Olfactory-Guided Behavior Uncovers Imaging and Molecular Signatures of Alzheimer’s Disease Risk
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Behavior Assessments
2.2.1. Odor Preference and Habituation–Dishabituation Test
2.2.2. Novel Odor Memory
2.3. Image Acquisition and Processing
2.4. Transcriptomics
2.5. Multivariate Modeling
2.6. Peripheral Molecular Mechanisms Associated with Brain Imaging Features
2.7. Statistical Analysis
3. Results
3.1. Olfactory Guided Behavior
3.1.1. Odor Salience
3.1.2. Anhedonia
3.1.3. Exploratory Dynamics
3.1.4. Habituation/Dishabituation
3.1.5. Odor Recognition Memory Across Delay Intervals
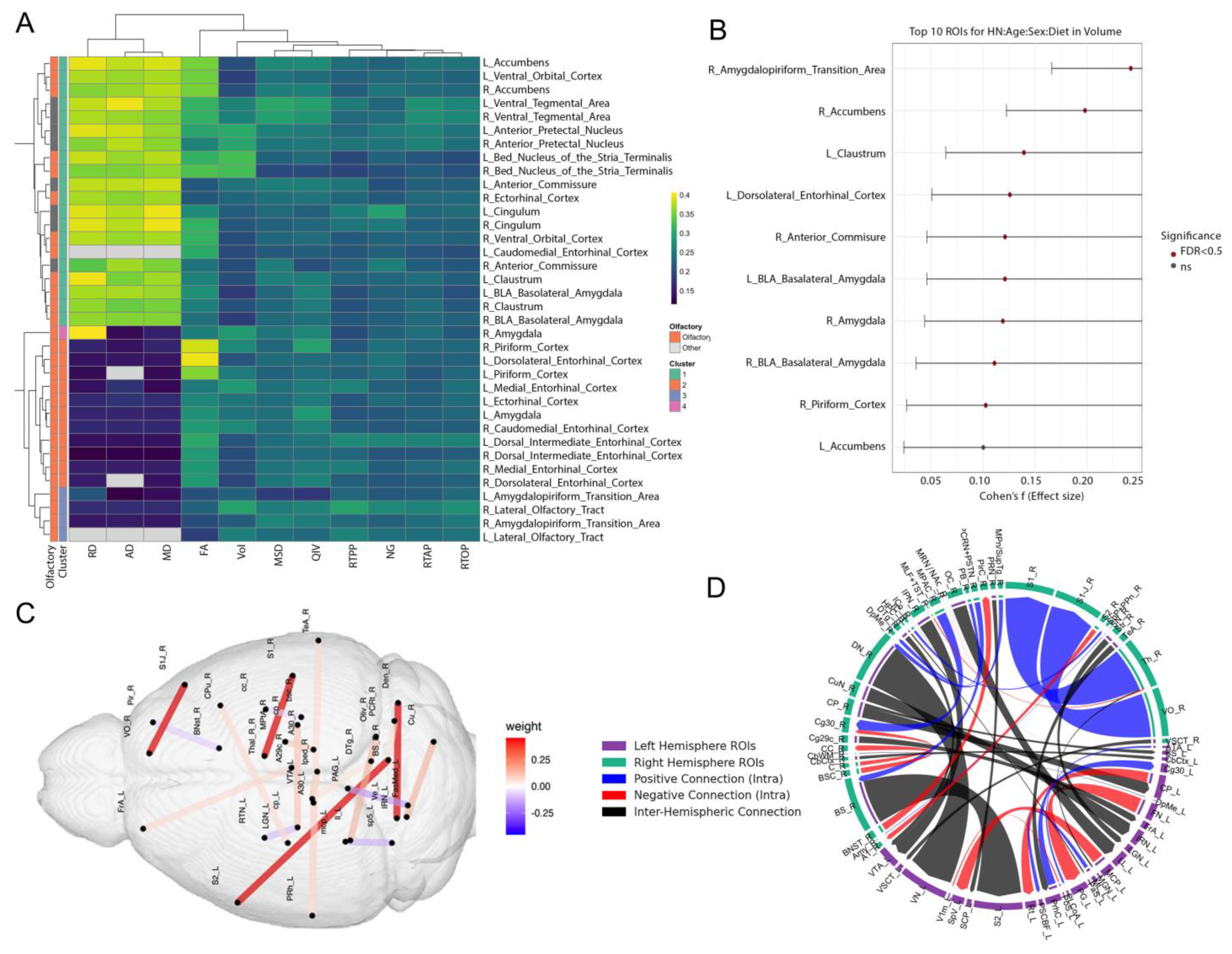
3.2. Multimodal Clustering Reveals Risk-Linked Olfactory–Limbic Networks
3.3. Transcriptomic Modules and Their Coupling with Brain Microstructure
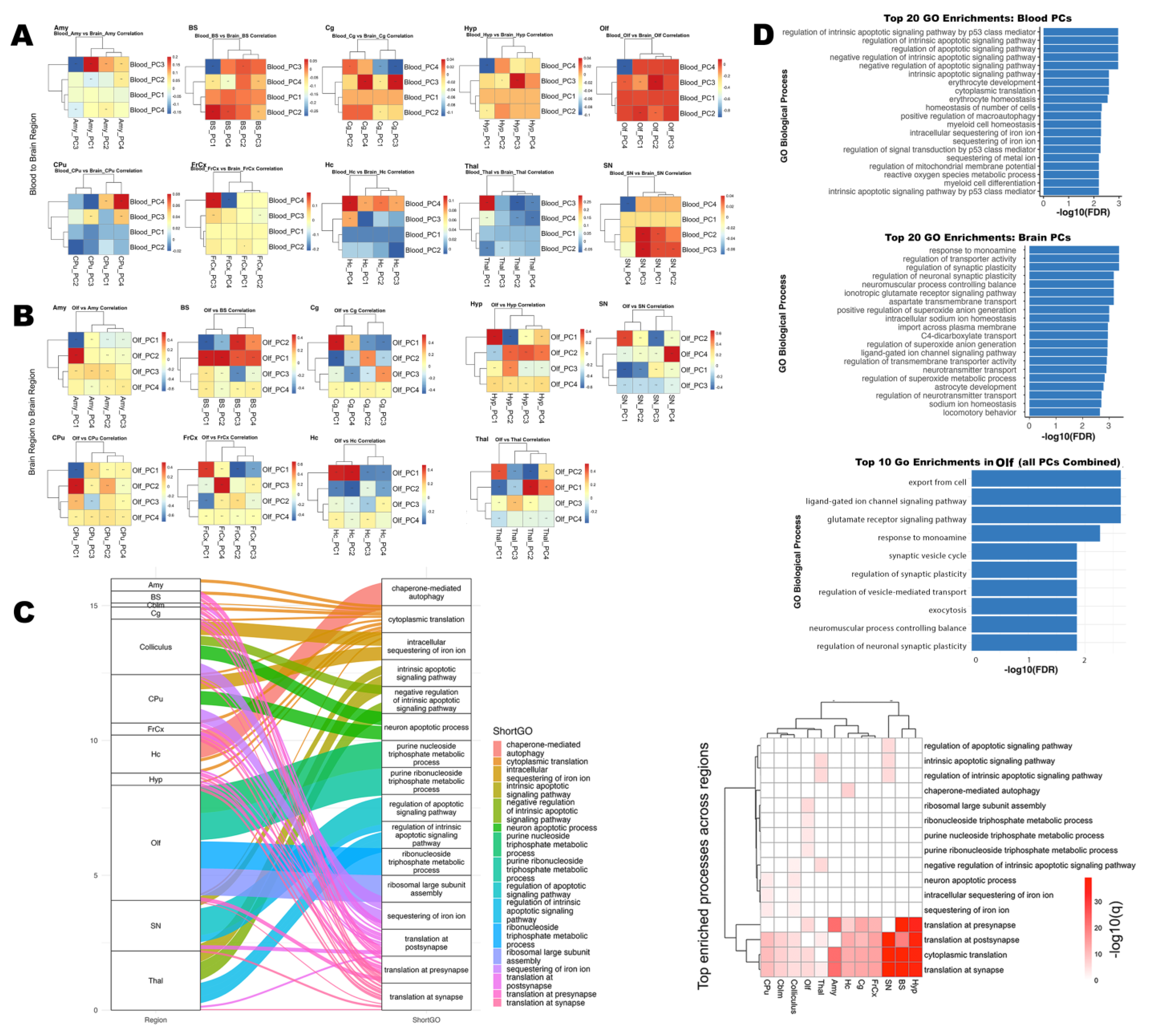
3.3.1. Functional Enrichment of Blood-Derived Gene Modules
3.3.2. Gene Modules Predict Regional Microstructure in Olfactory–Memory Circuits
3.3.3. Brain Regions–Blood Transcriptomic Coupling
3.3.4. High-Loading Genes Reveal Shared Molecular Drivers
3.3.5. Synaptic and Ion Regulation Pathways Shared by Blood and Olfactory Brain Regions
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AD | Alzheimer’s disease |
| MCCA | Multi-set Canonical Correlation |
| APOE | Apolipoprotein E. |
| PC | Principal component |
| PCA | Principal component analysis |
References
- Alzheimer and Association. Alzheimer’s Association 2024 Alzheimer’s Disease Facts and Figures; Alzheimer and Association: Chicago, IL, USA, 2024. [Google Scholar]
- Rajmohan, R.; Reddy, P.H. Amyloid Beta and Phosphorylated Tau Accumulations Cause Abnormalities at Synapses of Alzheimer’s disease Neurons. J. Alzheimer’s Dis. JAD 2017, 57, 975. [Google Scholar] [CrossRef]
- Deture, M.A.; Dickson, D.W. The neuropathological diagnosis of Alzheimer’s disease. Mol. Neurodegener. 2019, 14, 32. [Google Scholar] [CrossRef]
- Weintraub, S.; Wicklund, A.H.; Salmon, D.P. The Neuropsychological Profile of Alzheimer Disease. Cold Spring Harb. Perspect. Med. 2012, 2, a006171. [Google Scholar] [CrossRef]
- Creese, B.; Brooker, H.; Ismail, Z.; Wesnes, K.A.; Hampshire, A.; Khan, Z.; Megalogeni, M.; Corbett, A.; Aarsland, D.; Ballard, C. Preclinical, Prodromal, and Dementia Stages of Alzheimer’s Disease—PracticalNeurology. Am. J. Geriatr. Psychiatry 2019, 27, 823–834. [Google Scholar] [CrossRef]
- Sperling, R.A.; Aisen, P.S.; Beckett, L.A.; Bennett, D.A.; Craft, S.; Fagan, A.M.; Iwatsubo, T.; Jack, C.R.; Kaye, J.; Montine, T.J.; et al. Toward defining the preclinical stages of Alzheimer’s disease: Recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimer’s Dement. J. Alzheimer’s Assoc. 2011, 7, 280. [Google Scholar] [CrossRef] [PubMed]
- Zou, Y.M.; Lu, D.; Liu, L.P.; Zhang, H.H.; Zhou, Y.Y. Olfactory dysfunction in Alzheimer’s disease. Neuropsychiatr. Dis. Treat. 2016, 12, 869. [Google Scholar] [CrossRef] [PubMed]
