Hybrid Deep Learning Architecture with Adaptive Feature Fusion for Multi-Stage Alzheimer’s Disease Classification
Abstract
1. Introduction
Research Contributions
- •
- Developed a hybrid deep learning framework that optimally integrates ResNet50-based localized structural feature extraction with vision transformer (ViT)-based global connectivity modeling, significantly enhancing diagnostic precision for multi-stage Alzheimer’s disease (AD) classification.
- •
- Introduced a pivotal adaptive feature fusion layer that employs an attention mechanism to achieve robust integration of multi-scale features, yielding stage-specific representations and overcoming limitations of fragmented feature modeling.
- •
- Achieved a classification accuracy of 99.42% (precision: 99.55%; recall: 99.46%; F1-score: 99.50%) on the AD5C dataset, reducing the error rate to 0.58% and surpassing the prior benchmark of 98.24%, establishing a new standard for Alzheimer’s disease (AD) diagnostics.
- •
- Demonstrated robust generalizability through external validation on a four-class Alzheimer’s disease (AD) dataset, confirming the framework’s applicability across diverse imaging conditions and its potential for clinical integration.
2. Literature Review
2.1. Conventional Methods
2.2. Hybrid Deep Learning Methods
2.3. Emerging and Specialized Approaches
2.4. Handwriting Analysis for Alzheimer’s Disease (AD)
| Author(s) | Model Used | Methodology | Accuracy | Focus Area | Limitations |
|---|---|---|---|---|---|
| Gurrala et al. (2024) [16] | CNN | Web-based CNN for AD staging | 94.50% | Staging classification | Limited to CNN feature extraction |
