Biomolecular Predictors of Recurrence Patterns and Survival in IDH-Wild-Type Glioblastoma: A Retrospective Analysis of Patients Treated with Radiotherapy and Temozolomide
Abstract
1. Introduction
2. Study Design and Patient Selection
2.1. Data Collection and Treatment Approach
- EGFR-amplified tumors with EGFR protein overexpression.
- Non-amplified (wild-type) tumors with normal, weak, or absent EGFR expression.
2.2. Outcome Measures and Recurrence Classification
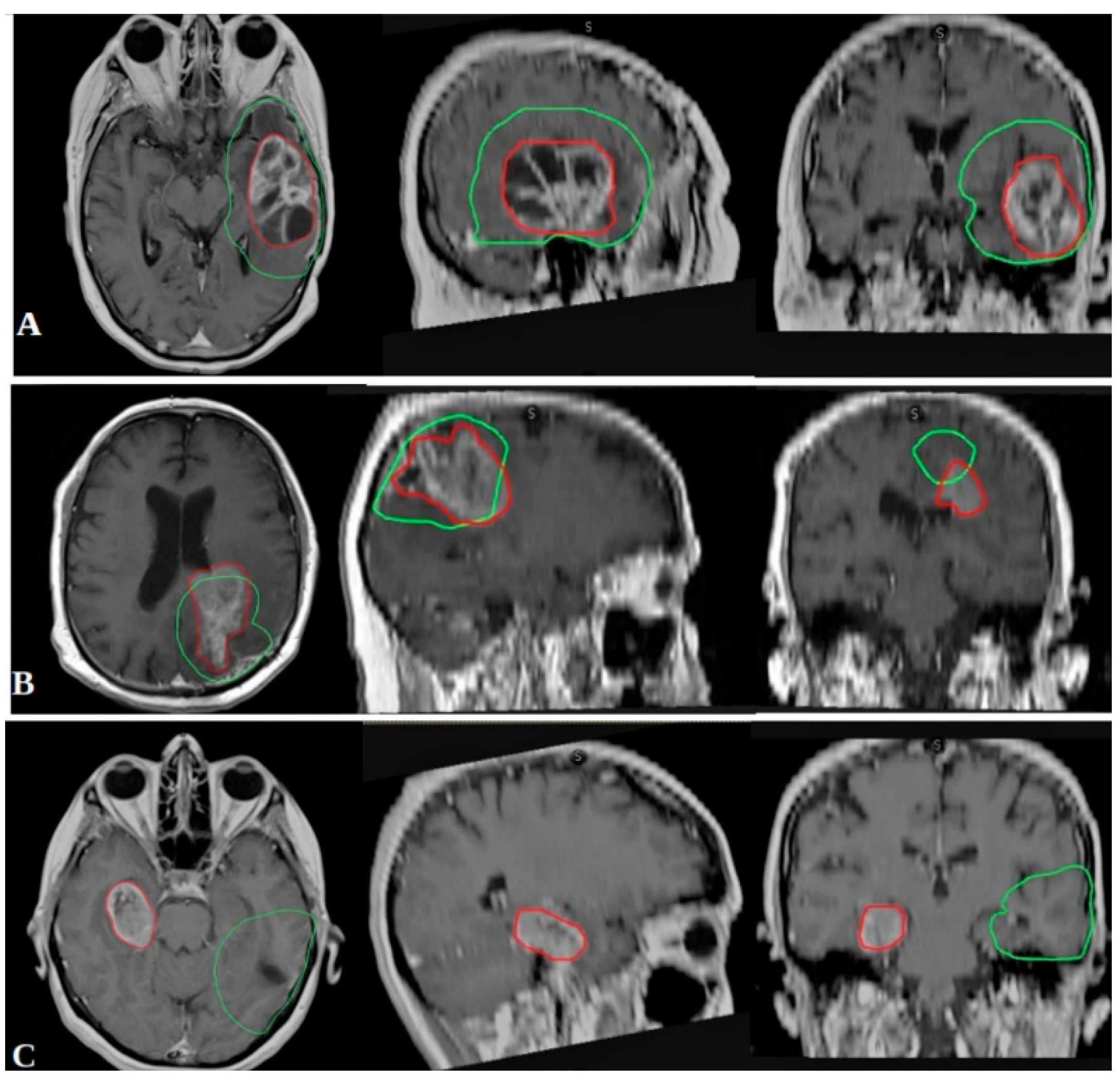
- In-field recurrence: ≥80% of the recurrent tumor volume within the 95% isodose line.