- Jobin, B.; Magdamo, C.; Delphus, D.; Runde, A.; Reineke, S.; Soto, A.A.; Ergun, B.; Mukhija, S.; Albers, A.D.; Albers, M.W. The AROMHA brain health test is a remote olfactory assessment to screen for cognitive impairment. Sci. Rep. 2025, 15, 9290. [Google Scholar] [CrossRef] [PubMed]
- Djordjevic, J.; Jones-Gotman, M.; De Sousa, K.; Chertkow, H. Olfaction in patients with mild cognitive impairment and Alzheimer’s disease. Neurobiol. Aging 2008, 29, 693–706. [Google Scholar] [CrossRef]
- Audronyte, E.; Pakulaite-Kazliene, G.; Sutnikiene, V.; Kaubrys, G. Odor Discrimination as a Marker of Early Alzheimer’s Disease. J. Alzheimer’s Dis. 2023, 94, 1169. [Google Scholar] [CrossRef]
- Diez, I.; Ortiz-Terán, L.; Ng, T.S.C.; Albers, M.W.; Marshall, G.; Orwig, W.; Kim, C.M.; Bueichekú, E.; Montal, V.; Olofsson, J.; et al. Tau propagation in the brain olfactory circuits is associated with smell perception changes in aging. Nat. Commun. 2024, 15, 4809. [Google Scholar] [CrossRef]
- Zhang, J.; Zhao, Z.; Sun, S.; Li, J.; Wang, Y.; Dong, J.; Yang, S.; Lou, Y.; Yang, J.; Li, W.; et al. Olfactory Evaluation in Alzheimer’s Disease Model Mice. Brain Sci. 2022, 12, 607. [Google Scholar] [CrossRef]
- Doty, R.L.; Shaman, P.; Kimmelman, C.P.; Dann, M.S. University of Pennsylvania Smell Identification Test: A rapid quantitative olfactory function test for the clinic. Laryngoscope 1984, 94 Pt 1, 176–178. [Google Scholar] [CrossRef]
- Kobal, G.; Hummel, T.; Sekinger, B.; Barz, S.; Roscher, S.; Wolf, S. “Sniffin’ sticks”: Screening of olfactory performance. Rhinology 1996, 34, 222–226. [Google Scholar]
- Devanand, D.P.; Lee, S.; Luchsinger, J.A.; Knopman, D.; Vassilaki, M.; Motter, J.N. Comparison of brief olfactory and cognitive assessments to neuroimaging biomarkers in the prediction of cognitive decline and dementia in the MCSA cohort. Alzheimer’s Dement. 2024, 20, 8346–8358. [Google Scholar] [CrossRef] [PubMed]
- Casadio, C.; Ballotta, D.; Ricci, F.; Zanelli, V.; Carpentiero, O.; Corni, M.G.; Bardi, E.; Filippini, N.; Lui, F.; Nichelli, P.F.; et al. Olfactory Testing and Gray Matter Volume: A Combined Approach to Predict the Conversion to Alzheimer. Brain Sci. 2025, 15, 310. [Google Scholar] [CrossRef] [PubMed]
- Cameron, E.L.; Köster, E.P.; Møller, P. Is novelty detection important in long-term odor memory? Brain Sci. 2021, 11, 1146. [Google Scholar] [CrossRef] [PubMed]
- Zhong, M.Z.; Peng, T.; Duarte, M.L.; Wang, M.; Cai, D. Updates on mouse models of Alzheimer’s disease. Mol. Neurodegener. 2024, 19, 23. [Google Scholar] [CrossRef]
- Yassine, H.N.; Finch, C.E. APOE Alleles and Diet in Brain Aging and Alzheimer’s Disease. Front. Aging Neurosci. 2020, 12, 544681. [Google Scholar] [CrossRef]
- Foley, K.E.; Hewes, A.A.; Garceau, D.T.; Kotredes, K.P.; Carter, G.W.; Sasner, M.; Howell, G.R. The APOEε3/ε4 Genotype Drives Distinct Gene Signatures in the Cortex of Young Mice. Front. Aging Neurosci. 2022, 14, 838436. [Google Scholar] [CrossRef]