| Arjaria et al. (2024) [14] | Digital platform | Cognitive, physiological monitoring | Not available | Progression tracking | Multi-modal data dependency |
| Bhattarai et al. (2024) [48] | Deep-SHAP | Explainable AI for biomarker-cognition mapping | Not available | Neuroimaging biomarkers | Requires robust validation for clinical use |
| Alatrany et al. (2024) [49] | ML algorithms | Explainable ML for AD classification | 89.20% | AD classification | Limited to explainable models |
| Zhou et al. (2024) [38] | Game app | Cognitive tests via app | Not available | Cognitive decline detection | Non-MRI specificity |
| Peng et al. (2024) [41] | SF-GCL | Stage-specific brain pattern analysis | 92.10% | Brain pattern analysis | Requires further validation |
| Anjali et al. (2024) [42] | STCNN | SMOTE-TOMEK for imbalance | 93.80% | Imbalanced classification | Limited to imbalanced data |
| Talha et al. (2024) [50] | DL models | Performance evaluation of DL models | 90.50% | AD detection | Broad evaluation lacks specificity |
| Bharath et al. (2024) [51] | ML algorithms | Predicting AD progression | 88.70% | Disease progression | Limited to ML approaches |
| Givian et al. (2025) [52] | ML algorithms | MRI analysis with ML | 91.30% | Early diagnosis | Limited generalizability |
| Alahmed et al. (2025) [53] | AlzONet | Optimized DL framework | 95.60% | Multi-class diagnosis | Requires high computational resources |
| Tenchov et al. (2024) [8] | Not specified | Exploring cognitive decline | Not available | Cognitive decline | Broad focus lacks specific metrics |
