Follicular lymphomas with and without translocation t(14;18) differ in gene expression profiles and genetic alterations. Blood 2009, 114, 826–834. [Google Scholar] [CrossRef] [PubMed]
- Pfeifer, D.; Pantic, M.; Skatulla, I.; Rawluk, J.; Kreutz, C.; Martens, U.M.; Fisch, P.; Timmer, J.; Veelken, H. Genome-wide analysis of DNA copy number changes and LOH in CLL using high-density SNP arrays. Blood 2007, 109, 1202–1210. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, S.; Ogawa, S.; Raynaud, S.D.; Sanada, M.; Nannya, Y.; Ticchioni, M.; Bastard, C.; Kawamata, N.; Koeffler, H.P. Molecular allelokaryotyping of early-stage, untreated chronic lymphocytic leukemia. Cancer 2008, 112, 1296–1305. [Google Scholar] [CrossRef] [PubMed]
- Edelmann, J.; Holzmann, K.; Miller, F.; Winkler, D.; Buhler, A.; Zenz, T.; Bullinger, L.; Kuhn, M.W.; Gerhardinger, A.; Bloehdorn, J.; et al. High-resolution genomic profiling of chronic lymphocytic leukemia reveals new recurrent genomic alterations. Blood 2012, 120, 4783–4794. [Google Scholar] [CrossRef] [PubMed]
- Gunnarsson, R.; Mansouri, L.; Isaksson, A.; Goransson, H.; Cahill, N.; Jansson, M.; Rasmussen, M.; Lundin, J.; Norin, S.; Buhl, A.M.; et al. Array-based genomic screening at diagnosis and during follow-up in chronic lymphocytic leukemia. Haematologica 2011, 96, 1161–1169. [Google Scholar] [CrossRef] [PubMed]
- Kujawski, L.; Ouillette, P.; Erba, H.; Saddler, C.; Jakubowiak, A.; Kaminski, M.; Shedden, K.; Malek, S.N. Genomic complexity identifies patients with aggressive chronic lymphocytic leukemia. Blood 2008, 112, 1993–2003. [Google Scholar] [CrossRef] [PubMed]
- Ouillette, P.; Collins, R.; Shakhan, S.; Li, J.; Peres, E.; Kujawski, L.; Talpaz, M.; Kaminski, M.; Li, C.; Shedden, K.; et al. Acquired genomic copy number aberrations and survival in chronic lymphocytic leukemia. Blood 2011, 118, 3051–3061. [Google Scholar] [CrossRef] [PubMed]
- Schweighofer, C.D.; Coombes, K.R.; Majewski, T.; Barron, L.L.; Lerner, S.; Sargent, R.L.; O’Brien, S.; Ferrajoli, A.; Wierda, W.G.; Czerniak, B.A.; et al. Genomic variation by whole-genome SNP mapping arrays predicts time-to-event outcome in patients with chronic lymphocytic leukemia: A comparison of CLL and hapmap genotypes. J. Mol. Diagn. 2013, 15, 196–209. [Google Scholar] [CrossRef] [PubMed]
- Mian, M.; Rinaldi, A.; Mensah, A.A.; Rossi, D.; Ladetto, M.; Forconi, F.; Marasca, R.; Uhr, M.; Stussi, G.; Kwee, I.; et al. Large genomic aberrations detected by SNP array are independent prognosticators of a shorter time to first treatment in chronic lymphocytic leukemia patients with normal fish. Ann. Oncol. 2013, 24, 1378–1384. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Znoyko, I.; Costa, L.J.; Conlin, L.K.; Daber, R.D.; Self, S.E.; Wolff, D.J. Clonal diversity analysis using SNP microarray: A new prognostic tool for chronic lymphocytic leukemia. Cancer Genet. 2011, 204, 654–665. [Google Scholar] [CrossRef] [PubMed]
- Ouillette, P.; Erba, H.; Kujawski, L.; Kaminski, M.; Shedden, K.; Malek, S.N. Integrated genomic profiling of chronic lymphocytic leukemia identifies subtypes of deletion 13q14. Cancer Res. 2008, 68, 1012–1021. [Google Scholar] [CrossRef] [PubMed]