- Williams, T.; Bathe, T.; Vo, Q.; Sacilotto, P.; McFarland, K.; Ruiz, A.J.; Hery, G.P.; Sullivan, P.; Borchelt, D.R.; Prokop, S.; et al. Humanized APOE genotypes influence lifespan independently of tau aggregation in the P301S mouse model of tauopathy. Acta Neuropathol. Commun. 2023, 11, 99. [Google Scholar] [CrossRef]
- Fernández-Calle, R.; Konings, S.C.; Frontiñán-Rubio, J.; García-Revilla, J.; Camprubí-Ferrer, L.; Svensson, M.; Martinson, I.; Boza-Serrano, A.; Venero, J.L.; Nielsen, H.M.; et al. APOE in the bullseye of neurodegenerative diseases: Impact of the APOE genotype in Alzheimer’s disease pathology and brain diseases. Mol. Neurodegener. 2022, 17, 62. [Google Scholar] [CrossRef]
- Peng, K.Y.; Mathews, P.M.; Levy, E.; Wilson, D.A. Apolipoprotein E4 causes early olfactory network abnormalities and short-term olfactory memory impairments. Neuroscience 2016, 343, 364. [Google Scholar] [CrossRef] [PubMed]
- Misiak, M.M.; Hipolito, M.S.; Ressom, H.W.; Obisesan, T.O.; Manaye, K.F.; Nwulia, E.A. Apo E4 Alleles and Impaired Olfaction as Predictors of Alzheimer’s Disease. Clin. Exp. Psychol. 2017, 3, 169. [Google Scholar] [CrossRef]
- Badea, A.; Wu, W.; Shuff, J.; Wang, M.; Anderson, R.J.; Qi, Y.; Johnson, G.A.; Wilson, J.G.; Koudoro, S.; Garyfallidis, E.; et al. Identifying Vulnerable Brain Networks in Mouse Models of Genetic Risk Factors for Late Onset Alzheimer’s Disease. Front. Neuroinform. 2019, 13, 483287. [Google Scholar] [CrossRef] [PubMed]
- Calabrese, E.; Badea, A.; Cofer, G.; Qi, Y.; Johnson, G.A. A Diffusion MRI Tractography Connectome of the Mouse Brain and Comparison with Neuronal Tracer Data. Cereb. Cortex 2015, 25, 4628–4637. [Google Scholar] [CrossRef] [PubMed]
- O’Leary, T.P.; Stover, K.R.; Mantolino, H.M.; Darvesh, S.; Brown, R.E. Intact olfactory memory in the 5xFAD mouse model of Alzheimer’s disease from 3 to 15 months of age. Behav. Brain Res. 2020, 393, 112731. [Google Scholar] [CrossRef]
- Winter, S.; Mahzarnia, A.; Anderson, R.J.; Han, Z.Y.; Tremblay, J.; Stout, J.A.; Moon, H.S.; Marcellino, D.; Dunson, D.B.; Badea, A. Brain network fingerprints of Alzheimer’s disease risk factors in mouse models with humanized APOE alleles. Magn. Reson. Imaging 2024, 114, 110251. [Google Scholar] [CrossRef]
- Moon, H.S.; Mahzarnia, A.; Stout, J.; Anderson, R.J.; Han, Z.Y.; Tremblay, J.T.; Badea, C.T.; Badea, A. Feature attention graph neural network for estimating brain age and identifying important neural connections in mouse models of genetic risk for Alzheimer’s disease. Imaging Neurosci. 2024, 2, 1–22. [Google Scholar] [CrossRef]
- Bridgeford, E.W.; Chung, J.; Anderson, R.J.; Mahzarnia, A.; Stout, J.A.; Moon, H.S.; Han, Z.Y.; Vogelstein, J.T.; Badea, A. Network Biomarkers of Alzheimer’s Disease Risk Derived from Joint Volume and Texture Covariance Patterns in Mouse Models. bioRxiv 2025. [Google Scholar] [CrossRef]
- Anderson, R.J.; Long, C.M.; Calabrese, E.D.; Robertson, S.H.; Johnson, G.A.; Cofer, G.P.; O’Brien, R.J.; Badea, A. Optimizing Diffusion Imaging Protocols for Structural Connectomics in Mouse Models of Neurological Conditions. Front. Phys. 2020, 8, 518252. [Google Scholar] [CrossRef]