- Marginal recurrence: 20–80% of the recurrent volume within the 95% isodose line.
- Out-field recurrence: <20% of the recurrent volume within the 95% isodose line.
2.3. Statistical Analysis
2.4. Ethical Considerations
3. Results
3.1. Patient Demographics and Clinical Characteristics
3.2. Biomolecular Markers and Grouping Strategy
- EGFR non-amplified/MGMT methylated (EGFR neg/MGMT met);
- EGFR non-amplified/MGMT unmethylated (EGFR neg/MGMT unm);
- EGFR amplified/MGMT methylated (EGFR iper/MGMT met);
- EGFR amplified/MGMT unmethylated (EGFR iper/MGMT unm).
3.3. Radiological Response and Disease Progression
3.4. Recurrence Patterns and Correlation with Biomolecular Markers
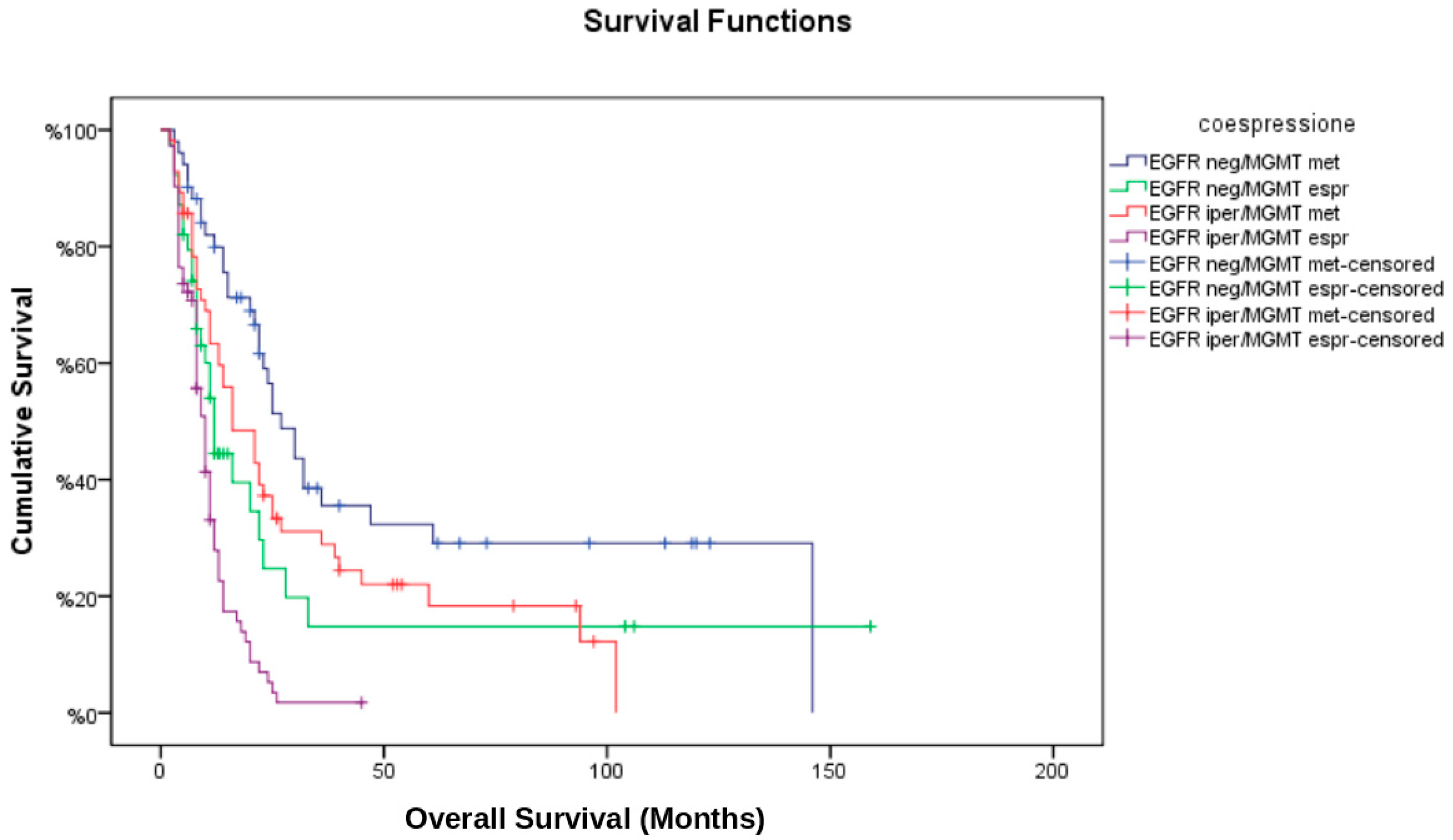
3.5. Multivariate Cox Regression Analysis
- MGMT methylation was significantly associated with improved OS (HR: 0.48; 95% CI: 0.33–0.69, p < 0.001) and PFS (HR: 0.54; 95% CI: 0.39–0.76, p = 0.001).
- EGFR amplification was independently associated with worse OS (HR: 1.57; 95% CI: 1.08–2.30, p = 0.02) and a higher likelihood of marginal recurrence (OR: 2.42; 95% CI: 1.22–4.81, p = 0.01).
- Gross total resection conferred a survival benefit (HR for OS: 0.67; 95% CI: 0.47–0.95, p = 0.03), whereas subtotal resection or biopsy was associated with earlier disease progression.
- The interaction term between EGFR amplification and MGMT methylation was not statistically significant (p = 0.09), suggesting that their effects on survival are largely independent.
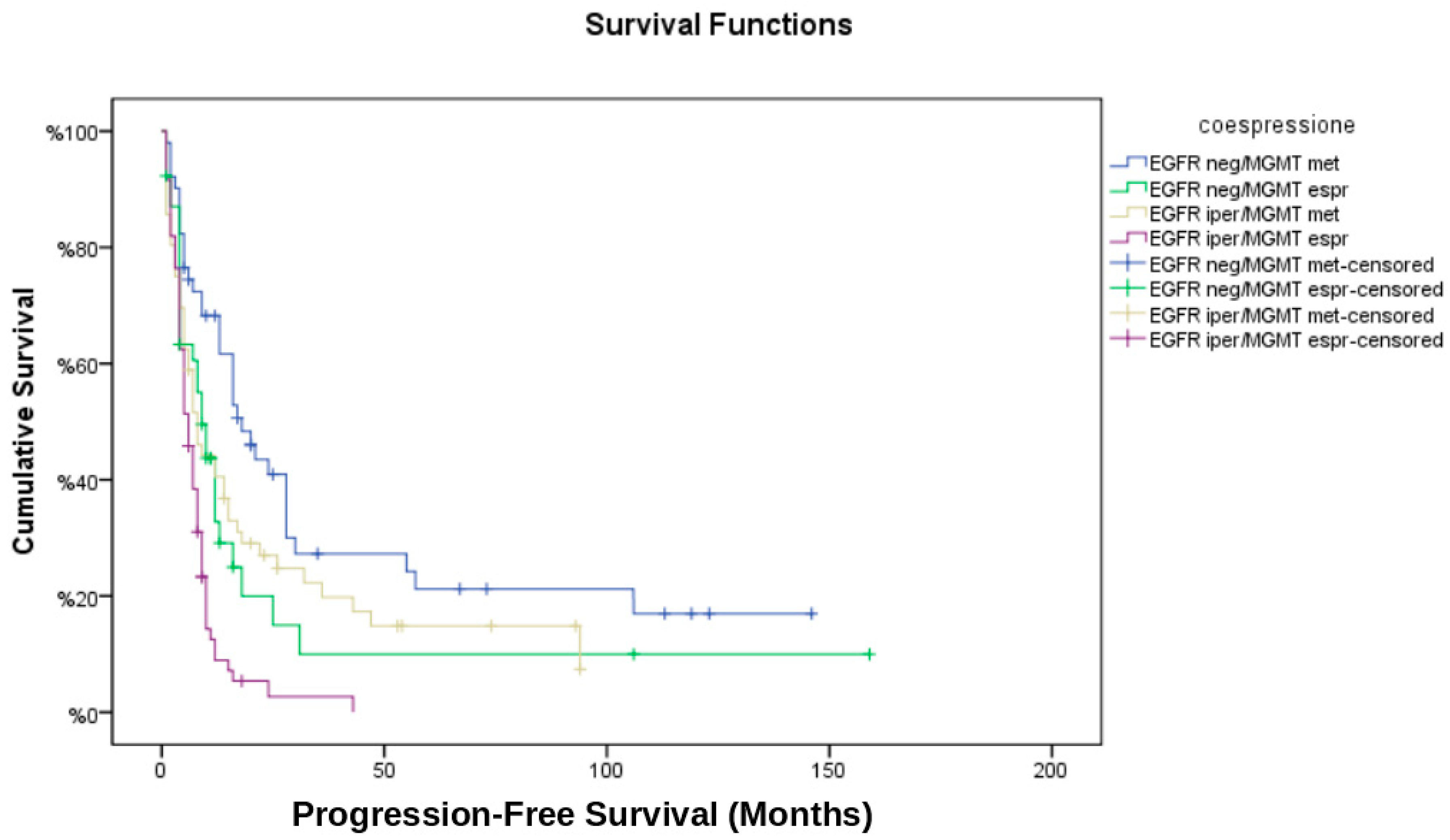
3.6. Temporal Analysis of Recurrence
- EGFR neg/MGMT met had the longest time to recurrence (median: 12.3 months).
- EGFR iper/MGMT unm had the shortest time to recurrence (median: 6.4 months).
3.7. Tumor Volume, Location and Biomolecular Markers
3.8. Recurrence Therapy and Second Surgery
4. Discussion
5. Limitations