- Ouillette, P.; Collins, R.; Shakhan, S.; Li, J.; Li, C.; Shedden, K.; Malek, S.N. The prognostic significance of various 13q14 deletions in chronic lymphocytic leukemia. Clin. Cancer Res. 2011, 17, 6778–6790. [Google Scholar] [CrossRef] [PubMed]
- Mosca, L.; Fabris, S.; Lionetti, M.; Todoerti, K.; Agnelli, L.; Morabito, F.; Cutrona, G.; Andronache, A.; Matis, S.; Ferrari, F.; et al. Integrative genomics analyses reveal molecularly distinct subgroups of B-cell chronic lymphocytic leukemia patients with 13q14 deletion. Clin. Cancer Res. 2010, 16, 5641–5653. [Google Scholar] [CrossRef] [PubMed]
- Gardiner, A.; Parker, H.; Glide, S.; Mould, S.; Robinson, H.; Tracy, I.; Stankovic, T.; Oscier, D.; Strefford, J. A new minimal deleted region at 11q22.3 reveals the importance of interpretation of diminished fish signals and the choice of probe for ATM deletion screening in chronic lymphocytic leukemia. Leuk. Res. 2012, 36, 307–310. [Google Scholar] [CrossRef] [PubMed]
- Kawamata, N.; Ogawa, S.; Gueller, S.; Ross, S.H.; Huynh, T.; Chen, J.; Chang, A.; Nabavi-Nouis, S.; Megrabian, N.; Siebert, R.; et al. Identified hidden genomic changes in mantle cell lymphoma using high-resolution single nucleotide polymorphism genomic array. Exp. Hematol. 2009, 37, 937–946. [Google Scholar] [CrossRef] [PubMed]
- Bea, S.; Salaverria, I.; Armengol, L.; Pinyol, M.; Fernandez, V.; Hartmann, E.M.; Jares, P.; Amador, V.; Hernandez, L.; Navarro, A.; et al. Uniparental disomies, homozygous deletions, amplifications, and target genes in mantle cell lymphoma revealed by integrative high-resolution whole-genome profiling. Blood 2009, 113, 3059–3069. [Google Scholar] [CrossRef] [PubMed]
- Flossbach, L.; Holzmann, K.; Mattfeldt, T.; Buck, M.; Lanz, K.; Held, M.; Moller, P.; Barth, T.F. High-resolution genomic profiling reveals clonal evolution and competition in gastrointestinal marginal zone B-cell lymphoma and its large cell variant. Int. J. Cancer 2013, 132, E116–E127. [Google Scholar] [CrossRef] [PubMed]
- Novak, U.; Rinaldi, A.; Kwee, I.; Nandula, S.V.; Rancoita, P.M.; Compagno, M.; Cerri, M.; Rossi, D.; Murty, V.V.; Zucca, E.; et al. The NF-κB negative regulator TNFAIP3 (A20) is inactivated by somatic mutations and genomic deletions in marginal zone lymphomas. Blood 2009, 113, 4918–4921. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, H.; Usui, Y.; Ueda, S.; Yamakawa, N.; Sato-Otsubo, A.; Sato, Y.; Ogawa, S.; Goto, H. Genome-wide analysis of ocular adnexal lymphoproliferative disorders using high-resolution single nucleotide polymorphism array. Investig. Ophthalmol. Vis. Sci. 2015, 56, 4156–4165. [Google Scholar] [CrossRef] [PubMed]
- Lundin, C.; Hjorth, L.; Behrendtz, M.; Ehinger, M.; Biloglav, A.; Johansson, B. Submicroscopic genomic imbalances in burkitt lymphomas/leukemias: Association with age and further evidence that 8q24/MYC translocations are not sufficient for leukemogenesis. Genes Chromosomes Cancer 2013, 52, 370–377. [Google Scholar] [CrossRef] [PubMed]