- Bouarab, C.; Thompson, B.; Polter, A.M. VTA GABA Neurons at the Interface of Stress and Reward. Front. Neural Circuits 2019, 13, 479774. [Google Scholar] [CrossRef]
- Barkus, C.; Sanderson, D.J.; Rawlins, J.N.P.; Walton, M.E.; Harrison, P.J.; Bannerman, D.M. What causes aberrant salience in schizophrenia? A role for impaired short-term habituation and the GRIA1 (GluA1) AMPA receptor subunit. Mol. Psychiatry 2014, 19, 1060. [Google Scholar] [CrossRef] [PubMed]
- Aqrabawi, A.J.; Kim, J.C. Behavioral Evaluation of Odor Memory in Mice. Bio-Protocol 2018, 8, e3023. [Google Scholar] [CrossRef] [PubMed]
- Tournier, J.D.; Smith, R.; Raffelt, D.; Tabbara, R.; Dhollander, T.; Pietsch, M.; Christiaens, D.; Jeurissen, B.; Yeh, C.H.; Connelly, A. MRtrix3: A fast, flexible and open software framework for medical image processing and visualisation. NeuroImage 2019, 202, 116137. [Google Scholar] [CrossRef] [PubMed]
- Garyfallidis, E.; Brett, M.; Amirbekian, B.; Rokem, A.; van der Walt, S.; Descoteaux, M.; Nimmo-Smith, I.; Dipy, C. Dipy, a library for the analysis of diffusion MRI data. Front. Neuroinform. 2014, 8, 8. [Google Scholar] [CrossRef]
- Anderson, R.J.; Cook, J.J.; Delpratt, N.; Nouls, J.C.; Gu, B.; McNamara, J.O.; Avants, B.B.; Johnson, G.A.; Badea, A. Small Animal Multivariate Brain Analysis (SAMBA)—A High Throughput Pipeline with a Validation Framework. Neuroinformatics 2019, 17, 451–472. [Google Scholar] [CrossRef]
- Winter, S.; Mahzarnia, A.; Anderson, R.J.; Yar Han, Z.; Tremblay, J.; Dunson, D.B.; Badea, A. Connectomes, APOE, Age, Sex, and Diet in Mouse Models of Aging; Zenodo: Geneva, Switzerland, 2023. [Google Scholar]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef]
- Moon, H.S.; Mahzarnia, A.; Stout, J.; Anderson, R.J.; Strain, M.; Tremblay, J.T.; Han, Z.Y.; Niculescu, A.; MacFarlane, A.; King, J.; et al. Multivariate investigation of aging in mouse models expressing the Alzheimer’s protective APOE2 allele: Integrating cognitive metrics, brain imaging, and blood transcriptomics. Brain Struct. Funct. 2024, 229, 231–249. [Google Scholar] [CrossRef]
- Zou, H.; Hastie, T. Regularization and Variable Selection Via the Elastic Net. J. R. Stat. Soc. Ser. B Stat. Methodol. 2005, 67, 301–320. [Google Scholar] [CrossRef]
- Arystarkhova, E.; Haq, I.U.; Luebbert, T.; Mochel, F.; Saunders-Pullman, R.; Bressman, S.B.; Feschenko, P.; Salazar, C.; Cook, J.F.; Demarest, S.; et al. Factors in the disease severity of ATP1A3 mutations: Impairment, misfolding, and allele competition. Neurobiol. Dis. 2019, 132, 104577. [Google Scholar] [CrossRef]
- Young-Pearse, T.L.; Chen, A.C.; Chang, R.; Marquez, C.; Selkoe, D.J. Secreted APP regulates the function of full-length APP in neurite outgrowth through interaction with integrin beta1. Neural Dev. 2008, 3, 15. [Google Scholar] [CrossRef]
- Stedehouder, J.; Couey, J.J.; Brizee, D.; Hosseini, B.; Slotman, J.A.; Dirven, C.M.F.; Shpak, G.; Houtsmuller, A.B.; Kushner, S.A. Fast-spiking Parvalbumin Interneurons are Frequently Myelinated in the Cerebral Cortex of Mice and Humans. Cereb. Cortex 2017, 27, 5001–5013. [Google Scholar] [CrossRef] [PubMed]