| Bortty et al. (2025) [54] | ViT-B16, CNNs | Weighted ensemble with GOA | 97.31% | Multi-class classification | Computational intensity |
| Fujita et al. (2024) [55] | Not specified | Brain volume changes analysis | Not available | Normal cognition | Limited to normal cognition focus |
3. Materials and Methods
3.1. Dataset and Preprocessing
3.2. Augmentation and Summary
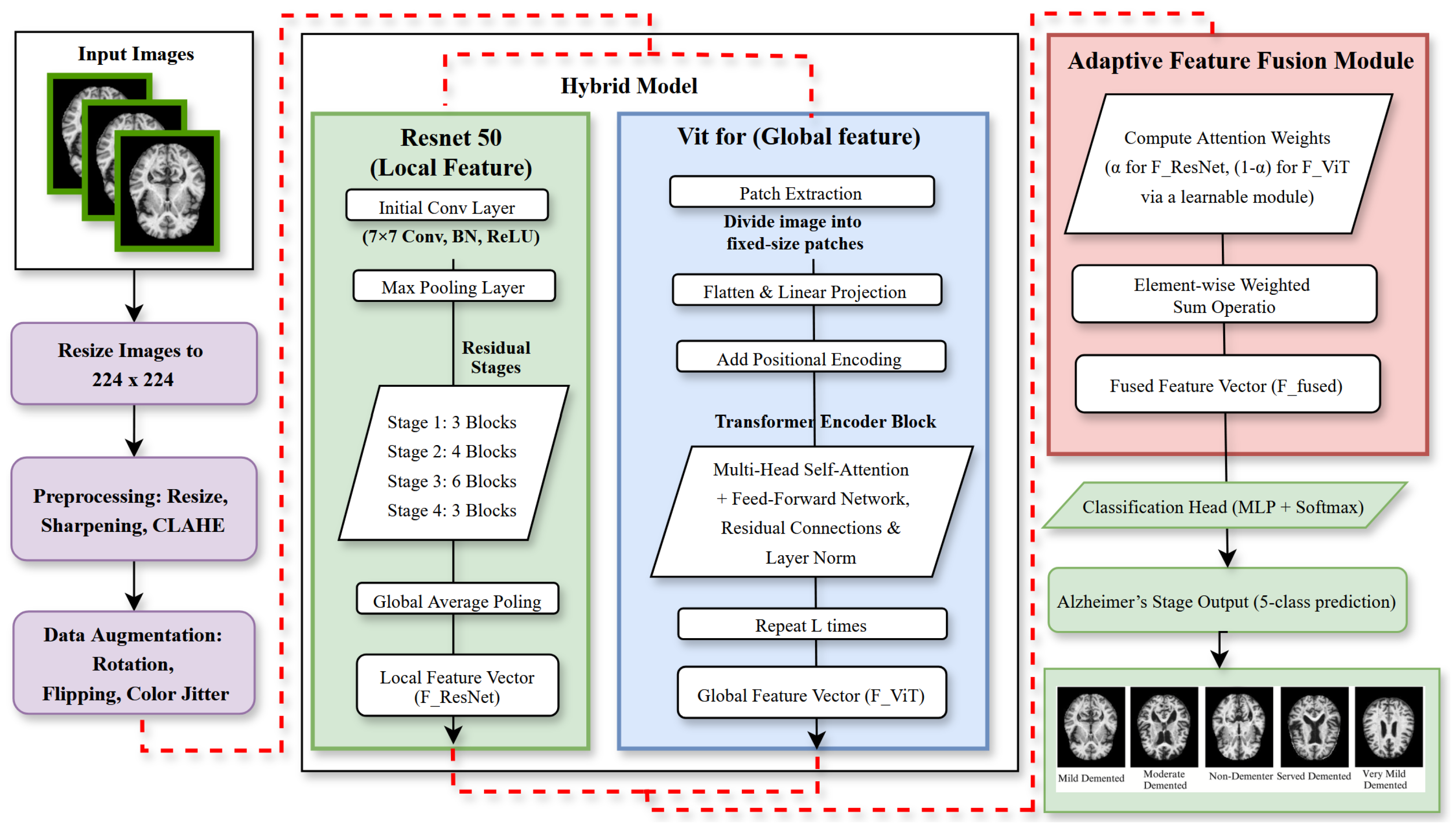
3.3. Model Architecture
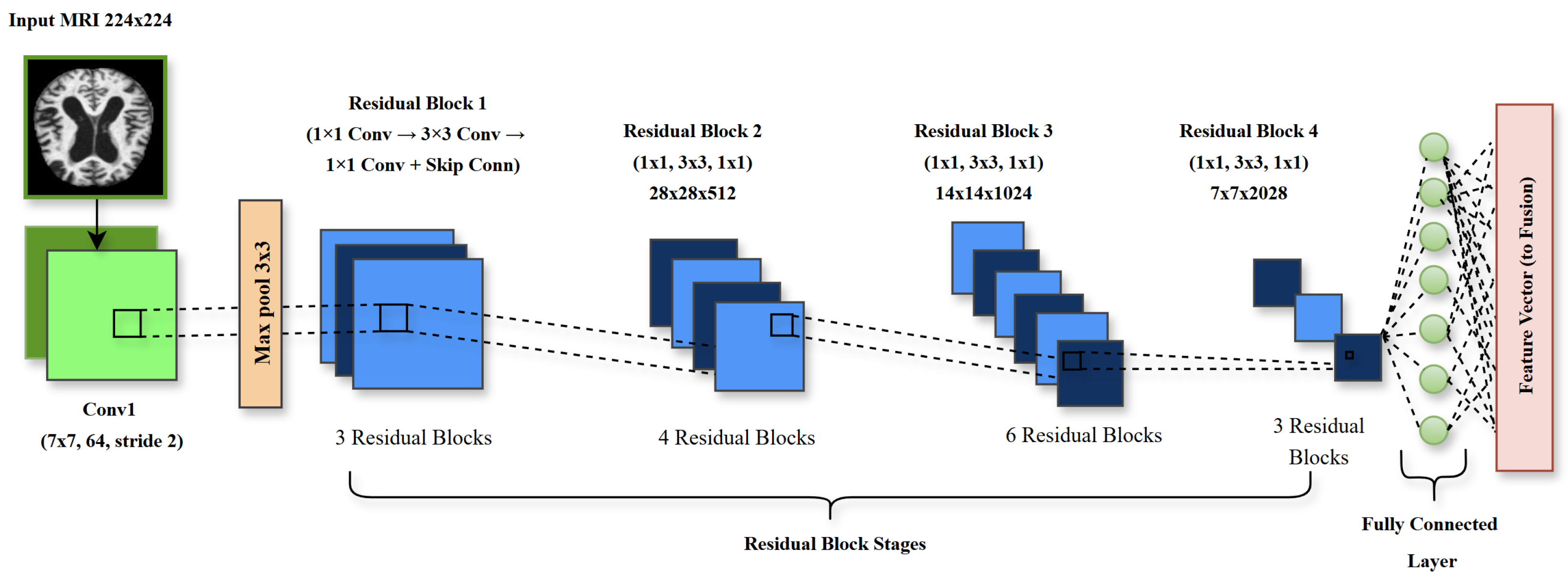
3.3.1. ResNet50 for Local Feature Extraction
3.3.2. Vision Transformer for Global Feature Extraction
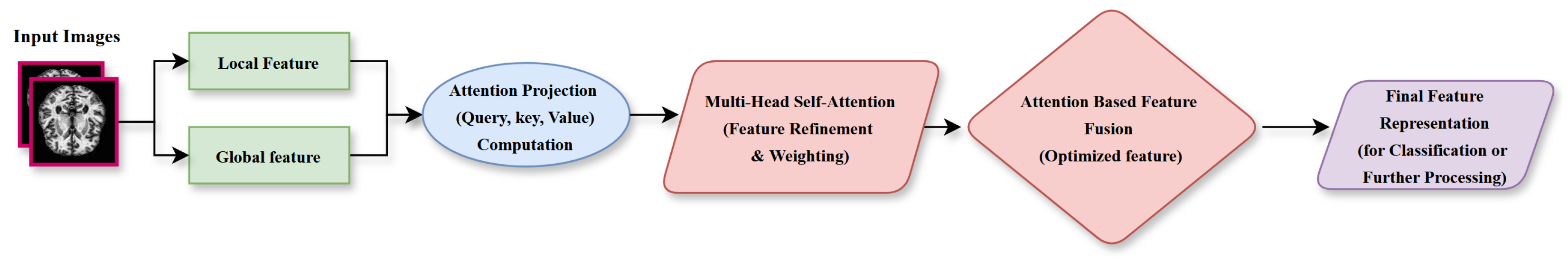
3.3.3. Adaptive Feature Fusion Layer
3.4. Proposed Algorithm for Alzheimer’s Disease (AD) Classification
| Algorithm 1 Deep Learning Framework Training and Testing Steps |
|
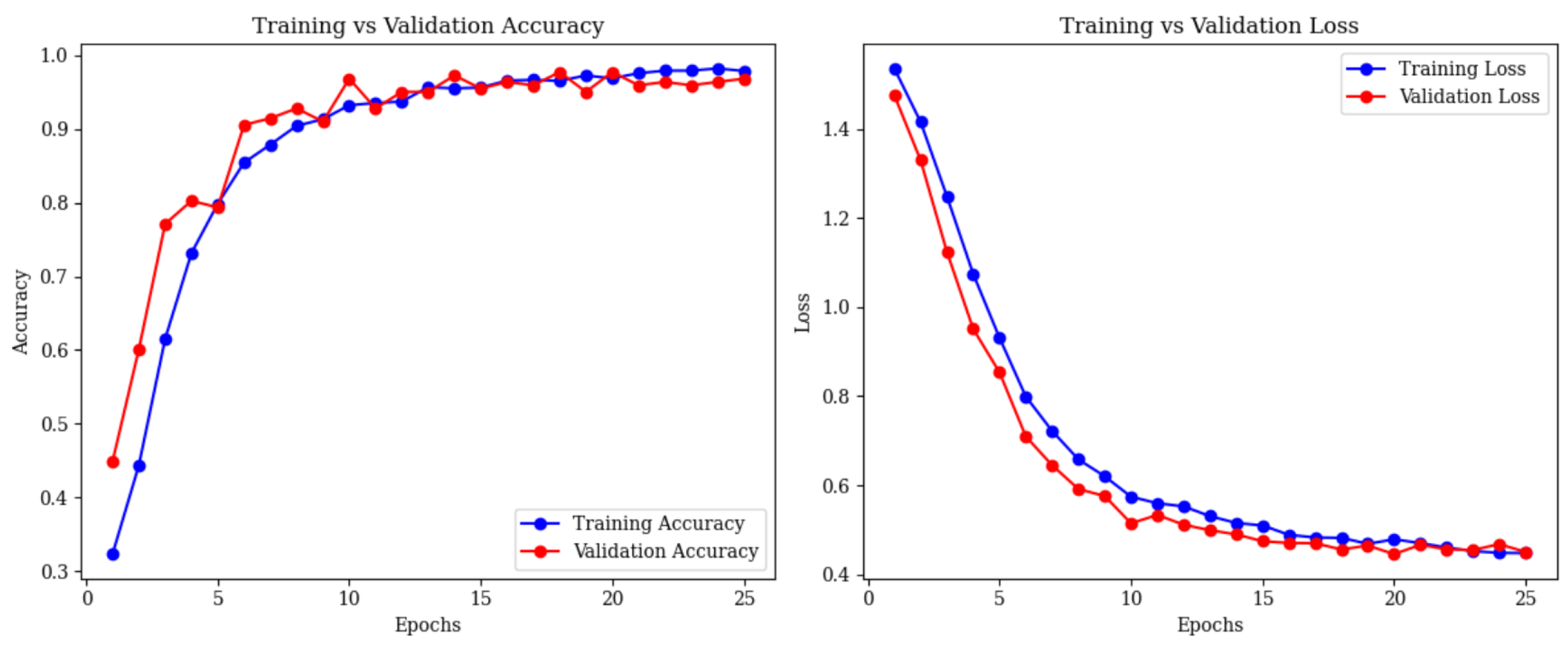
4. Experimental Results
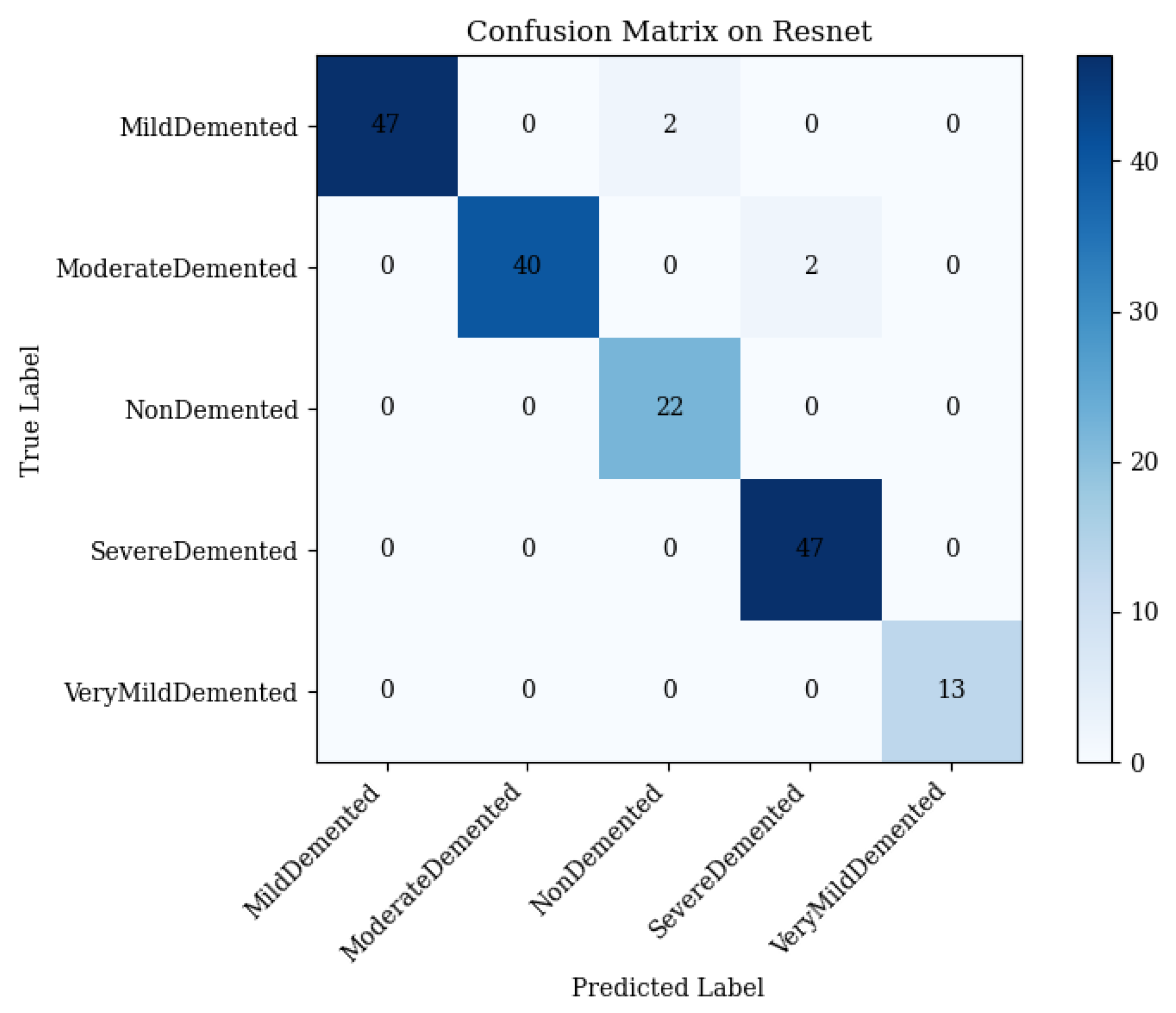
4.1. ResNet50 Performance
- •
- Mild Demented: 47 correct; 2 misclassified as Non-Demented.
- •
- Moderate Demented: 40 correct; 2 misclassified as Severe Demented.
- •
- Non-Demented: 22 correct; 0 misclassified.
- •
- Severe Demented: 47 correct; 0 misclassified.
- •
- Very Mild Demented: 13 correct; 0 misclassified.
4.2. Vision Transformer Performance
- •
- Mild Demented: 47 correct; 2 misclassified as Non-Demented.
- •
- Moderate Demented: 40 correct; 2 misclassified as Severe Demented.
- •
- Non-Demented: 21 correct; 1 misclassified as Mild Demented.
- •
- Severe Demented: 47 correct; 0 misclassified.
- •
- Very Mild Demented: 13 correct; 0 misclassified.