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ostrom, Q.T.; Price, M.; Neff, C.; Cioffi, G.; Waite, K.A.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2015–2019. Neuro-Oncology 2022, 24 (Suppl. 5), v1–v95. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.K.; Pfister, S.M.; Reifenberger, G.; et al. The 2021 WHO Classification of Tumors of the Central Nervous System: A summary. Neuro-Oncology 2021, 23, 1231–1251. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ostrom, Q.T.; Bauchet, L.; Davis, F.G.; Deltour, I.; Fisher, J.L.; Langer, C.E.; Pekmezci, M.; Schwartzbaum, J.A.; Turner, M.C.; Walsh, K.M.; et al. The epidemiology of glioma in adults: A “state of the science” review. Neuro-Oncology 2014, 16, 896–913. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Stupp, R.; Hegi, M.E.; Mason, W.P.; van den Bent, M.J.; Taphoorn, M.J.; Janzer, R.C.; Ludwin, S.K.; Allgeier, A.; Fisher, B.; Belanger, K.; et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009, 10, 459–466. [Google Scholar] [CrossRef] [PubMed]
- Hegi, M.E.; Liu, L.; Herman, J.G.; Stupp, R.; Wick, W.; Weller, M.; Mehta, M.P.; Gilbert, M.R. Correlation of O6-methylguanine methyltransferase (MGMT) promoter methylation with clinical outcomes in glioblastoma and clinical strategies to modulate MGMT activity. J. Clin. Oncol. 2008, 26, 4189–4199. [Google Scholar] [CrossRef] [PubMed]
- Niyazi, M.; Andratschke, N.; Bendszus, M.; Chalmers, A.J.; Erridge, S.C.; Galldiks, N.; Lagerwaard, F.J.; Navarria, P.; Munck Af Rosenschöld, P.; Ricardi, U.; et al. ESTRO-EANO guideline on target delineation and radiotherapy details for glioblastoma. Radiother. Oncol. 2023, 184, 109663. [Google Scholar] [CrossRef] [PubMed]
- Stupp, R.; Taillibert, S.; Kanner, A.A.; Kesari, S.; Steinberg, D.M.; Toms, S.A.; Taylor, L.P.; Lieberman, F.; Silvani, A.; Fink, K.L.; et al. Maintenance therapy with tumor-treating fields plus temozolomide vs temozolomide alone for glioblastoma. JAMA 2015, 314, 2535–2543. [Google Scholar] [CrossRef]