- Xue, M.; Reim, K.; Chen, X.; Chao, H.T.; Deng, H.; Rizo, J.; Brose, N.; Rosenmund, C. Distinct domains of complexin I differentially regulate neurotransmitter release. Nat. Struct. Mol. Biol. 2007, 14, 949–958. [Google Scholar] [CrossRef] [PubMed]
- Doty, R.L. Age-Related Deficits in Taste and Smell. Otolaryngol. Clin. N. Am. 2018, 51, 815–825. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez Echandía, E.L.; Fóscolo, M.; Broitman, S.T. Preferential nesting in lemon-scented environment in rats reared on lemon-scented bedding from birth to weaning. Physiol. Behav. 1982, 29, 47–49. [Google Scholar] [CrossRef]
- Schellinck, H.M.; Forestell, C.A.; LoLordo, V.M. A Simple and Reliable Test of Olfactory Learning and Memory in Mice. Chem. Senses 2001, 26, 663–672. [Google Scholar] [CrossRef]
- Janssen, C.I.F.; Jansen, D.; Mutsaers, M.P.C.; Dederen, P.J.W.C.; Geenen, B.; Mulder, M.T.; Kiliaan Kiliaan, A.J. The Effect of a High-Fat Diet on Brain Plasticity, Inflammation and Cognition in Female ApoE4-Knockin and ApoE-Knockout Mice. PLoS ONE 2016, 11, e0155307. [Google Scholar] [CrossRef]
- Li, Z.; Shue, F.; Zhao, N.; Shinohara, M.; Bu, G. APOE2: Protective mechanism and therapeutic implications for Alzheimer’s disease. Mol. Neurodegener. 2020, 15, 63. [Google Scholar] [CrossRef]
- O’Shea, D.M.; Zhang, A.S.; Rader, K.; Shakour, R.L.; Besser, L.; Galvin, J.E. APOE ε4 carrier status moderates the effect of lifestyle factors on cognitive reserve. Alzheimer’s Dement. 2024, 20, 8062. [Google Scholar] [CrossRef]
- Ning, Z.; Liu, Y.; Wan, M.; Zuo, Y.; Chen, S.; Shi, Z.; Xu, Y.; Li, H.; Ko, H.; Zhang, J.; et al. APOE2 protects against Aβ pathology by improving neuronal mitochondrial function through ERRα signaling. Cell. Mol. Biol. Lett. 2024, 29, 87. [Google Scholar] [CrossRef]
- Di Battista, M.A.; MHeinsinger, N.; William Rebeck, G. Alzheimer’s Disease Genetic Risk Factor APOE-ε4 Also Affects Normal Brain Function. Curr. Alzheimer Res. 2016, 13, 1200. [Google Scholar] [CrossRef]
- Har-Paz, I.; Arieli, E.; Moran, A. ApoE4 attenuates cortical neuronal activity in young behaving apoE4 rats. Neurobiol. Dis. 2021, 155, 105373. [Google Scholar] [CrossRef] [PubMed]
- Goodman, T.; Trouche, S.; Massou, I.; Verret, L.; Zerwas, M.; Roullet, P.; Rampon, C. Young hippocampal neurons are critical for recent and remote spatial memory in adult mice. Neuroscience 2010, 171, 769–778. [Google Scholar] [CrossRef] [PubMed]
- Guskjolen, A.; Josselyn, S.A.; Frankland, P.W. Age-dependent changes in spatial memory retention and flexibility in mice. Neurobiol. Learn. Mem. 2017, 143, 59–66. [Google Scholar] [CrossRef] [PubMed]
- Hunter, C.L.; Bimonte, H.A.; Granholm, A.C.E. Behavioral comparison of 4 and 6 month-old Ts65Dn mice:: Age-related impairments in working and reference memory. Behav. Brain Res. 2003, 138, 121–131. [Google Scholar] [CrossRef]