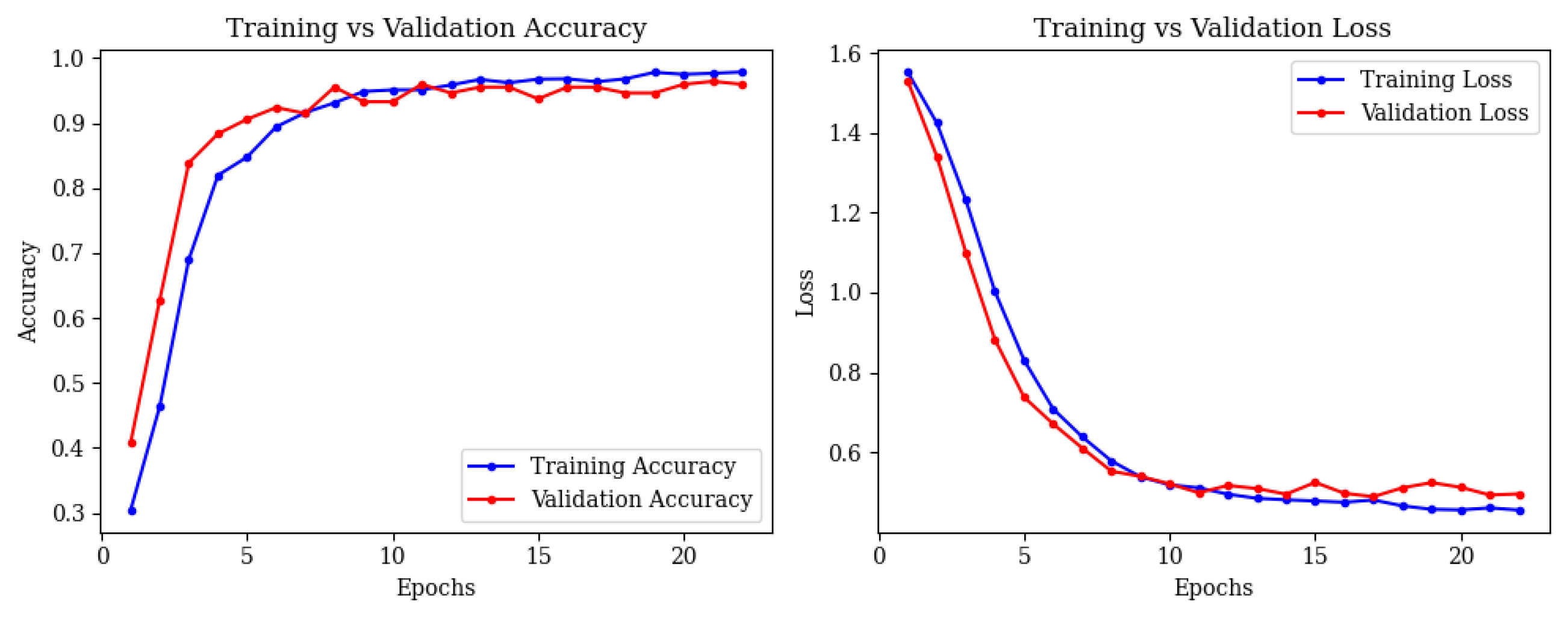
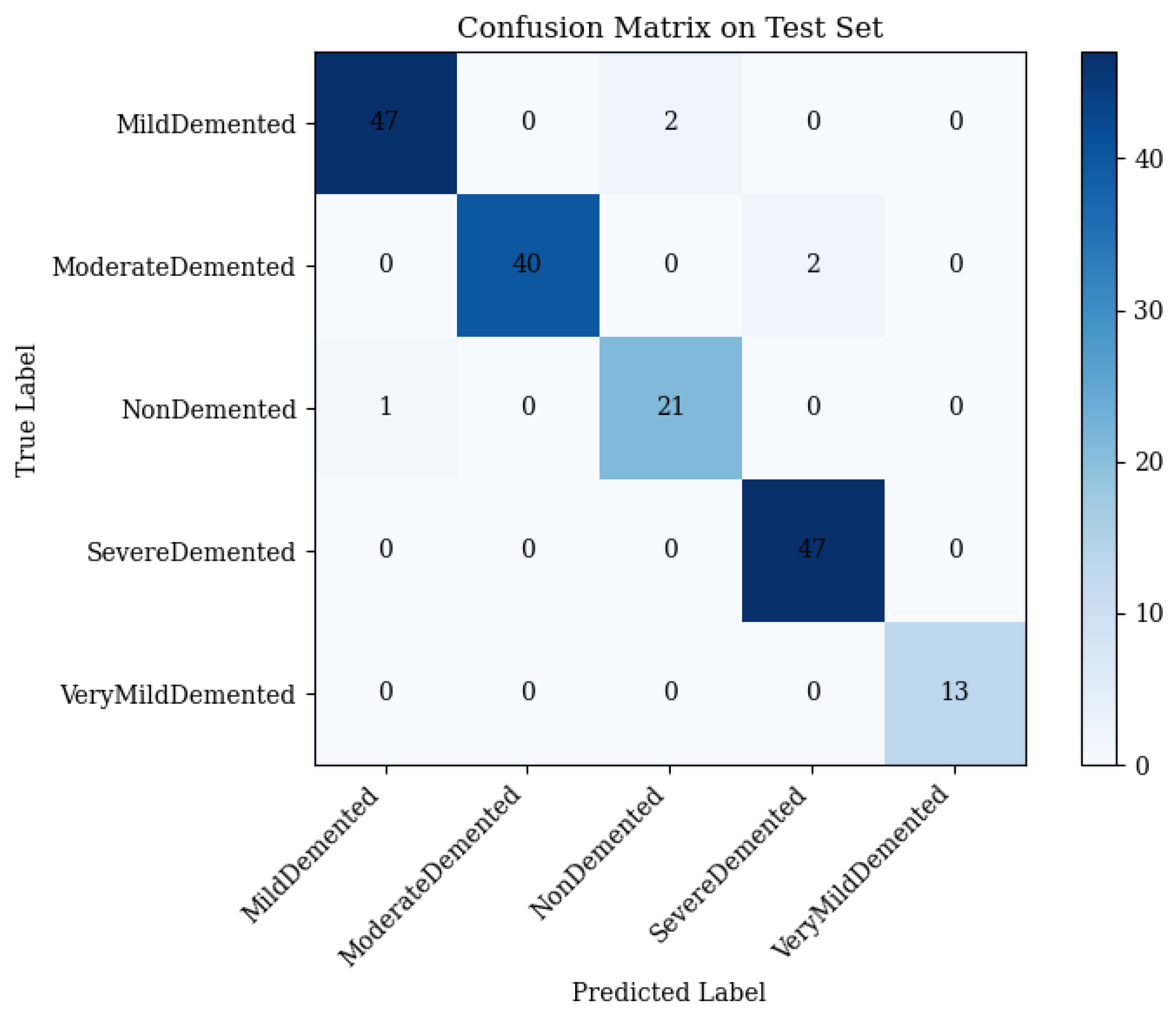
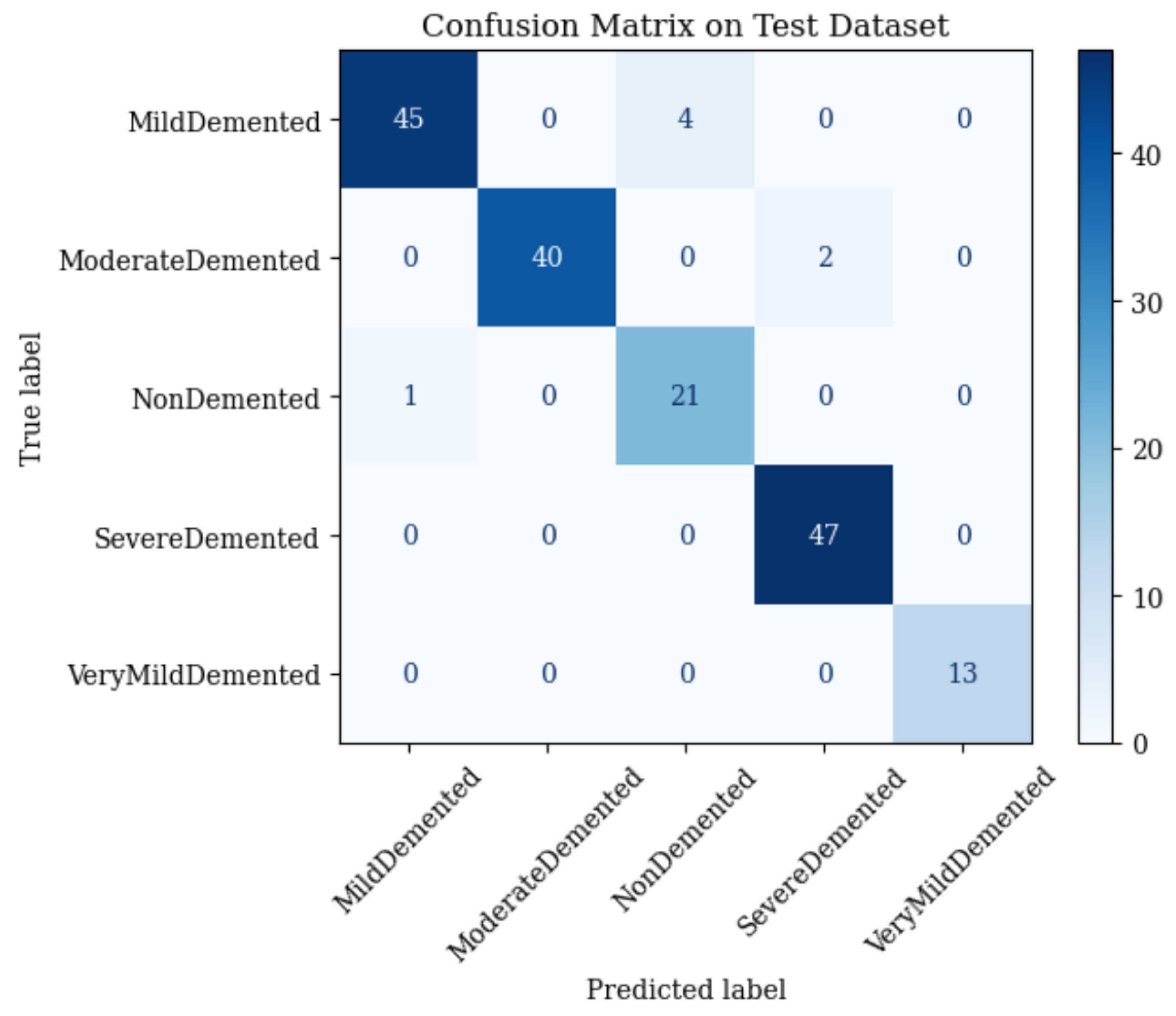
4.3. Combined ResNet50 and Vision Transformer (ViT)
- •
- Mild Demented: 45 correct; 4 misclassified as Non-Demented.