- Forconi, F.; Poretti, G.; Kwee, I.; Sozzi, E.; Rossi, D.; Rancoita, P.M.; Capello, D.; Rinaldi, A.; Zucca, E.; Raspadori, D.; et al. High density genome-wide DNA profiling reveals a remarkably stable profile in hairy cell leukaemia. Br. J. Haematol. 2008, 141, 622–630. [Google Scholar] [CrossRef] [PubMed]
- Rinaldi, A.; Kwee, I.; Young, K.H.; Zucca, E.; Gaidano, G.; Forconi, F.; Bertoni, F. Genome-wide high resolution DNA profiling of hairy cell leukaemia. Br. J. Haematol. 2013, 162, 566–569. [Google Scholar] [CrossRef] [PubMed]
- Walker, B.A.; Leone, P.E.; Jenner, M.W.; Li, C.; Gonzalez, D.; Johnson, D.C.; Ross, F.M.; Davies, F.E.; Morgan, G.J. Integration of global SNP-based mapping and expression arrays reveals key regions, mechanisms, and genes important in the pathogenesis of multiple myeloma. Blood 2006, 108, 1733–1743. [Google Scholar] [CrossRef] [PubMed]
- Agnelli, L.; Mosca, L.; Fabris, S.; Lionetti, M.; Andronache, A.; Kwee, I.; Todoerti, K.; Verdelli, D.; Battaglia, C.; Bertoni, F.; et al. A SNP microarray and fish-based procedure to detect allelic imbalances in multiple myeloma: An integrated genomics approach reveals a wide gene dosage effect. Genes Chromosomes Cancer 2009, 48, 603–614. [Google Scholar] [CrossRef] [PubMed]
- Avet-Loiseau, H.; Li, C.; Magrangeas, F.; Gouraud, W.; Charbonnel, C.; Harousseau, J.L.; Attal, M.; Marit, G.; Mathiot, C.; Facon, T.; et al. Prognostic significance of copy-number alterations in multiple myeloma. J. Clin. Oncol. 2009, 27, 4585–4590. [Google Scholar] [CrossRef] [PubMed]
- Walker, B.A.; Leone, P.E.; Chiecchio, L.; Dickens, N.J.; Jenner, M.W.; Boyd, K.D.; Johnson, D.C.; Gonzalez, D.; Dagrada, G.P.; Protheroe, R.K.; et al. A compendium of myeloma-associated chromosomal copy number abnormalities and their prognostic value. Blood 2010, 116, e56–e65. [Google Scholar] [CrossRef] [PubMed]
- Kamada, Y.; Sakata-Yanagimoto, M.; Sanada, M.; Sato-Otsubo, A.; Enami, T.; Suzukawa, K.; Kurita, N.; Nishikii, H.; Yokoyama, Y.; Okoshi, Y.; et al. Identification of unbalanced genome copy number abnormalities in patients with multiple myeloma by single-nucleotide polymorphism genotyping microarray analysis. Int. J. Hematol. 2012, 96, 492–500. [Google Scholar] [CrossRef] [PubMed]
- Jenner, M.W.; Leone, P.E.; Walker, B.A.; Ross, F.M.; Johnson, D.C.; Gonzalez, D.; Chiecchio, L.; Dachs Cabanas, E.; Dagrada, G.P.; Nightingale, M.; et al. Gene mapping and expression analysis of 16q loss of heterozygosity identifies WWOX and CYLD as being important in determining clinical outcome in multiple myeloma. Blood 2007, 110, 3291–3300. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Lee, S.H.; Kim, J.; Lee, S.E.; Kim, Y.J.; Min, C.K. Copy number variations could predict the outcome of bortezomib plus melphalan and prednisone for initial treatment of multiple myeloma. Genes Chromosomes Cancer 2015, 54, 20–27. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Corral, L.; Sarasquete, M.E.; Bea, S.; Garcia-Sanz, R.; Mateos, M.V.; Corchete, L.A.; Sayagues, J.M.; Garcia, E.M.; Blade, J.; Oriol, A.; et al. SNP-based mapping arrays reveal high genomic complexity in monoclonal gammopathies, from MGUS to myeloma status. Leukemia 2012, 26, 2521–2529. