GAH-TNet: A Graph Attention-Based Hierarchical Temporal Network for EEG Motor Imagery Decoding
Abstract
1. Introduction
- A novel deep neural network model, GAH-TNet, is proposed for MI decoding tasks. It integrates graph attention–based spatial structure and multi-level temporal feature encoding to achieve collaborative modeling of the spatial topology and dynamic temporal evolution of EEG signals.
- The GATE module is designed to capture spatial dependencies and local temporal dynamic across EEG channels, enabling collaborative encoding of spatiotemporal features and enhancing both the structural integrity and discriminative power of feature representations.
- We develop the HADTE module, which employs hierarchical attention guidance for cross-scale deep temporal modeling, significantly improving the ability of the model to perceive multi-stage deep temporal patterns during motor imagery.
- We perform systematic experimental verification on the public BCI IV 2a and 2b datasets, where GAH-TNet consistently outperforms existing state-of-the-art models, demonstrating strong stability, generalization, and achieving SOTA performance.

2. Materials and Methods
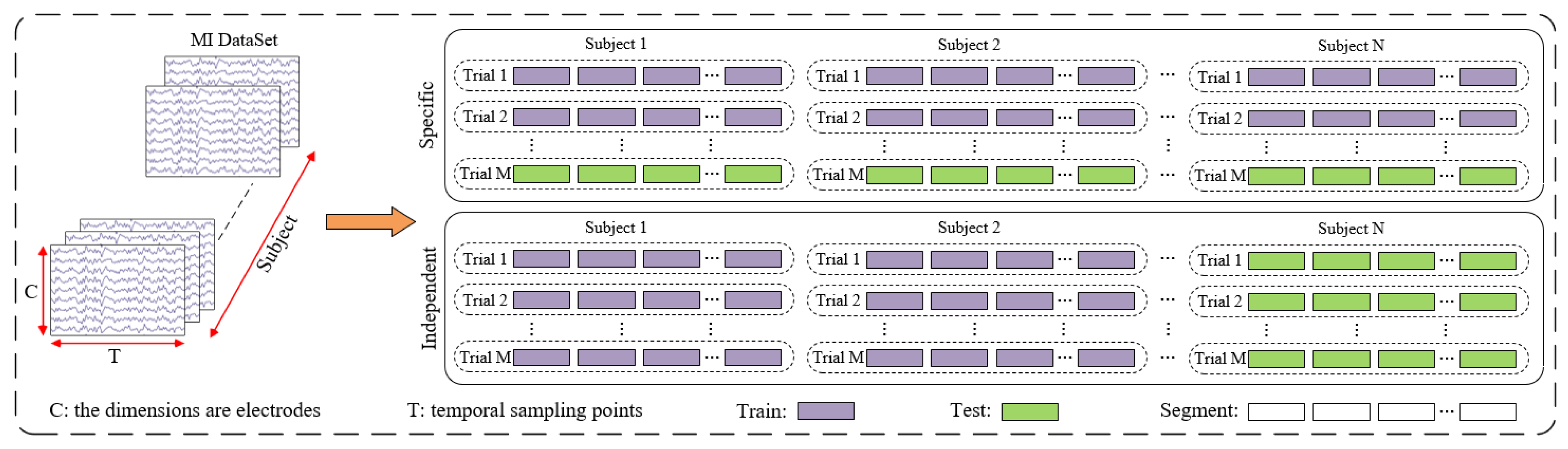
2.1. Dataset
2.1.1. BCI Competition IV Dataset 2a
2.1.2. BCI Competition IV Dataset 2b
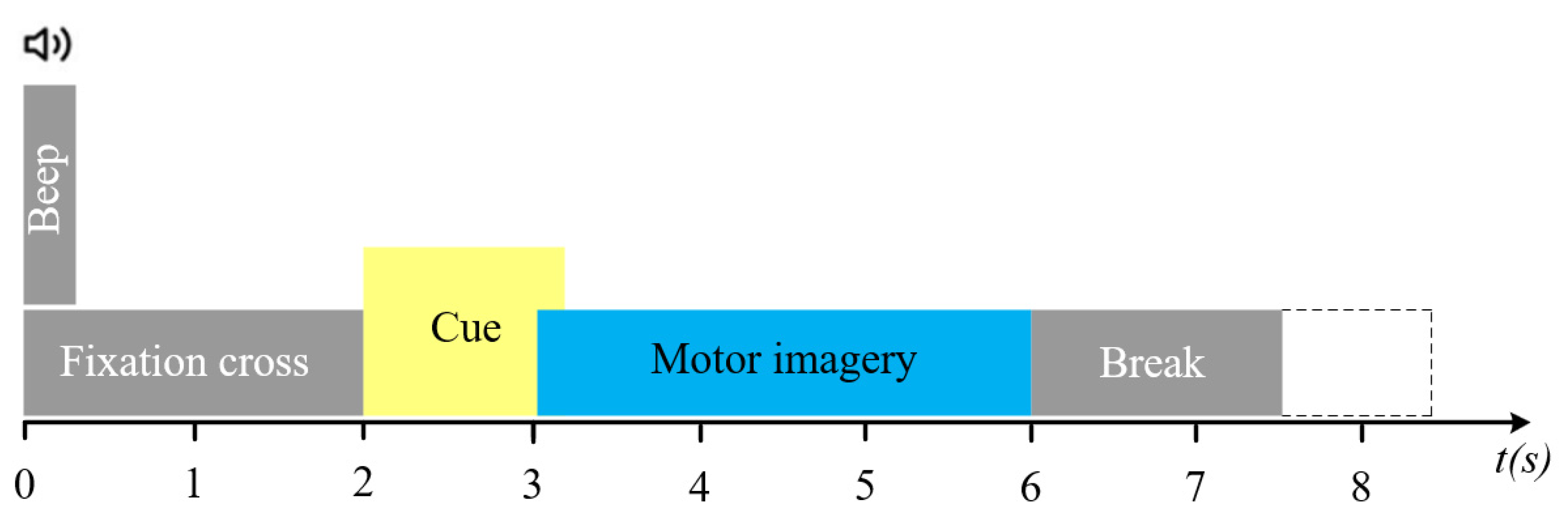
2.2. Data Pre-Processing
2.3. Model Architecture
2.3.1. Graph-Attentional Temporal Encoding Block
- (1)
- Graph Construction and Normalization
- (2)
- Chebyshev Graph Convolution
- (3)
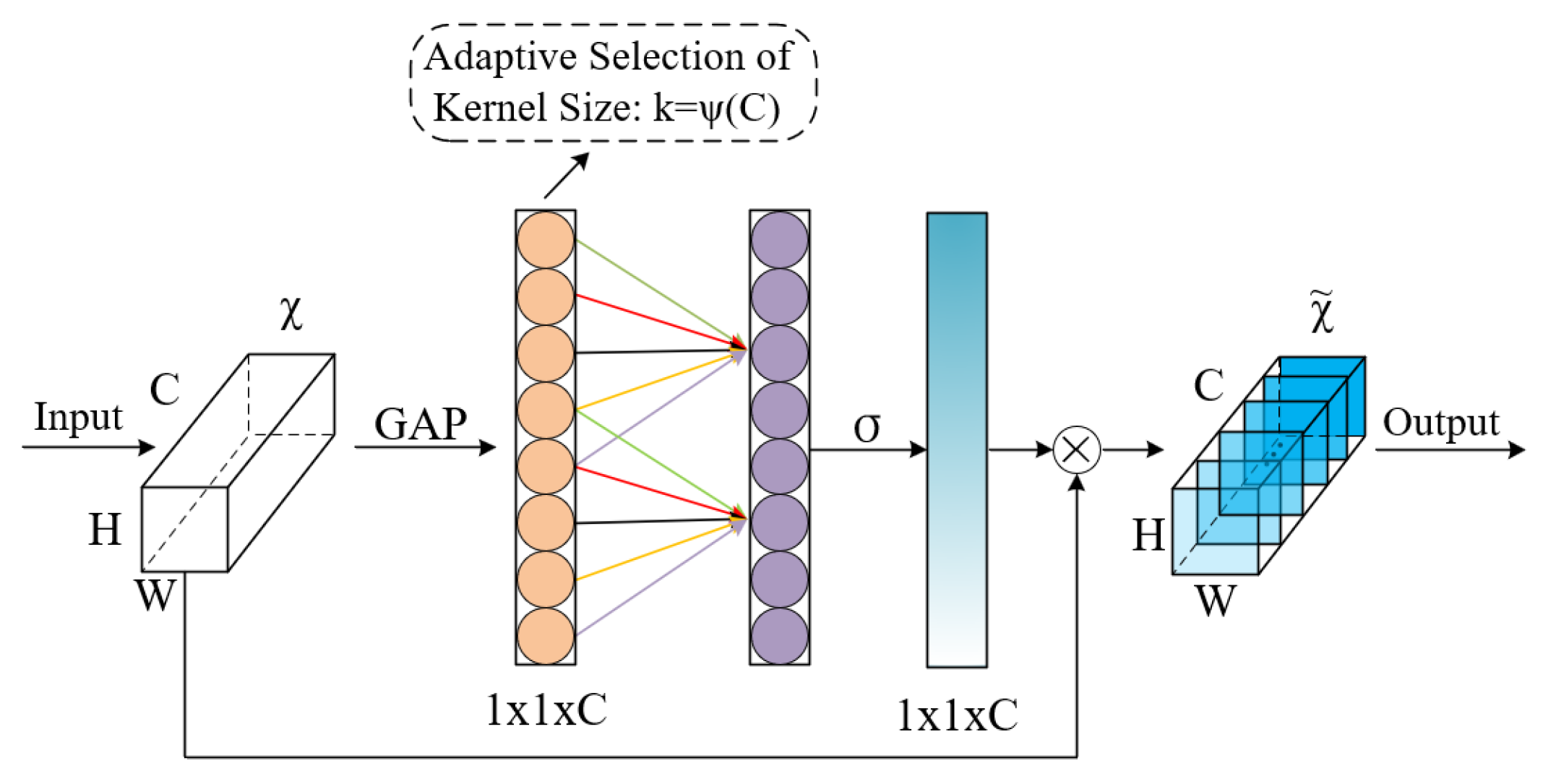
- Channel Attention
- (4)
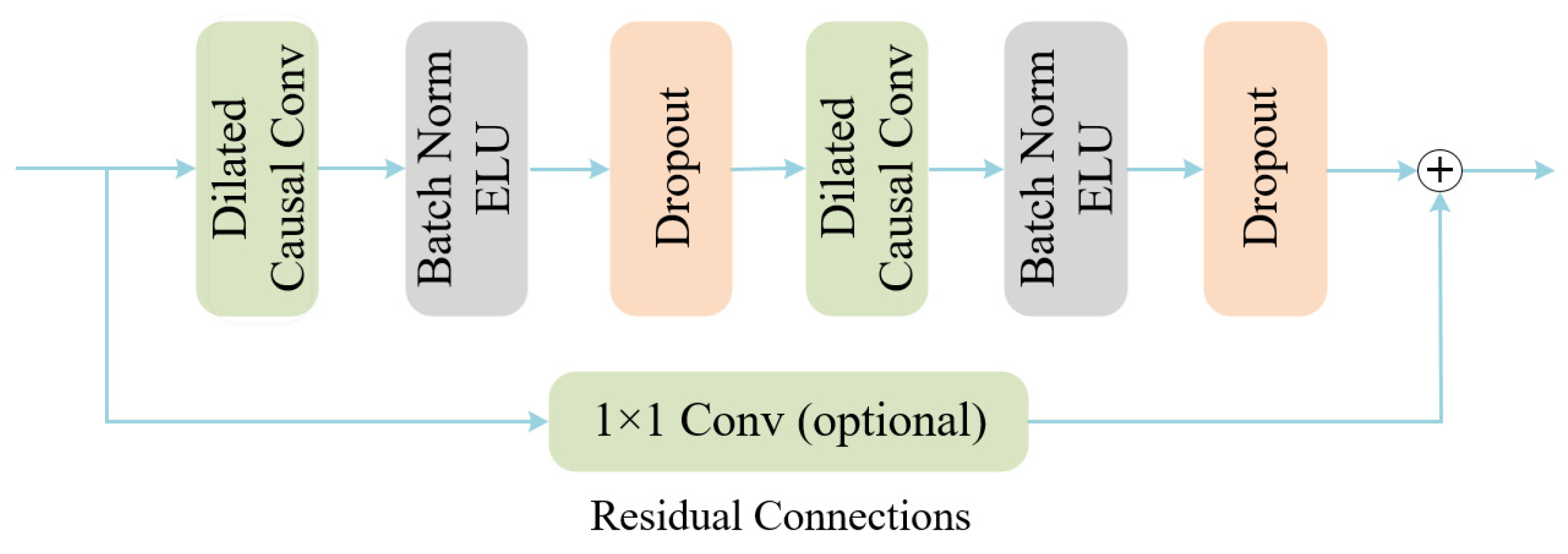
- Temporal–Spatial Convolutional Encoder
2.3.2. Hierarchical Attention-Guided Deep Temporal Feature Encoding Block
- (1)
- Residual-ECA
- (2)