- •
- Moderate Demented: 40 correct; 2 misclassified as Severe Demented.
- •
- Non-Demented: 21 correct; 1 misclassified as Mild Demented.
- •
- Severe Demented: 47 correct; 0 misclassified.
- •
- Very Mild Demented: 13 correct; 0 misclassified.
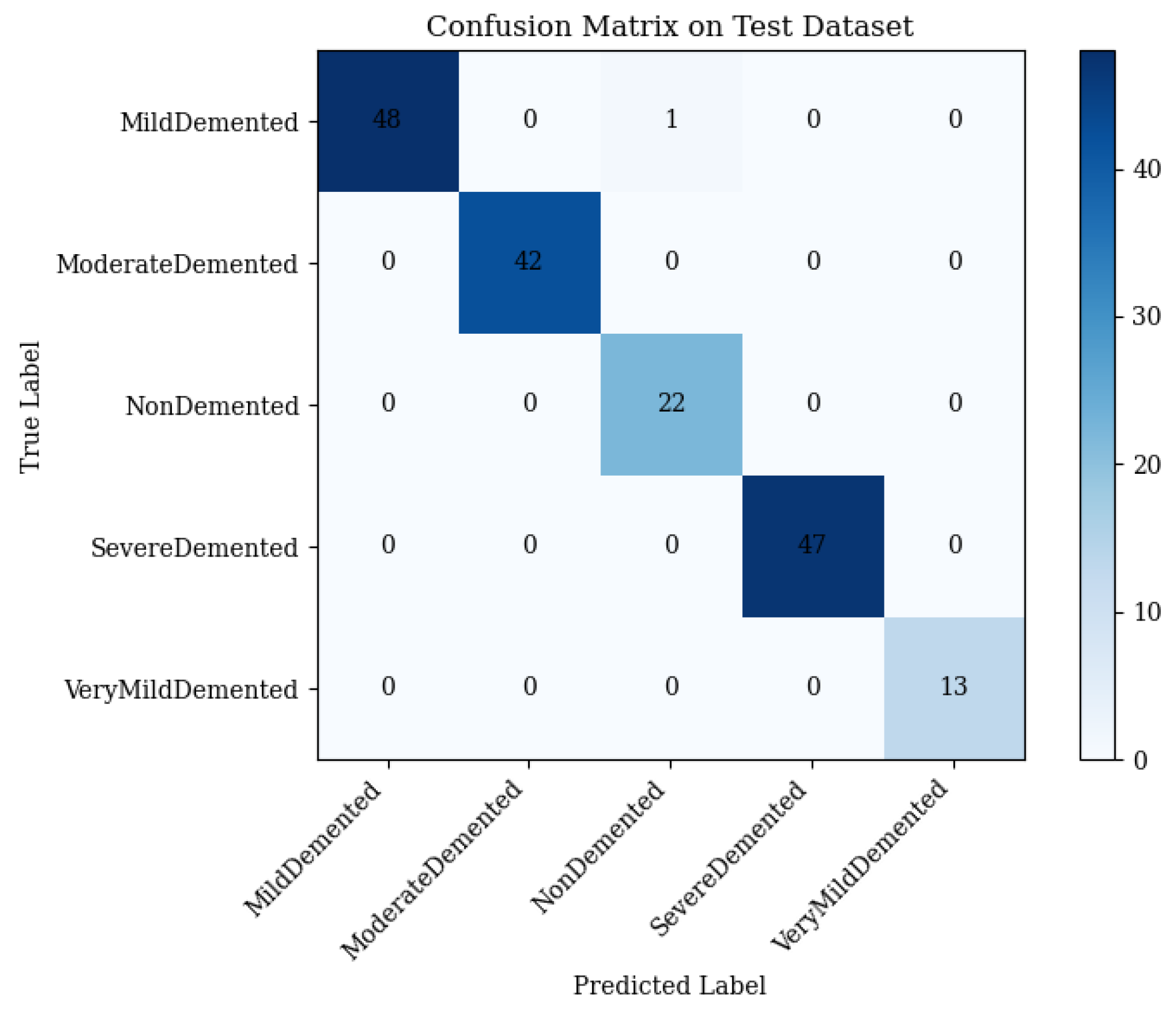
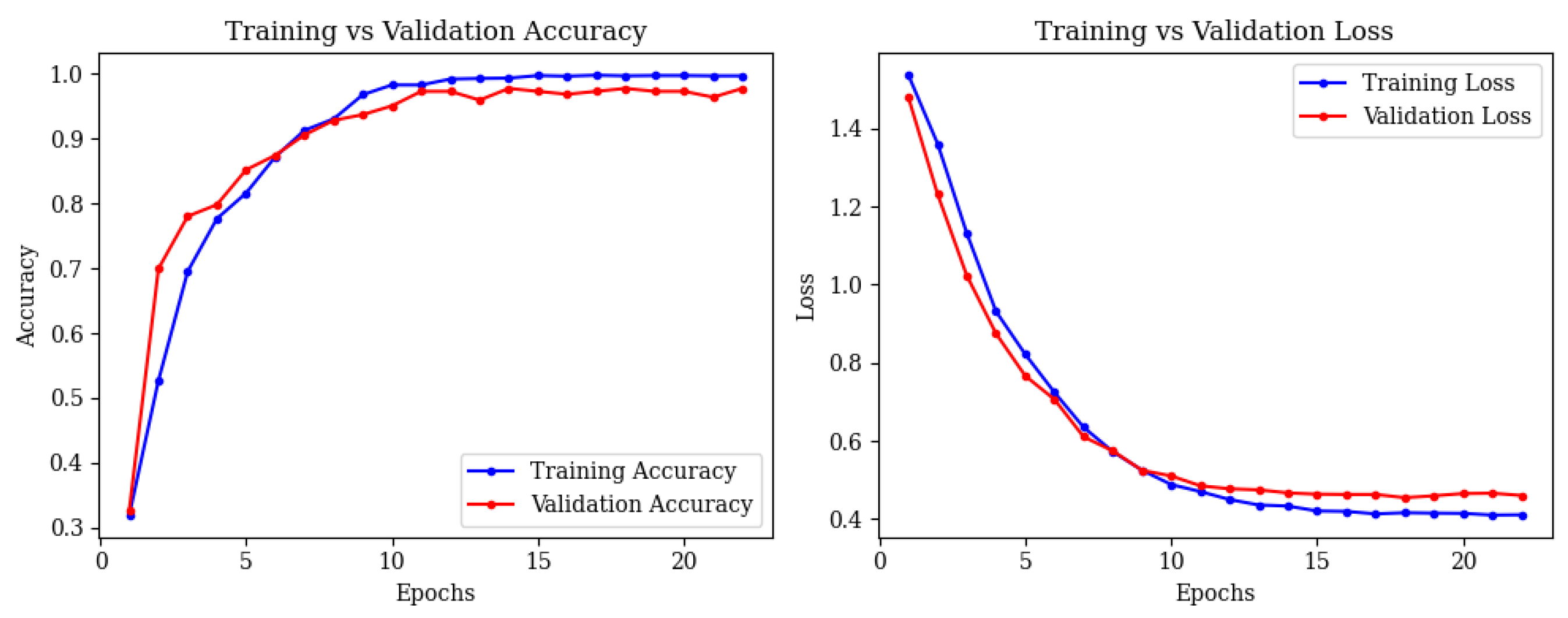
4.4. Full Framework with Adaptive Feature Fusion
- •
- Mild Demented: 48 correct; 1 misclassified as Non-Demented.