- Louis, D.N.; Perry, A.; Reifenberger, G.; Von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A summary. Acta Neuropathol. 2016, 131, 803–820. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 2008, 455, 1061–1068. [Google Scholar] [CrossRef]
- Brennan, C.W.; Verhaak, R.G.; McKenna, A.; Campos, B.; Noushmehr, H.; Salama, S.R.; Zheng, S.; Chakravarty, D.; Sanborn, J.Z.; Berman, S.H.; et al. The somatic genomic landscape of glioblastoma. Cell 2013, 155, 462–477. [Google Scholar] [CrossRef]
- Pearson, J.R.; Regad, T. Targeting cellular pathways in glioblastoma multiforme. Signal Transduct. Target. Ther. 2017, 2, 17040. [Google Scholar] [CrossRef]
- Zhu, F.; Qiu, J.; Ye, H.; Su, W.; Wang, R.; Fu, Y. The Prognostic Significance of Epidermal Growth Factor Receptor Amplification and Epidermal Growth Factor Receptor Variant III Mutation in Glioblastoma: A Systematic Review and Meta-Analysis with Implications for Targeted Therapy. Int. J. Mol. Sci. 2025, 26, 3539. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wick, W.; Weller, M.; van den Bent, M.; Sanson, M.; Weiler, M.; Von Deimling, A.; Plass, C.; Hegi, M.; Platten, M.; Reifenberger, G. MGMT testing—The challenges for biomarker-based glioma treatment. Nat. Rev. Neurol. 2014, 10, 372–385. [Google Scholar] [CrossRef] [PubMed]
- Gan, H.K.; Cvrljevic, A.N.; Johns, T.G. The epidermal growth factor receptor variant III (EGFRvIII): Where wild things are altered. FEBS J. 2013, 280, 5350–5370. [Google Scholar] [CrossRef] [PubMed]
- Mangiola, A.; Anile, C.; Pompucci, A.; Capone, G.; Rigante, L.; De Bonis, P. Glioblastoma therapy: Going beyond Hercules Columns. Expert. Rev. Neurother. 2010, 10, 507–514. [Google Scholar] [CrossRef] [PubMed]
- Pham, J.; Cote, D.J.; Kang, K.; Briggs, R.G.; Gomez, D.; Prasad, A.; Daggupati, S.; Sisti, J.; Chow, F.; Attenello, F.; et al. Treatment practices and survival outcomes for IDH-wildtype glioblastoma patients according to MGMT promoter methylation status: Insights from the U.S. National Cancer Database. J. Neurooncol. 