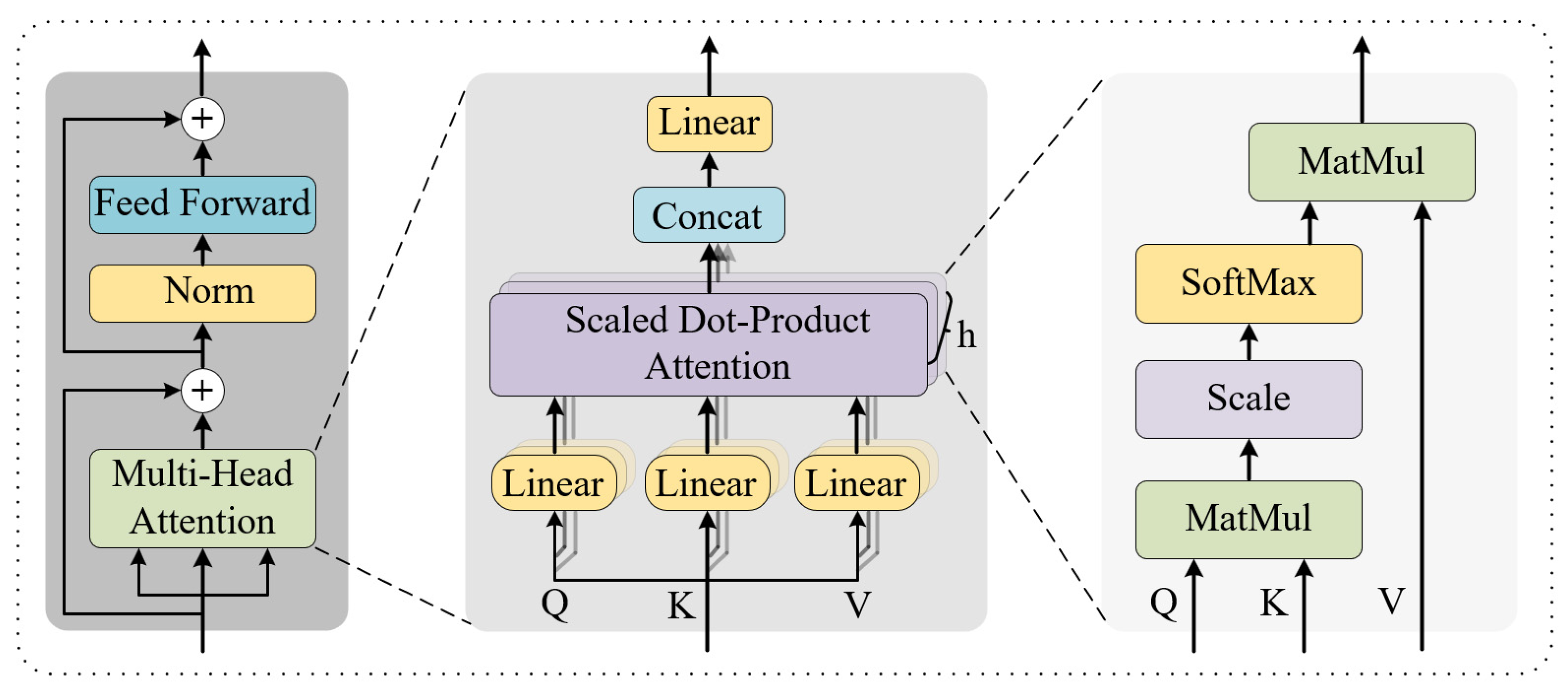
- Local Masked Multi-Head Attention
- (3)
- Global Multi-Head Attention
- (4)
- Deep temporal modeling
2.3.3. Classifier Module
2.4. Evaluation Metrics
3. Experiments and Results
3.1. Experiment Details
3.2. Results
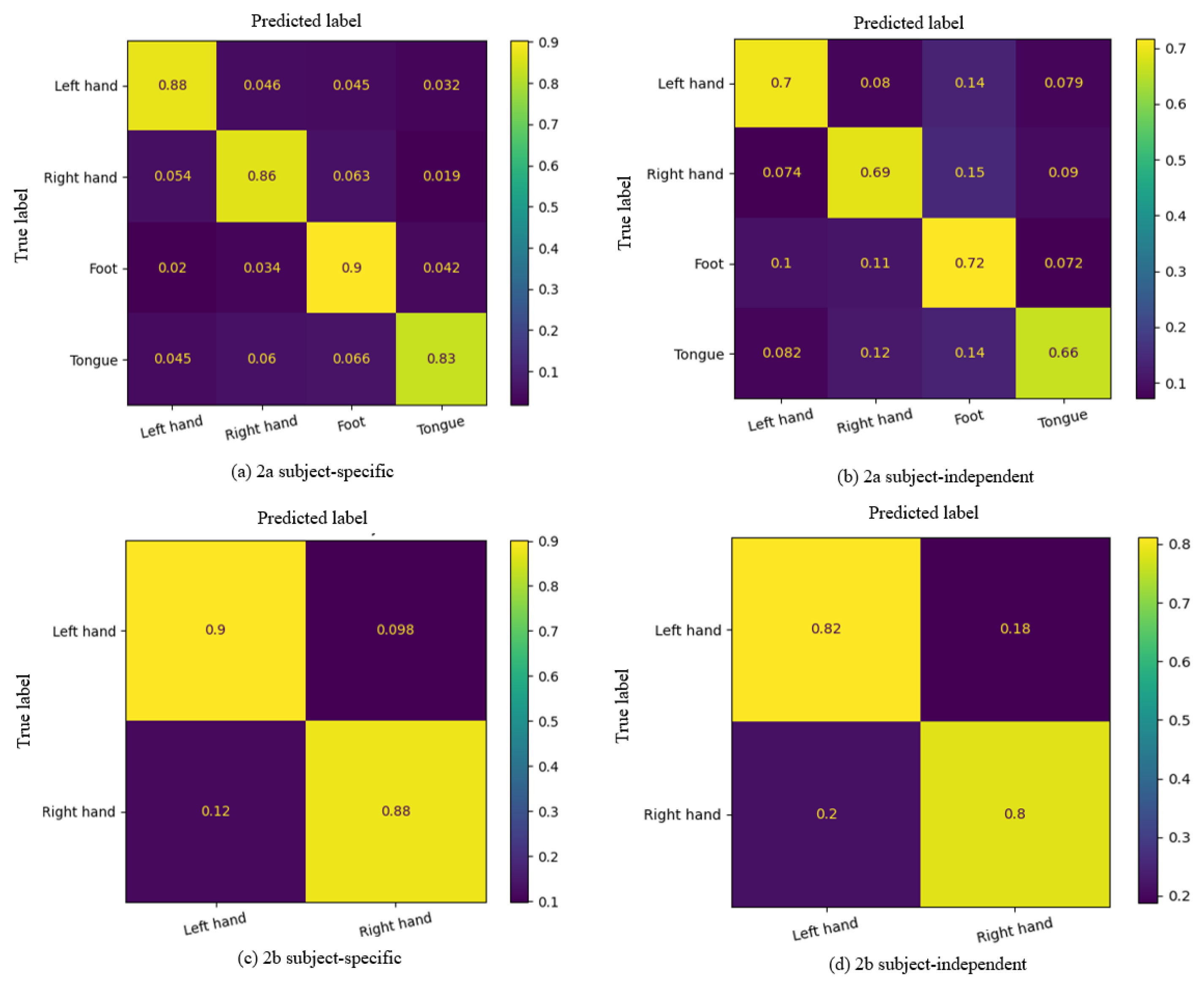
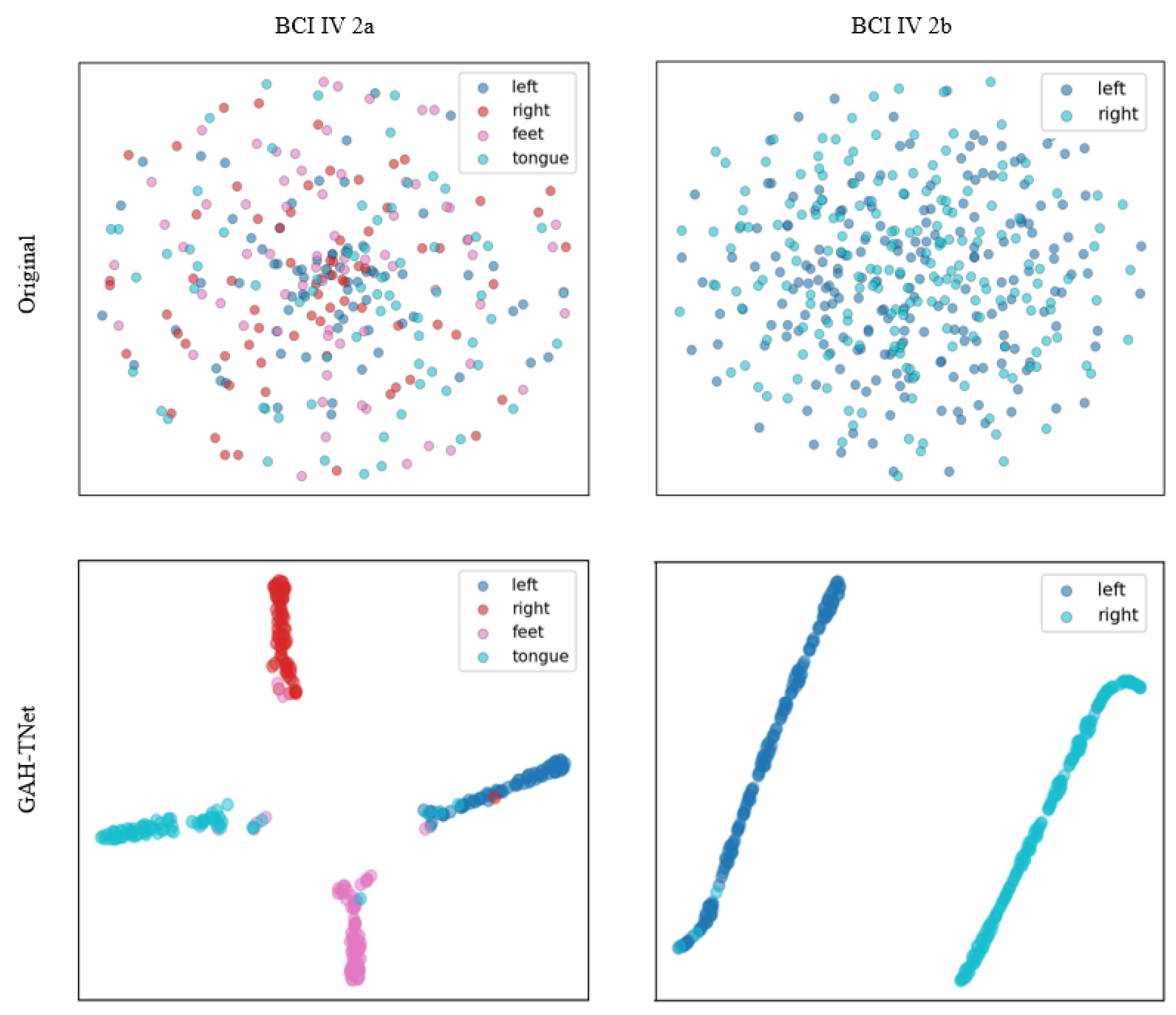
3.2.1. Experimental Results of MI-EEG Decoding
3.2.2. Ablation Experiment
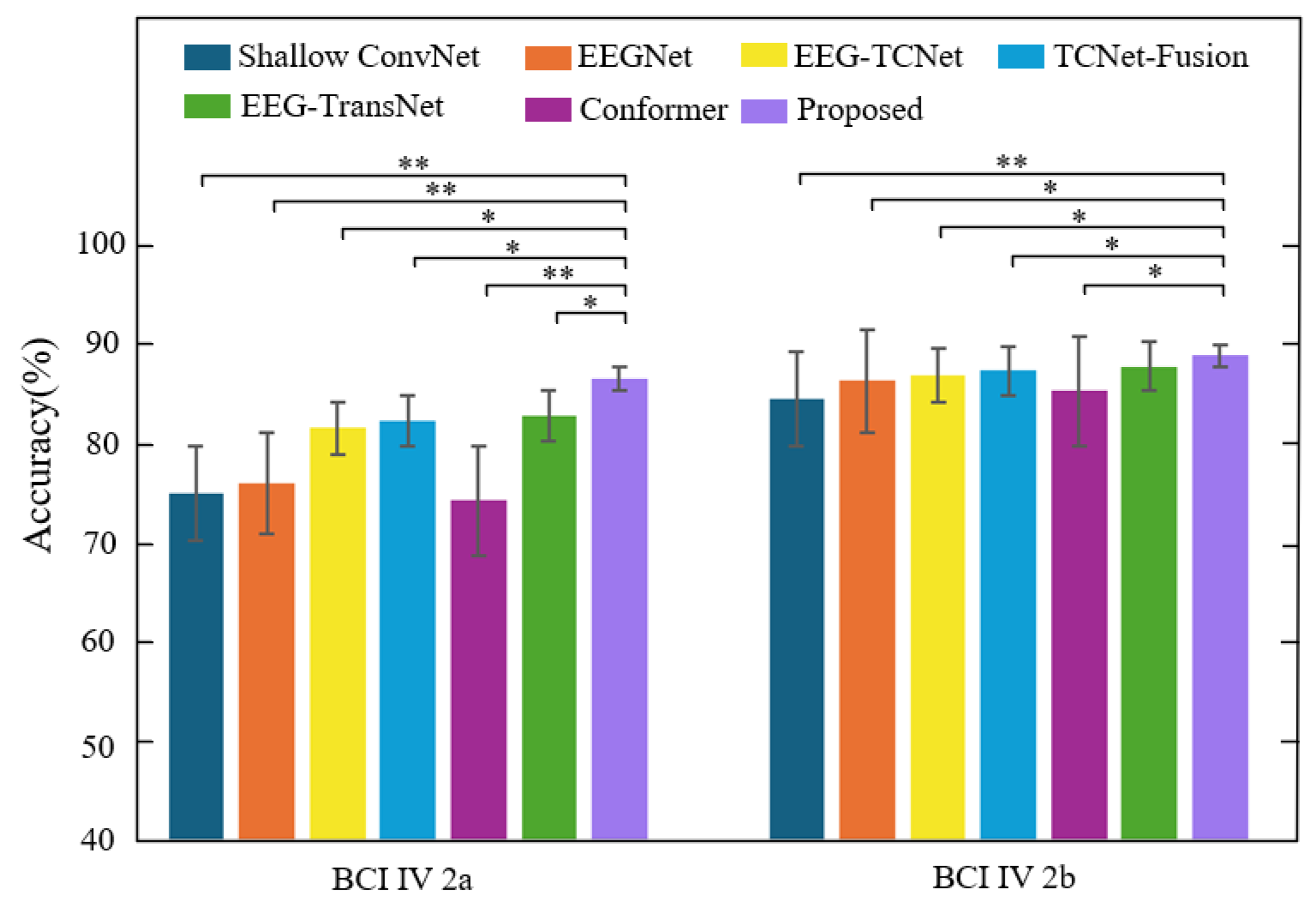
3.2.3. Comparison of GAH-TNet with Other Models
- •
- Advantages over traditional CNN models in spatial dependency modeling: Shallow ConvNet and EEGNet primarily rely on convolution operations to extract local features, making it difficult to model non-local spatial dependencies between channels. In contrast, GAH-TNet introduces a GCN module via GATE, effectively leveraging the topological structure of EEG electrodes to model cross-channel spatial