- Mahzarnia, A.; Stout, J.A.; Anderson, R.J.; Moon, H.S.; Yar Han, Z.; Beck, K.; Browndyke, J.N.; Dunson, D.B.; Johnson, K.G.; O’Brien, R.J.; et al. Identifying vulnerable brain networks associated with Alzheimer’s disease risk. Cereb. Cortex 2022, 33, 5307. [Google Scholar] [CrossRef]
- Schoenbaum, G.; Roesch, M.R.; Stalnaker, T.A.; Takahashi, Y.K. Orbitofrontal Cortex and Outcome Expectancies: Optimizing Behavior and Sensory Perception. In Neurobiology of Sensation and Reward; CRC Press: Boca Raton, FL, USA, 2011; pp. 349–370. [Google Scholar]
- Rolls, E.T.; Cheng, W.; Feng, J. The orbitofrontal cortex: Reward, emotion and depression. Brain Commun. 2020, 2, fcaa196. [Google Scholar] [CrossRef]
- Lebow, M.A.; Chen, A. Overshadowed by the amygdala: The bed nucleus of the stria terminalis emerges as key to psychiatric disorders. Mol. Psychiatry 2016, 21, 450–463. [Google Scholar] [CrossRef]
- Blazing, R.M.; Franks, K.M. Odor coding in piriform cortex: Mechanistic insights into distributed coding. Curr. Opin. Neurobiol. 2020, 64, 96. [Google Scholar] [CrossRef]
- Matsukawa, M.; Yoshikawa, M.; Katsuyama, N.; Aizawa, S.; Sato, T. The Anterior Piriform Cortex and Predator Odor Responses: Modulation by Inhibitory Circuits. Front. Behav. Neurosci. 2022, 16, 896525. [Google Scholar] [CrossRef]
- Yu, M.; Wang, S.-M. Neuroanatomy, Nucleus Fastigial; StatPearls: Petersburg, FL, USA, 2023. [Google Scholar]
- Brandão, M.L.; Lovick, T.A. Role of the dorsal periaqueductal gray in posttraumatic stress disorder: Mediation by dopamine and neurokinin. Transl. Psychiatry 2019, 9, 232. [Google Scholar] [CrossRef] [PubMed]
- Cacciola, A.; Bertino, S.; Basile, G.A.; Di Mauro, D.; Calamuneri, A.; Chillemi, G.; Duca, A.; Bruschetta, D.; Flace, P.; Favaloro, A.; et al. Mapping the structural connectivity between the periaqueductal gray and the cerebellum in humans. Brain Struct. Funct. 2019, 224, 2153. [Google Scholar] [CrossRef] [PubMed]
- Ables, J.L.; Park, K.; Ibañez-Tallon, I. Understanding the Habenula: A Major Node in Circuits Regulating Emotion and Motivation. Pharmacol. Res. 2023, 190, 106734. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.Y.; Peng, X.L.; Deng, Q.S.; Chen, M.j.; Du, J.L.; Zhang, B.B. Role of Olfactorily Responsive Neurons in the Right Dorsal Habenula–Ventral Interpeduncular Nucleus Pathway in Food-Seeking Behaviors of Larval Zebrafish. Neuroscience 2019, 404, 259–267. [Google Scholar] [CrossRef]
- Molas, S.; Zhao-Shea, R.; Freels, T.G.; Tapper, A.R. Viral Tracing Confirms Paranigral Ventral Tegmental Area Dopaminergic Inputs to the Interpeduncular Nucleus Where Dopamine Release Encodes Motivated Exploration. eNeuro 2023, 10. [Google Scholar] [CrossRef]
- Xu, H.; Geng, C.; Hua, X.; Liu, P.; Xu, J.; Li, A. Distinct Characteristics of Odor-evoked Calcium and Electrophysiological Signals in Mitral/Tufted Cells in the Mouse Olfactory Bulb. Neurosci. Bull. 2021, 37, 959. [Google Scholar] [CrossRef]
- Poplawsky, A.J.; Cover, C.; Reddy, S.; Chishti, H.B.; Vazquez, A.; Fukuda, M. Odor-evoked layer-specific fMRI activities in the awake mouse olfactory bulb. NeuroImage 2023, 274, 120121. [Google Scholar] [CrossRef]