2025, 172, 655–665. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Yuan, M.; Hu, X.; Yang, Z.; Cheng, J.; Leng, H.; Zhou, Z. Identification of Recurrence-associated Gene Signatures and Machine Learning-based Prediction in IDH-Wildtype Histological Glioblastoma. J. Mol. Neurosci. 2025, 75, 48. [Google Scholar] [CrossRef] [PubMed]
- Paulsson, A.K.; McMullen, K.P.; Peiffer, A.M.; Hinson, W.H.; Kearns, W.T.; Johnson, A.J.; Lesser, G.J.; Ellis, T.L.; Tatter, S.B.; Debinski, W.; et al. Limited margins using modern radiotherapy techniques does not increase marginal failure rate of glioblastoma. Am. J. Clin. Oncol. 2014, 37, 177–181. [Google Scholar] [CrossRef]
- Minniti, G.; Tini, P.; Giraffa, M.; Capone, L.; Raza, G.; Russo, I.; Cinelli, E.; Gentile, P.; Bozzao, A.; Paolini, S.; et al. Feasibility of clinical target volume reduction for glioblastoma treated with standard chemoradiation based on patterns of failure analysis. Radiother. Oncol. 2023, 181, 109435. [Google Scholar] [CrossRef] [PubMed]
- van den Elshout, R.; Ariëns, B.; Blaauboer, J.; Meijer, F.J.A.; van der Kolk, A.G.; Esmaeili, M.; Scheenen, T.W.J.; Henssen, D.J.H.A. Quantification of perineural satellitosis in pretreatment glioblastoma with structural MRI and a diffusion tensor imaging template. Neurooncol. Adv. 2023, 6, vdad168. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Leao, D.J.; Craig, P.G.; Godoy, L.F.; Leite, C.D.C.; Policeni, B. Response Assessment in Neuro-Oncology Criteria for Gliomas: Practical Approach Using Conventional and Advanced Techniques. AJNR Am. J. Neuroradiol. 2020, 41, 10–20. [Google Scholar] [CrossRef]
- Hegi, M.E.; Diserens, A.C.; Gorlia, T.; Hamou, M.F.; de Tribolet, N.; Weller, M.; Kros, J.M.; Hainfellner, J.A.; Mason, W.; Mariani, L.; et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N. Engl. J. Med. 2005, 352, 997–1003. [Google Scholar] [CrossRef] [PubMed]