relationships, enabling the model to uncover latent connections from structured graph spaces. In cross-subject experiments, GAH-TNet outperformed EEGNet by 12.6 percentage points on the 2a dataset, validating the critical role of its spatial modeling capabilities in generalization performance.
- •
- Advantages over Transformer models in hierarchical temporal modeling mechanisms: EEG-Conformer and EEG-TransNet often emphasize global attention or single-time-domain modeling, which may overlook critical local time segments. GAH-TNet introduces local-aware masked attention in the HADTE module, highlighting local dynamic patterns by restricting the attention window while simultaneously achieving joint modeling of global semantics. In the subject-specific setting of the 2b dataset, GAH-TNet achieves a 1.05 percentage point improvement over EEG-TransNet at near-saturated performance levels, indicating that its hierarchical temporal modeling mechanism can capture more discriminative features in critical time segments.
- •
- Advantages over fusion models in spatiotemporal synergy: TCNet Fusion and EEG-TCNet address feature diversity to some extent but still lack effective modeling of spatio-temporal dependencies. Under the subject-independent setting of the 2a dataset, GAH-TNet achieved an accuracy of 69.39%, representing a 6.1 percentage point improvement over TCNet Fusion (63.30%), and on the 2b dataset, it maintained a 3.5 percentage point advantage. These results indicate that GAH-TNet not only leverages the long-range temporal modeling capability of TCN but also enhances feature representation through the synergistic effects of graph attention and spatiotemporal convolutional encoding, thereby improving the spatiotemporal resolution and decoding performance of EEG signals.