- Grant, W.B. A Brief History of the Progress in Our Understanding of Genetics and Lifestyle, Especially Diet, in the Risk of Alzheimer’s Disease. J. Alzheimer’s Dis. 2024, 100 (Suppl. 1), S165. [Google Scholar] [CrossRef]
- Miura, E.; Hasegawa, T.; Konno, M.; Suzuki, M.; Sugeno, N.; Fujikake, N.; Geisler, S.; Tabuchi, M.; Oshima, R.; Kikuchi, A.; et al. VPS35 dysfunction impairs lysosomal degradation of α-synuclein and exacerbates neurotoxicity in a Drosophila model of Parkinson’s disease. Neurobiol. Dis. 2014, 71, 1–13. [Google Scholar] [CrossRef]
- Coultrap, S.J.; Bayer, K.U. CaMKII regulation in information processing and storage. Trends Neurosci. 2012, 35, 607–618. [Google Scholar] [CrossRef]
- Adams, J.; Crosbie, J.; Wigg, K.; Ickowicz, A.; Pathare, T.; Roberts, W.; Malone, M.; Schachar, R.; Tannock, R.; Kennedy, J.L.; et al. Glutamate receptor, ionotropic, N-methyl D-aspartate 2A (GRIN2A) gene as a positional candidate for attention-deficit/hyperactivity disorder in the 16p13 region. Mol. Psychiatry 2004, 9, 494–499. [Google Scholar] [CrossRef]
- Du, Z.; Song, Y.; Chen, X.; Zhang, W.; Zhang, G.; Li, H.; Chang, L.; Wu, Y. Knockdown of astrocytic Grin2a aggravates β-amyloid-induced memory and cognitive deficits through regulating nerve growth factor. Aging Cell 2021, 20, e13437. [Google Scholar] [CrossRef] [PubMed]
- Svenningsson, P.; Nishi, A.; Fisone, G.; Girault, J.A.; Nairn, A.C.; Greengard, P. DARPP-32: An integrator of neurotransmission. Annu. Rev. Pharmacol. Toxicol. 2004, 44, 269–296. [Google Scholar] [CrossRef] [PubMed]
- Costa-Mattioli, M.; Sossin, W.S.; Klann, E.; Sonenberg, N. Translational control of long-lasting synaptic plasticity and memory. Neuron 2009, 61, 10–26. [Google Scholar] [CrossRef]
- Grimm, A.; Eckert, A. Brain aging and neurodegeneration: From a mitochondrial point of view. J. Neurochem. 2017, 143, 418–431. [Google Scholar] [CrossRef]
- Santello, M.; Toni, N.; Volterra, A. Astrocyte function from information processing to cognition and cognitive impairment. Nat. Neurosci. 2019, 22, 154–166. [Google Scholar] [CrossRef]
- Fields, R.D. White matter in learning, cognition and psychiatric disorders. Trends Neurosci. 2008, 31, 361–370. [Google Scholar] [CrossRef]
- Swarup, V.; Hinz, F.I.; Rexach, J.E.; Noguchi, K.I.; Toyoshiba, H.; Oda, A.; Hirai, K.; Sarkar, A.; Seyfried, N.T.; Cheng, C.; et al. Identification of evolutionarily conserved gene networks mediating neurodegenerative dementia. Nat. Med. 2019, 25, 152–164. [Google Scholar] [CrossRef]



| APOE | Diet | Female | Male | 18 Months | 12 Months | mNOS2 | HN | Total |
|---|---|---|---|---|---|---|---|---|
| APOE2 | Control | 46 | 48 | 51 | 43 | 48 | 46 | 94 |
| APOE2 | HFD | 37 | 34 | 31 | 40 | 29 | 42 | 71 |
| APOE3 | Control | 45 | 41 | 41 | 45 | 43 | 43 | 86 |
| APOE3 | HFD | 35 | 33 | 32 | 36 | 30 | 38 | 68 |
| APOE4 | Control | 41 | 35 | 28 | 48 | 43 | 33 | 76 |
| APOE4 | HFD | 39 | 31 | 23 | 47 | 34 | 36 | 70 |
| Eigengene Module | Top Enriched Pathway | p-Value | Notes |