- •
- Moderate Demented: 42 correct; 0 misclassified.
- •
- Non-Demented: 22 correct; 0 misclassified.
- •
- Severe Demented: 47 correct; 0 misclassified.
- •
- Very Mild Demented: 13 correct; 0 misclassified.
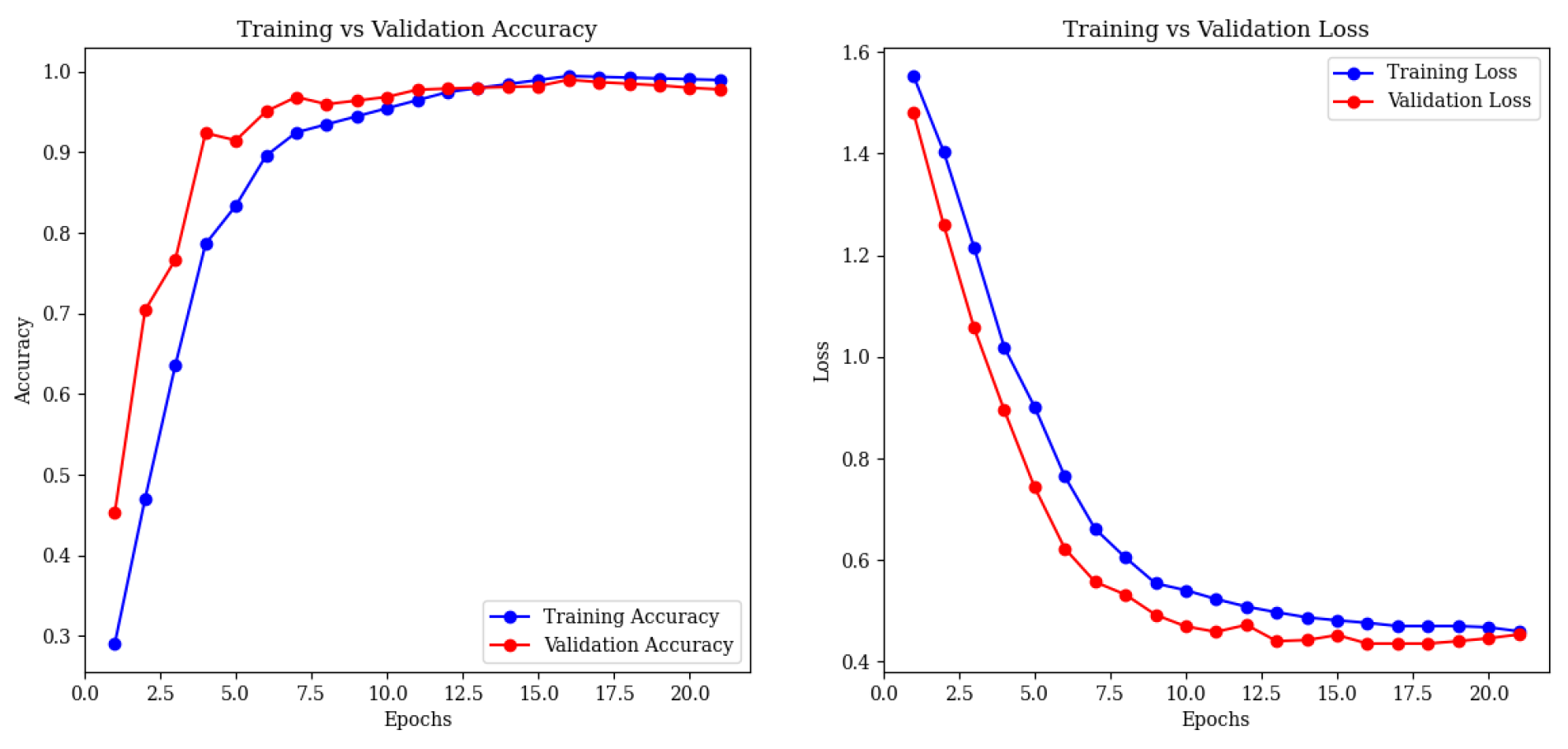
4.5. Error Analysis
4.6. Component Ablation
Analysis of Component Synergies
4.7. Classical Machine Learning Baselines
4.8. External Dataset Validation
5. Comparison with State-of-the-Art Methods
6. Discussion
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AD | Alzheimer’s Disease |
| AD5C | Alzheimer’s 5-Class Dataset |
| CNN | Convolutional Neural Network |
References
- Gustavsson, A.; Norton, N.; Fast, T.; Frölich, L.; Georges, J.; Holzapfel, D.; Kirabali, T.; Krolak-Salmon, P.; Rossini, P.M.; Ferretti, M.T.; et al. Global estimates on the number of persons across the Alzheimer’s disease continuum. Alzheimers Dement. 2023, 19, 658–670. [Google Scholar] [CrossRef] [PubMed]
- Tahami Monfared, A.A.; Byrnes, M.J.; White, L.A.; Zhang, Q. Alzheimer’s Disease: Epidemiology and Clinical Progression. Neurol. Ther. 2022, 11, 553–569. [Google Scholar] [CrossRef] [PubMed]
- Wilson, P.; Clark, E.; Harris, J. Brain Connectivity Disruptions in Alzheimer’s Disease. Brain Sci. 2024, 14, 890. [Google Scholar] [CrossRef]
- Sehar, U.; Rawat, P.; Reddy, A.P.; Kopel, J.; Reddy, P.H. Amyloid Beta in Aging and Alzheimer’s Disease. Int. J. Mol. Sci. 2022, 23, 12924. [Google Scholar] [CrossRef]
- Al-Shoukry, S.; Rassem, T.H.; Makbol, N.M. Alzheimer’s diseases detection by using deep learning algorithms: A mini-review. IEEE Access 2020, 8, 77131–77141. [Google Scholar] [CrossRef]
- Rahman, M.M.; Lendel, C. Extracellular protein components of amyloid plaques and their roles in Alzheimer’s disease pathology. Mol. Neurodegener. 2021, 16, 59. [Google Scholar] [CrossRef]
- Zhang, H.; Wei, W.; Zhao, M.; Ma, L.; Jiang, X.; Pei, H.; Cao, Y.; Li, H. Interaction between Aβ and tau in the pathogenesis of Alzheimer’s disease. Int. J. Biol. Sci. 2021, 17, 2181–2192. [Google Scholar] [CrossRef]
- Tenchov, R.; Sasso, J.M.; Zhou, Q.A. Alzheimer’s Disease: Exploring the Landscape of Cognitive Decline. ACS Chem. Neurosci. 2024, 15, 3800–3827. [Google Scholar] [CrossRef]
- Pradhan, A.; Mishra, D.; Mishra, K.; Panda, S. Detection of Alzheimer’s Disease in MRI Images Using Deep Learning. Int. J. Eng. Res. Technol. 2021, 10, 580–585. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar] [CrossRef]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An Image is Worth 16x16 Words: Transformers for Image Recognition at Scale. arXiv 2021, arXiv:2010.11929. [Google Scholar] [CrossRef]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, L.; Polosukhin, I. Attention is All You Need. arXiv 2017, arXiv:1706.03762. [Google Scholar] [CrossRef]
- Zia-ur-Rehman; Awang, M.K.; Rashid, J.; Ali, G.; Hamid, M.; Mahmoud, S.F.; Saleh, D.I.; Ahmad, H.I. Classification of Alzheimer Disease Using DenseNet-201 Based on Deep Transfer Learning Technique. PLoS ONE 2024, 19, e0297858. [Google Scholar] [CrossRef]
- Arjaria, S.K.; Rathore, A.S.; Bisen, D.; Bhattacharyya, S. Performances of Machine Learning Models for Diagnosis of Alzheimer’s Disease. Ann. Data Sci. 2024, 11, 307–335. [Google Scholar] [CrossRef]
- Alshammari, M.; Mezher, M. A Modified Convolutional Neural Networks for MRI-based Images for Detection and Stage Classification of Alzheimer Disease. In Proceedings of the 2021 National Computing Colleges Conference (NCCC), Taif, Saudi Arabia, 27–28 March 2021. [Google Scholar] [CrossRef]
- Gurrala, V.K.; Talasila, S.; Medikonda, N.R.; Challa, S.; Sohail, S.; Siddiq, M.A.B. A Web-Based Interface for Comprehensive Staging Classification of Alzheimer’s Disease Diagnosis through Convolutional Neural Networks. In Proceedings of the 2024 5th International Conference for Emerging Technology (INCET), Belgaum, India, 24–26 May 2024; pp. 1–5. [Google Scholar] [CrossRef]
- Kumar, S.; Singh, N.P.; Brahma, B. AI-Based Model for Detection and Classification of Alzheimer Disease. In Proceedings of the 2023 IEEE International Conference on Computer Vision and Machine Intelligence (CVMI), Gwalior, India, 10–11 December 2023; pp. 1–6. [Google Scholar] [CrossRef]
- Archana, B.; Kalirajan, K. Alzheimer’s Disease Classification using Convolutional Neural Networks. In Proceedings of the 2023 International Conference on Innovative Data Communication Technologies and Application (ICIDCA), Uttarakhand, India, 14–16 March 2023; pp. 1044–1048. [Google Scholar] [CrossRef]
- Prabha, C. Classification and Detection of Alzheimer’s Disease: A Brief Analysis. In Proceedings of the 2023 International Conference on Self Sustainable Artificial Intelligence Systems (ICSSAS), Erode, India, 18–20 October 2023; pp. 777–782. [Google Scholar] [CrossRef]