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhao, W.; Jiang, X.; Zhang, B.; Xiao, S.; Weng, S. CTNet: A Convolutional Transformer Network for EEG-Based Motor Imagery Classification. Sci. Rep. 2024, 14, 20237. [Google Scholar] [CrossRef]
- Liao, W.; Miao, Z.; Liang, S.; Zhang, L.; Li, C. A Composite Improved Attention Convolutional Network for Motor Imagery EEG Classification. Front. Neurosci. 2025, 19, 1543508. [Google Scholar] [CrossRef]
- Machado, S.; Araújo, F.; Paes, F.; Velasques, B.; Cunha, M.; Budde, H.; Basile, L.F.; Anghinah, R.; Arias-Carrión, O.; Cagy, M. EEG-Based Brain-Computer Interfaces: An Overview of Basic Concepts and Clinical Applications in Neurorehabilitation. Rev. Neurosci. 2010, 21, 451–468. [Google Scholar] [CrossRef]
- Ma, Z.-Z.; Wu, J.-J.; Cao, Z.; Hua, X.-Y.; Zheng, M.-X.; Xing, X.-X.; Ma, J.; Xu, J.-G. Motor Imagery-Based Brain–Computer Interface Rehabilitation Programs Enhance Upper Extremity Performance and Cortical Activation in Stroke Patients. J. Neuroeng. Rehabil. 2024, 21, 91. [Google Scholar] [CrossRef]
- Liu, H.; Wei, P.; Wang, H.; Lv, X.; Duan, W.; Li, M.; Zhao, Y.; Wang, Q.; Chen, X.; Shi, G. An EEG Motor Imagery Dataset for Brain Computer Interface in Acute Stroke Patients. Sci. Data 2024, 11, 131. [Google Scholar] [CrossRef]
- Naser, M.Y.; Bhattacharya, S. Towards Practical BCI-Driven Wheelchairs: A Systematic Review Study. IEEE Trans. Neural Syst. Rehabil. Eng. 2023, 31, 1030–1044. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, A.; Suhag, A.K.; Kumar, N.; Bhardwaj, A. Use of BCI Systems in the Analysis of EEG Signals for Motor and Speech Imagery Task: A SLR. ACM Comput. Surv. 2025. [Google Scholar] [CrossRef]
- Jeong, H.; Song, M.; Jang, S.-H.; Kim, J. Investigating the Cortical Effect of False Positive Feedback on Motor Learning in Motor Imagery Based Rehabilitative BCI Training. J. NeuroEngineering Rehabil. 2025, 22, 61. [Google Scholar]
- Chen, S.; Yao, L.; Cao, L.; Caimmi, M.; Jia, J. Exploration of the Non-Invasive Brain-Computer Interface and Neurorehabilitation; Frontiers Media SA: Lausanne, Switzerland, 2024; Volume 18, p. 1377665. ISBN 1662-453X. [Google Scholar]
- Brunner, I.; Lundquist, C.B.; Pedersen, A.R.; Spaich, E.G.; Dosen, S.; Savic, A. Brain Computer Interface Training with Motor Imagery and Functional Electrical Stimulation for Patients with Severe Upper Limb Paresis after Stroke: A Randomized Controlled Pilot Trial. J. Neuroeng. Rehabil. 2024, 21, 10. [Google Scholar] [CrossRef]
- Hameed, I.; Khan, D.M.; Ahmed, S.M.; Aftab, S.S.; Fazal, H. Enhancing Motor Imagery EEG Signal Decoding through Machine Learning: A Systematic Review of Recent Progress. Comput. Biol. Med. 2025, 185, 109534. [Google Scholar] [CrossRef]
- Sharma, N.; Sharma, M.; Singhal, A.; Vyas, R.; Malik, H.; Afthanorhan, A.; Hossaini, M.A. Recent Trends in EEG-Based Motor Imagery Signal Analysis and Recognition: A Comprehensive Review. IEEE Access 2023, 11, 80518–80542. [Google Scholar] [CrossRef]
- Gong, S.; Xing, K.; Cichocki, A.; Li, J. Deep Learning in EEG: Advance of the Last Ten-Year Critical Period. IEEE Trans. Cogn. Dev. Syst. 2021, 14, 348–365. [Google Scholar] [CrossRef]
- Shiam, A.A.; Hassan, K.M.; Islam, M.R.; Almassri, A.M.; Wagatsuma, H.; Molla, M.K.I. Motor Imagery Classification Using Effective Channel Selection of Multichannel EEG. Brain Sci. 2024, 14, 462. [Google Scholar] [CrossRef]
- Hooda, N.; Kumar, N. Cognitive Imagery Classification of EEG Signals Using CSP-Based Feature Selection Method. IETE Tech. Rev. 2020, 37, 315–326. [Google Scholar] [CrossRef]
- Rithwik, P.; Benzy, V.; Vinod, A. High Accuracy Decoding of Motor Imagery Directions from EEG-Based Brain Computer Interface Using Filter Bank Spatially Regularised Common Spatial Pattern Method. Biomed. Signal Process. Control 2022, 72, 103241. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, W.; Lin, C.-L.; Pei, Z.; Chen, J.; Chen, Z. Boosting-LDA Algriothm with Multi-Domain Feature Fusion for Motor Imagery EEG Decoding. Biomed. Signal Process. Control 2021, 70, 102983. [Google Scholar] [CrossRef]
- Fu, R.; Tian, Y.; Bao, T.; Meng, Z.; Shi, P. Improvement Motor Imagery EEG Classification Based on Regularized Linear Discriminant Analysis. J. Med. Syst. 2019, 43, 169. [Google Scholar] [CrossRef]
- Altaheri, H.; Muhammad, G.; Alsulaiman, M.; Amin, S.U.; Altuwaijri, G.A.; Abdul, W.; Bencherif, M.A.; Faisal, M. Deep Learning Techniques for Classification of Electroencephalogram (EEG) Motor Imagery (MI) Signals: A Review. Neural Comput. Appl. 2023, 35, 14681–14722. [Google Scholar] [CrossRef]