|---|---|---|---|
| Eigengene 1 | Pattern recognition receptor signaling | 0.003 | Strong immune/sensory relevance |
| Eigengene 2 | Phagocytosis, recognition | 0.007 | Sensory clearance processes |
| Eigengene 3 | Learning or memory | 0.002 | Direct cognitive link (memory formation) |
| Eigengene 4 | Neuron recognition | 0.010 | Supports neuronal differentiation |
| Eigengene 5 | Pattern recognition receptor activity | 0.015 | Supports sensory input mechanisms |
| Eigengene 6 | Cell surface receptor signaling | 0.018 | Moderate sensory/cognitive relevance |
| Eigengene 7 | Cell–cell recognition | 0.022 | Intercellular signaling |
| Eigengene 8 | Cell adhesion involved in neuron migration | 0.029 | Neuronal development process |
| Structure | Metric | Eigengene | r | p_FDR | R2 |
|---|---|---|---|---|---|
| Parasubiculum | AD | Eigengene 2 | −0.4919 | 1.6 × 10−32 | 0.24 |
| Parasubiculum | MD | Eigengene 2 | −0.4870 | 8.6 × 10−32 | 0.24 |
| Parasubiculum | RD | Eigengene 2 | −0.4836 | 3.2 × 10−31 | 0.23 |
| Hippocampus | AD | Eigengene 2 | −0.4780 | 7.7 × 10−31 | 0.23 |
| Postsubiculum | AD | Eigengene 2 | −0.4777 | 8.1 × 10−31 | 0.23 |
| Piriform Cortex | MSD | Eigengene 8 | 0.3636 | 2.1 × 10−12 | 0.14 |
| Piriform Cortex | FA | Eigengene 2 | 0.3176 | 6.8 × 10−13 | 0.10 |
| Piriform Cortex | NG | Eigengene 5 | −0.3237 | 2.8 × 10−11 | 0.13 |
| Piriform Cortex | RTOP | Eigengene 8 | −0.3102 | 2.4 × 10−9 | 0.10 |
| Piriform Cortex | RTOP | Eigengene 4 | −0.3089 | 5.9 × 10−11 | 0.12 |
| Hippocampus | MD | Eigengene 2 | −0.4758 | 1.5 × 10−30 | 0.23 |
| Hippocampus | RD | Eigengene 2 | −0.4738 | 2.8 × 10−30 | 0.22 |
| Hippocampus | FA | Eigengene 2 | 0.3689 | 2.7 × 10−17 | 0.14 |
| Hippocampus | QIV | Eigengene 8 | 0.3643 | 2.1 × 10−12 | 0.14 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moon, H.S.; Han, Z.Y.; Anderson, R.J.; Mahzarnia, A.; Stout, J.A.; Niculescu, A.R.; Tremblay, J.T.; Badea, A. Olfactory-Guided Behavior Uncovers Imaging and Molecular Signatures of Alzheimer’s Disease Risk. Brain Sci. 2025, 15, 863. https://doi.org/10.3390/brainsci15080863
Moon HS, Han ZY, Anderson RJ, Mahzarnia A, Stout JA, Niculescu AR, Tremblay JT, Badea A. Olfactory-Guided Behavior Uncovers Imaging and Molecular Signatures of Alzheimer’s Disease Risk. Brain Sciences. 2025; 15(8):863. https://doi.org/10.3390/brainsci15080863
Chicago/Turabian StyleMoon, Hae Sol, Zay Yar Han, Robert J. Anderson, Ali Mahzarnia, Jacques A. Stout, Andrei R. Niculescu, Jessica T. Tremblay, and Alexandra Badea. 2025. "Olfactory-Guided Behavior Uncovers Imaging and Molecular Signatures of Alzheimer’s Disease Risk" Brain Sciences 15, no. 8: 863. https://doi.org/10.3390/brainsci15080863
APA StyleMoon, H. S., Han, Z. Y., Anderson, R. J., Mahzarnia, A., Stout, J. A., Niculescu, A. R., Tremblay, J. T., & Badea, A. (2025). Olfactory-Guided Behavior Uncovers Imaging and Molecular Signatures of Alzheimer’s Disease Risk. Brain Sciences, 15(8), 863. https://doi.org/10.3390/brainsci15080863