- Tini, P.; Nardone, V.; Pastina, P.; Battaglia, G.; Miracco, C.; Carbone, S.F.; Sebaste, L.; Rubino, G.; Cerase, A.; Pirtoli, L. Epidermal Growth Factor Receptor Expression Predicts Time and Patterns of Recurrence in Patients with Glioblastoma After Radiotherapy and Temozolomide. World Neurosurg. 2018, 109, e662–e668. [Google Scholar] [CrossRef] [PubMed]
- Tini, P.; Nardone, V.; Pirtoli, L. Is there a potential role for EGFR expression to lead margin reduction in glioblastoma? J. Neurooncol. 2017, 133, 203–205. [Google Scholar] [CrossRef] [PubMed]
- Le Rhun, E.; Preusser, M.; Roth, P.; Reardon, D.A.; van den Bent, M.; Wen, P.; Reifenberger, G.; Weller, M. Molecular targeted therapy of glioblastoma. Cancer Treat. Rev. 2019, 80, 101896. [Google Scholar] [CrossRef] [PubMed]
| Variable | N of Patients (%) |
|---|---|
| Total patients | 218 |
| Age, mean (SD), years | 61.8 (11.16) |
| Sex, male | 141 (64.7%) |
| Sex, female | 77 (35.3%) |
| Karnofsky Performance Status ≥80 | 193 (88.5%) |
| Karnofsky Performance Status <80 | 25 (11.5%) |
| Extent of resection: Gross Total | 62 (28.4%) |
| Extent of resection: Subtotal | 103 (47.2%) |
| Extent of resection: Biopsy | 53 (24.3) |
| EGFR amplification (positive) | 128 (58.7%) |
| EGFR amplification (negative) | 90 (41.3%) |
| MGMT methylation (positive) | 107 (49.1%) |
| MGMT methylation (negative) | 111 (50.9%) |
| Biomarker Group (EGFR/MGMT) | CR (%) | PR (%) | SD (%) | PD (%) | TOT |
|---|---|---|---|---|---|
| EGFR neg/MGMT met | 16 (32.7%) | 11 (22.4%) | 10 (20.4%) | 12 (24.5%) | 49 (100%) |
| EGFR neg/MGMT unm | 4 (10.3%) | 11 (28.2%) | 3 (7.7%) | 21 (53.8%) | 39 (100%) |
| EGFR iper/MGMT met | 11 (23.9%) | 9 (19.6%) | 10 (21.7%) | 26 (56.5%) | 46 (100%) |
| EGFR iper/MGMT unm | 8 (11.1%) | 12 (16.7%) | 9 (12.5%) | 43 (59.7%) | 72 (100%) |
| Biomarker Group (EGFR/MGMT) | In-Field Recurrence (%) | Marginal Recurrence (%) | Out-Field Recurrence (%) | TOT |
|---|---|---|---|---|
| EGFR neg/MGMT met | 30 (81.1%) | 1 (2.7%) | 6 (16.2%) | 37 (100%) |