- Das, R.; Kalita, S. Classification of Alzheimer’s Disease Stages Through Volumetric Analysis of MRI Data. In Proceedings of the 2022 IEEE Calcutta Conference (CALCON), Kolkata, India, 10–11 December 2022; pp. 165–169. [Google Scholar] [CrossRef]
- Kayalvizhi, M.; Senthil Kumar, G.; Tushal, V.; Yashvanth, M.; Santhosh, G. Deep Learning-Based Severity Detection in Alzheimer’s Disease: A Comprehensive Study on Cognitive Impairment. In Proceedings of the 2023 International Conference on Data Science, Agents & Artificial Intelligence (ICDSAAI), Chennai, India, 21–23 December 2023; pp. 1–6. [Google Scholar] [CrossRef]
- Jansi, R.; Gowtham, N.; Ramachandran, S.; Sai Praneeth, V. Revolutionizing Alzheimer’s Disease Prediction using InceptionV3 in Deep Learning. In Proceedings of the 2023 7th International Conference on Electronics, Communication and Aerospace Technology (ICECA), Coimbatore, India, 22–24 November 2023; pp. 1155–1160. [Google Scholar] [CrossRef]
- Sushmitha, S.; Chitrakala, S.; Bharathi, U. sMRI Classification of Alzheimer’s Disease Using Genetic Algorithm and Multi-Instance Learning (GA+MIL). In Proceedings of the 2023 International Conference on Electrical, Electronics, Communication and Computers (ELEXCOM), Roorkee, India, 26–27 August 2023; pp. 1–4. [Google Scholar] [CrossRef]
- Qu, Z.; Yao, T.; Liu, X.; Wang, G. A Graph Convolutional Network Based on Univariate Neurodegeneration Biomarker for Alzheimer’s Disease Diagnosis. IEEE J. Transl. Eng. Health Med. 2023, 11, 405–416. [Google Scholar] [CrossRef]
- Tushar; Patel, R.K.; Aggarwal, E.; Solanki, K.; Dahiya, O.; Yadav, S.A. A Logistic Regression and Decision Tree Based Hybrid Approach to Predict Alzheimer’s Disease. In Proceedings of the 2023 International Conference on Computational Intelligence and Sustainable Engineering Solutions (CISES), Greater Noida, India, 28–30 April 2023; pp. 722–726. [Google Scholar] [CrossRef]
- Liu, B. Alzheimer’s disease classification using hippocampus and improved DenseNet. In Proceedings of the 2023 International Conference on Image Processing, Computer Vision and Machine Learning (ICICML), Chengdu, China, 3–5 November 2023; pp. 451–454. [Google Scholar] [CrossRef]
- Sanjeev Kumar, K.; Reddy, B.S.; Ravichandran, M. Alzheimer’s Disease Detection Using Transfer Learning: Performance Analysis of InceptionResNetV2 and ResNet50 Models. In Proceedings of the 2023 Seventh International Conference on Image Information Processing (ICIIP), Solan, India, 22–24 November 2023; pp. 832–837. [Google Scholar] [CrossRef]
- Lu, P.; Tan, Y.; Xing, Y.; Liang, Q.; Yan, X.; Zhang, G. An Alzheimer’s disease classification method based on ConvNeXt. In Proceedings of the 2023 3rd International Symposium on Computer Technology and Information Science (ISCTIS), Chengdu, China, 7–9 July 2023; pp. 884–888. [Google Scholar] [CrossRef]
- Neetha, P.U.; Simran, S.; Sunilkumar, G.; Pushpa, C.N.; Thriveni, J.; Venugopal, K.R. Borderline-DEMNET for Multi-Class Alzheimer’s Disease Classification. In Proceedings of the 2023 IEEE 5th International Conference on Cybernetics, Cognition and Machine Learning Applications (ICCCMLA), Hamburg, Germany, 7–8 October 2023; pp. 192–197. [Google Scholar] [CrossRef]
- Tripathy, S.K.; Singh, D.; Jaiswal, A. Multi-Layer Feature Fusion-based Deep Multi-layer Depth Separable Convolution Neural Network for Alzheimer’s Disease Detection. In Proceedings of the 2023 International Conference on IoT, Communication and Automation Technology (ICICAT), Gorakhpur, India, 23–24 June 2023; pp. 1–5. [Google Scholar] [CrossRef]
- Yin, Y.; Jin, W.; Bai, J.; Liu, R.; Zhen, H. SMIL-DeiT: Multiple Instance Learning and Self-supervised Vision Transformer network for Early Alzheimer’s disease classification. In Proceedings of the 2022 International Joint Conference on Neural Networks (IJCNN), Padua, Italy, 18–23 July 2022; pp. 1–6. [Google Scholar] [CrossRef]
- Bushra, U.H.; Priya, F.C.; Patwary, M.J.A. Fuzziness-Based Semi-Supervised Learning for Early Detection of Alzheimer’s Disease using MRI data. In Proceedings of the 2023 26th International Conference on Computer and Information Technology (ICCIT), Cox’s Bazar, Bangladesh, 13–15 December 2023; pp. 1–6. [Google Scholar] [CrossRef]
- Thatere, A.; Verma, P.; Reddy, K.T.V.; Umate, L. A Short Survey on Alzheimer’s Disease: Recent Diagnosis and Obstacles. In Proceedings of the 2023 1st DMIHER International Conference on Artificial Intelligence in Education and Industry 4.0 (IDICAIEI), Wardha, India, 27–28 November 2023; pp. 1–6. [Google Scholar] [CrossRef]