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, S.; Gesk, S.; Scholtysik, R.; Kreuz, M.; Bug, S.; Vater, I.; Doring, C.; Cogliatti, S.; Parrens, M.; Merlio, J.P.; et al. High resolution SNP array genomic profiling of peripheral T cell lymphomas, not otherwise specified, identifies a subgroup with chromosomal aberrations affecting the REL locus. Br. J. Haematol. 2010, 148, 402–412. [Google Scholar] [CrossRef] [PubMed]
- Fujiwara, S.I.; Yamashita, Y.; Nakamura, N.; Choi, Y.L.; Ueno, T.; Watanabe, H.; Kurashina, K.; Soda, M.; Enomoto, M.; Hatanaka, H.; et al. High-resolution analysis of chromosome copy number alterations in angioimmunoblastic T-cell lymphoma and peripheral T-cell lymphoma, unspecified, with single nucleotide polymorphism-typing microarrays. Leukemia 2008, 22, 1891–1898. [Google Scholar] [CrossRef] [PubMed]
- Hidaka, T.; Nakahata, S.; Hatakeyama, K.; Hamasaki, M.; Yamashita, K.; Kohno, T.; Arai, Y.; Taki, T.; Nishida, K.; Okayama, A.; et al. Down-regulation of TCF8 is involved in the leukemogenesis of adult T-cell leukemia/lymphoma. Blood 2008, 112, 383–393. [Google Scholar] [CrossRef] [PubMed]
- Nakahata, S.; Saito, Y.; Hamasaki, M.; Hidaka, T.; Arai, Y.; Taki, T.; Taniwaki, M.; Morishita, K. Alteration of enhancer of polycomb 1 at 10p11.2 is one of the genetic events leading to development of adult T-cell leukemia/lymphoma. Genes Chromosomes Cancer 2009, 48, 768–776. [Google Scholar] [CrossRef] [PubMed]
- Durig, J.; Bug, S.; Klein-Hitpass, L.; Boes, T.; Jons, T.; Martin-Subero, J.I.; Harder, L.; Baudis, M.; Duhrsen, U.; Siebert, R. Combined single nucleotide polymorphism-based genomic mapping and global gene expression profiling identifies novel chromosomal imbalances, mechanisms and candidate genes important in the pathogenesis of T-cell prolymphocytic leukemia with INV(14)(q11q32). Leukemia 2007, 21, 2153–2163. [Google Scholar] [CrossRef] [PubMed]
- Nowak, D.; le Toriellec, E.; Stern, M.H.; Kawamata, N.; Akagi, T.; Dyer, M.J.; Hofmann, W.K.; Ogawa, S.; Koeffler, H.P. Molecular allelokaryotyping of T-cell prolymphocytic leukemia cells with high density single nucleotide polymorphism arrays identifies novel common genomic lesions and acquired uniparental disomy. Haematologica 2009, 94, 518–527. [Google Scholar] [CrossRef] [PubMed]
- Mao, X.; Chaplin, T.; Young, B.D. Integrated genomic analysis of sezary syndrome. Genet. Res. Int. 2011, 2011. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, J.; Shao, H. SNP Array in Hematopoietic Neoplasms: A Review. Microarrays 2016, 5, 1. https://doi.org/10.3390/microarrays5010001
Song J, Shao H. SNP Array in Hematopoietic Neoplasms: A Review. Microarrays. 2016; 5(1):1. https://doi.org/10.3390/microarrays5010001
Chicago/Turabian StyleSong, Jinming, and Haipeng Shao. 2016. "SNP Array in Hematopoietic Neoplasms: A Review" Microarrays 5, no. 1: 1. https://doi.org/10.3390/microarrays5010001
APA StyleSong, J., & Shao, H. (2016). SNP Array in Hematopoietic Neoplasms: A Review. Microarrays, 5(1), 1. https://doi.org/10.3390/microarrays5010001