| EGFR neg/MGMT unm | 23 (82.1%) | 2 (7.2%) | 3 (10.7%) | 28 (100%) |
| EGFR iper/MGMT met | 26 (56.5%) | 6 (13.1%) | 14 (30.4%) | 46 (100%) |
| EGFR iper/MGMT unm | 47 (71.2%) | 16 (24.2%) | 3 (4.6%) | 66 (100%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tini, P.; Donnini, F.; Marampon, F.; Vannini, M.; Carfagno, T.; Pastina, P.; Rubino, G.; Chibbaro, S.; Cerase, A.; Bagnacci, G.; et al. Biomolecular Predictors of Recurrence Patterns and Survival in IDH-Wild-Type Glioblastoma: A Retrospective Analysis of Patients Treated with Radiotherapy and Temozolomide. Brain Sci. 2025, 15, 713. https://doi.org/10.3390/brainsci15070713
Tini P, Donnini F, Marampon F, Vannini M, Carfagno T, Pastina P, Rubino G, Chibbaro S, Cerase A, Bagnacci G, et al. Biomolecular Predictors of Recurrence Patterns and Survival in IDH-Wild-Type Glioblastoma: A Retrospective Analysis of Patients Treated with Radiotherapy and Temozolomide. Brain Sciences. 2025; 15(7):713. https://doi.org/10.3390/brainsci15070713
Chicago/Turabian StyleTini, Paolo, Flavio Donnini, Francesco Marampon, Marta Vannini, Tommaso Carfagno, Pierpaolo Pastina, Giovanni Rubino, Salvatore Chibbaro, Alfonso Cerase, Giulio Bagnacci, and et al. 2025. "Biomolecular Predictors of Recurrence Patterns and Survival in IDH-Wild-Type Glioblastoma: A Retrospective Analysis of Patients Treated with Radiotherapy and Temozolomide" Brain Sciences 15, no. 7: 713. https://doi.org/10.3390/brainsci15070713
APA StyleTini, P., Donnini, F., Marampon, F., Vannini, M., Carfagno, T., Pastina, P., Rubino, G., Chibbaro, S., Cerase, A., Bagnacci, G., Perrella, A., Mazzei, M. A., Pascucci, A., D’Alonzo, V., Giacomo, A. M. D., & Minniti, G. (2025). Biomolecular Predictors of Recurrence Patterns and Survival in IDH-Wild-Type Glioblastoma: A Retrospective Analysis of Patients Treated with Radiotherapy and Temozolomide. Brain Sciences, 15(7), 713. https://doi.org/10.3390/brainsci15070713