- Echtioui, A.; Zouch, W.; Ghorbel, M.; Mhiri, C. Convolutional Neural Network with Support Vector Machine for Motor Imagery EEG Signal Classification. Multimed. Tools Appl. 2023, 82, 45891–45911. [Google Scholar] [CrossRef]
- Candelori, B.; Bardella, G.; Spinelli, I.; Ramawat, S.; Pani, P.; Ferraina, S.; Scardapane, S. Spatio-Temporal Transformers for Decoding Neural Movement Control. J. Neural Eng. 2025, 22, 016023. [Google Scholar] [CrossRef]
- Liu, F.; Meamardoost, S.; Gunawan, R.; Komiyama, T.; Mewes, C.; Zhang, Y.; Hwang, E.; Wang, L. Deep Learning for Neural Decoding in Motor Cortex. J. Neural Eng. 2022, 19, 056021. [Google Scholar] [CrossRef]
- Schirrmeister, R.T.; Springenberg, J.T.; Fiederer, L.D.J.; Glasstetter, M.; Eggensperger, K.; Tangermann, M.; Hutter, F.; Burgard, W.; Ball, T. Deep Learning with Convolutional Neural Networks for EEG Decoding and Visualization. Hum. Brain Mapp. 2017, 38, 5391–5420. [Google Scholar] [CrossRef]
- Lawhern, V.J.; Solon, A.J.; Waytowich, N.R.; Gordon, S.M.; Hung, C.P.; Lance, B.J. EEGNet: A Compact Convolutional Neural Network for EEG-Based Brain–Computer Interfaces. J. Neural Eng. 2018, 15, 056013. [Google Scholar] [CrossRef]
- Bai, S.; Kolter, J.Z.; Koltun, V. An Empirical Evaluation of Generic Convolutional and Recurrent Networks for Sequence Modeling. arXiv 2018, arXiv:1803.01271. [Google Scholar] [CrossRef]
- Musallam, Y.K.; AlFassam, N.I.; Muhammad, G.; Amin, S.U.; Alsulaiman, M.; Abdul, W.; Altaheri, H.; Bencherif, M.A.; Algabri, M. Electroencephalography-Based Motor Imagery Classification Using Temporal Convolutional Network Fusion. Biomed. Signal Process. Control 2021, 69, 102826. [Google Scholar] [CrossRef]
- Ingolfsson, T.M.; Hersche, M.; Wang, X.; Kobayashi, N.; Cavigelli, L.; Benini, L. EEG-TCNet: An Accurate Temporal Convolutional Network for Embedded Motor-Imagery Brain–Machine Interfaces. In Proceedings of the 2020 IEEE International Conference on Systems, Man, and Cybernetics (SMC), Toronto, ON, Canada, 11–14 October 2020; IEEE: New York, NY, USA, 2020; pp. 2958–2965. [Google Scholar]
- Zhao, Q.; Zhu, W. TMSA-Net: A Novel Attention Mechanism for Improved Motor Imagery EEG Signal Processing. Biomed. Signal Process. Control 2025, 102, 107189. [Google Scholar] [CrossRef]
- Sun, H.; Ding, Y.; Bao, J.; Qin, K.; Tong, C.; Jin, J.; Guan, C. Leveraging Temporal Dependency for Cross-Subject-MI BCIs by Contrastive Learning and Self-Attention. Neural Netw. 2024, 178, 106470. [Google Scholar] [CrossRef]
- Song, Y.; Zheng, Q.; Liu, B.; Gao, X. EEG Conformer: Convolutional Transformer for EEG Decoding and Visualization. IEEE Trans. Neural Syst. Rehabil. Eng. 2022, 31, 710–719. [Google Scholar] [CrossRef]
- Altaheri, H.; Muhammad, G.; Alsulaiman, M. Physics-Informed Attention Temporal Convolutional Network for EEG-Based Motor Imagery Classification. IEEE Trans. Ind. Inform. 2022, 19, 2249–2258. [Google Scholar] [CrossRef]
- Ma, X.; Chen, W.; Pei, Z.; Zhang, Y.; Chen, J. Attention-Based Convolutional Neural Network with Multi-Modal Temporal Information Fusion for Motor Imagery EEG Decoding. Comput. Biol. Med. 2024, 175, 108504. [Google Scholar] [CrossRef] [PubMed]
- Leng, J.; Gao, L.; Jiang, X.; Lou, Y.; Sun, Y.; Wang, C.; Li, J.; Zhao, H.; Feng, C.; Xu, F. A Multi-feature Fusion Graph Attention Network for Decoding Motor Imagery Intention in Spinal Cord Injury Patients. J. Neural Eng. 2024, 21, 066044. [Google Scholar] [CrossRef] [PubMed]
- Tan, X.; Wang, D.; Xu, M.; Chen, J.; Wu, S. Efficient Multi-View Graph Convolutional Network with Self-Attention for Multi-Class Motor Imagery Decoding. Bioengineering 2024, 11, 926. [Google Scholar] [CrossRef]
- Patel, R.; Zhu, Z.; Bryson, B.; Carlson, T.; Jiang, D.; Demosthenous, A. Advancing EEG Classification for Neurodegenerative Conditions Using BCI: A Graph Attention Approach with Phase Synchrony. Neuroelectronics 2025, 2, 1. [Google Scholar] [CrossRef]
- Brunner, C.; Leeb, R.; Müller-Putz, G.; Schlögl, A.; Pfurtscheller, G. BCI Competition 2008–Graz Data Set A. Inst. Knowl. Discov. Lab. Brain Comput. Interfaces Graz Univ. Technol. 2008, 16, 34. [Google Scholar]
- Leeb, R.; Brunner, C.; Müller-Putz, G.; Schlögl, A.; Pfurtscheller, G. BCI Competition 2008–Graz Data Set B. Graz Univ. Technol. Austria 2008, 16, 1–6. [Google Scholar]
- Cao, Y.; Xu, J.; Lin, S.; Wei, F.; Hu, H. Gcnet: Non-Local Networks Meet Squeeze-Excitation Networks and Beyond. In Proceedings of the IEEE/CVF International Conference on Computer Vision Workshops, Seoul, Republic of Korea, 27–28 October 2019. [Google Scholar]
- Wang, Q.; Wu, B.; Zhu, P.; Li, P.; Zuo, W.; Hu, Q. ECA-Net: Efficient Channel Attention for Deep Convolutional Neural Networks. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 13–19 June 2020; pp. 11534–11542. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention Is All You Need. Adv. Neural Inf. Process. Syst. 2017, 30. [Google Scholar]
- van der Maaten, L.; Hinton, G. Visualizing Data Using T-SNE. J. Mach. Learn. Res. 2008, 9, 2579–2605. [Google Scholar]
| Subject | BCI IV 2a | BCI IV 2b | ||||||
|---|---|---|---|---|---|---|---|---|
| GAH-TNet | GAH-TNet (LOSO) | GAH-TNet | GAH-TNet (LOSO) | |||||
| % | k | % | k | % | k | % | k | |
| A01 | 90.97 | 0.88 | 76.74 | 0.69 | 80.94 | 0.62 | 80.28 | 0.60 |
| A02 | 76.39 | 0.69 | 53.30 | 0.38 | 73.57 | 0.48 | 74.71 | 0.49 |
| A03 | 97.22 | 0.96 | 86.11 | 0.81 | 88.44 | 0.76 | 67.64 | 0.35 |
| A04 | 84.38 | 0.79 | 63.02 | 0.50 | 97.81 | 0.95 | 86.76 | 0.74 |
| A05 | 81.25 | 0.75 | 59.38 | 0.46 | 94.69 | 0.89 | 87.97 | 0.76 |
| A06 | 75.69 | 0.67 | 53.82 | 0.38 | 90.31 | 0.81 | 83.47 | 0.66 |
| A07 | 96.18 | 0.95 | 72.05 | 0.63 | 92.81 | 0.85 | 85.83 | 0.71 |
| A08 | 88.19 | 0.84 | 82.99 | 0.77 | 95.31 | 0.91 | 82.50 | 0.65 |
| A09 | 91.32 | 0.88 | 77.08 | 0.69 | 88.44 | 0.76 | 79.86 | 0.60 |
| Mean | 86.84 | 0.82 | 69.39 | 0.59 | 89.15 | 0.78 | 81.00 | 0.62 |
| Dataset | Method | Accuracy (%) | Kappa |
|---|---|---|---|
| BCI IV 2a | w/o Channel Attention | 85.22 | 0.79 |
| w/o Temporal-Spatial Convolutional Encoder | 79.68 | 0.72 | |
| w/o Residual-ECA | 85.53 | 0.80 | |
| w/o Local Masked Multi-Head Attention | 83.65 | 0.77 | |
| w/o Global Multi-Head Attention | 84.47 | 0.79 | |
| w/o Local Masked Multi-Head Attention and Global Multi-Head Attention | 82.64 | 0.76 | |
| w/o HADTE | 80.29 | 0.73 | |
| GAH-TNet | 86.84 | 0.82 | |
| BCI IV 2b | w/o Channel Attention | 87.85 | 0.76 |
| w/o Temporal-Spatial Convolutional Encoder | 81.04 | 0.63 | |
| w/o Residual-ECA | 87.44 | 0.74 | |
| w/o Local Masked Multi-Head Attention | 86.31 | 0.72 | |
| w/o Global Multi-Head Attention | 87.54 | 0.75 | |
| w/o Local Masked Multi-Head Attention and Global Multi-Head Attention | 84.66 | 0.70 | |
| w/o HADTE | 83.25 | 0.68 | |
| GAH-TNet | 89.15 | 0.78 |
| Dataset | Method | Accuracy (%) | Kappa |
|---|---|---|---|
| Subject-specific | Shallow ConvNet | 75.25 | 0.66 |
| EEGNet | 76.25 | 0.68 | |
| Conformer | 74.51 | 0.65 | |
| EEG-TCNet | 81.84 | 0.75 | |
| TCNet Fusion | 82.53 | 0.76 | |
| EEG-TransNet | 83.03 | 0.78 | |
| GAH-TNet | 86.84 | 0.82 | |
| Subject-independent | Shallow ConvNet | 55.03 | 0.40 |
| EEGNet | 56.80 | 0.42 | |
| Conformer | 55.25 | 0.40 | |
| EEG-TCNet | 62.26 | 0.49 | |
| TCNet Fusion | 63.30 | 0.49 | |
| EEG-TransNet | 64.53 | 0.50 | |
| GAH-TNet | 69.39 | 0.59 |
| Dataset | Method | Accuracy (%) | Kappa |
|---|---|---|---|
| Subject-specific | Shallow ConvNet | 84.82 | 0.70 |
| EEGNet | 86.63 | 0.71 | |
| Conformer | 85.63 | 0.70 | |
| EEG-TCNet | 87.20 | 0.74 | |
| TCNet Fusion | 87.61 | 0.74 | |
| EEG-TransNet | 88.10 | 0.76 | |
| GAH-TNet | 89.15 | 0.78 | |
| Subject-independent | Shallow ConvNet | 75.48 | 0.51 |
| EEGNet | 76.33 | 0.53 | |
| Conformer | 73.26 | 0.46 | |
| EEG-TCNet | 77.08 | 0.52 | |
| TCNet Fusion | 77.53 | 0.53 | |
| EEG-TransNet | 79.68 | 0.58 | |
| GAH-TNet | 81.00 | 0.62 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, Q.; Sun, Y.; Ye, H.; Song, Z.; Zhao, J.; Shi, L.; Kuang, Z. GAH-TNet: A Graph Attention-Based Hierarchical Temporal Network for EEG Motor Imagery Decoding. Brain Sci. 2025, 15, 883. https://doi.org/10.3390/brainsci15080883
Han Q, Sun Y, Ye H, Song Z, Zhao J, Shi L, Kuang Z. GAH-TNet: A Graph Attention-Based Hierarchical Temporal Network for EEG Motor Imagery Decoding. Brain Sciences. 2025; 15(8):883. https://doi.org/10.3390/brainsci15080883
Chicago/Turabian StyleHan, Qiulei, Yan Sun, Hongbiao Ye, Ze Song, Jian Zhao, Lijuan Shi, and Zhejun Kuang. 2025. "GAH-TNet: A Graph Attention-Based Hierarchical Temporal Network for EEG Motor Imagery Decoding" Brain Sciences 15, no. 8: 883. https://doi.org/10.3390/brainsci15080883
APA StyleHan, Q., Sun, Y., Ye, H., Song, Z., Zhao, J., Shi, L., & Kuang, Z. (2025). GAH-TNet: A Graph Attention-Based Hierarchical Temporal Network for EEG Motor Imagery Decoding. Brain Sciences, 15(8), 883. https://doi.org/10.3390/brainsci15080883