- Alatrany, A.S.; Hussain, A.; Alatrany, S.S.J.; Mustafina, J.; Al-Jumeily, D. Comparison of Machine Learning Algorithms for classification of Late Onset Alzheimer’s disease. In Proceedings of the 2023 15th International Conference on Developments in eSystems Engineering (DeSE), Baghdad & Anbar, Iraq, 9–12 January 2023; pp. 60–64. [Google Scholar] [CrossRef]
- Bhargavi, M.S.; Prabhakar, B. Deep Learning Approaches for Early Detection of Alzheimer’s Disease using MRI Neuroimaging. In Proceedings of the 2022 International Conference on Connected Systems & Intelligence (CSI), Trivandrum, India, 31 August–2 September 2022; pp. 1–6. [Google Scholar] [CrossRef]
- Pallawi, S.; Singh, D.K. Detection of Alzheimer’s Disease Stages Using Pre-Trained Deep Learning Approaches. In Proceedings of the 2023 IEEE 5th International Conference on Cybernetics, Cognition and Machine Learning Applications (ICCCMLA), Hamburg, Germany, 7–8 October 2023; pp. 252–256. [Google Scholar] [CrossRef]
- Islam, J.; Furqon, E.N.; Farady, I.; Lung, C.W.; Lin, C.Y. Early Alzheimer’s Disease Detection Through YOLO-Based Detection of Hippocampus Region in MRI Images. In Proceedings of the 2023 Sixth International Symposium on Computer, Consumer and Control (IS3C), Taichung, Taiwan, 30 June–3 July 2023; pp. 32–35. [Google Scholar] [CrossRef]
- Zhou, Y.; Gao, C.; Zhang, X.; Zhang, W.; Wan, S.; Liu, Y. Early Detection and Intervention of Alzheimer’s disease Based on Game APP. In Proceedings of the 2024 5th International Conference on Information Science, Parallel and Distributed Systems (ISPDS), Guangzhou, China, 31 May–2 June 2024; Volume 2017, pp. 182–188. [Google Scholar] [CrossRef]
- Jiang, Y.; Yu, Z.; Yin, X.; Guo, H. Early Diagnosis and Progression of Alzheimer’s Disease Based on Long Short-Term Memory Model. In Proceedings of the 2023 5th International Conference on Robotics, Intelligent Control and Artificial Intelligence (RICAI), Hangzhou, China, 1–3 December 2023; pp. 620–624. [Google Scholar] [CrossRef]
- Subha, R.; Nayana, B.R.; Selvadas, M. Hybrid Machine Learning Model Using Particle Swarm Optimization for Effectual Diagnosis of Alzheimer’s Disease from Handwriting. In Proceedings of the 2022 4th International Conference on Circuits, Control, Communication and Computing (I4C), Bangalore, India, 21–23 December 2022; pp. 491–495. [Google Scholar] [CrossRef]
- Peng, C.; Liu, M.; Meng, C.; Xue, S.; Keogh, K.; Xia, F. Stage-aware Brain Graph Learning for Alzheimer’s Disease. In Proceedings of the 2024 IEEE Conference on Artificial Intelligence (CAI), Singapore, 25–27 June 2024; pp. 1346–1349. [Google Scholar] [CrossRef]
- Anjali; Singh, D.; Pandey, O.J.; Dai, H.N. STCNN: Combining SMOTE-TOMEK with CNN for Imbalanced Classification of Alzheimer’s Disease. IEEE Sens. Lett. 2024, 8, 6002104. [Google Scholar] [CrossRef]
- Impedovo, D.; Pirlo, G.; Vessio, G.; Angelillo, M.T. A Handwriting-Based Protocol for Assessing Neurodegenerative Dementia. Cogn. Comput. 2019, 11, 576–586. [Google Scholar] [CrossRef]
- Impedovo, D.; Pirlo, G. Dynamic Handwriting Analysis for the Assessment of Neurodegenerative Diseases: A Pattern Recognition Perspective. IEEE Rev. Biomed. Eng. 2019, 12, 209–220. [Google Scholar] [CrossRef]
- Vessio, G. Dynamic Handwriting Analysis for Neurodegenerative Disease Assessment: A Literary Review. Appl. Sci. 2019, 9, 4666. [Google Scholar] [CrossRef]
- D’Alessandro, T.; De Stefano, C.; Fontanella, F.; Nardone, E.; Scotto di Freca, A. Feature Evaluation in Handwriting Analysis for Alzheimer’s Disease Using Bayesian Network. In Graphonomics in Human Body Movement—Bridging Research and Practice from Motor Control to Handwriting Analysis and Recognition, Proceedings of the 21st International Conference of the International Graphonomics Society, IGS 2023, Évora, Portugal, 16–19 October 2023; Springer: Cham, Switzerland, 2023; pp. 122–135. [Google Scholar] [CrossRef]
- D’Alessandro, T.; De Stefano, C.; Fontanella, F.; Nardone, E.; Pace, C.D. From Handwriting Analysis to Alzheimer’s Disease Prediction: An Experimental Comparison of Classifier Combination Methods. In Document Analysis and Recognition—ICDAR 2024, Proceedings of the 18th International Conference, Athens, Greece, 30 August–4 September 2024; Part II; Springer: Cham, Switzerland, 2024; pp. 334–351. [Google Scholar] [CrossRef]
- Bhattarai, P.; Thakuri, D.S.; Nie, Y.; Chand, G.B. Explainable AI-based Deep-SHAP for mapping the multivariate relationships between regional neuroimaging biomarkers and cognition. Eur. J. Radiol. 2024, 174, 111403. [Google Scholar] [CrossRef]
- Alatrany, A.S.; Khan, W.; Hussain, A.; Kolivand, H.; Al-Jumeily, D. An explainable machine learning approach for Alzheimer’s disease classification. Sci. Rep. 2024, 14, 2637. [Google Scholar] [CrossRef]
- Talha, A.; Dhanasree, C.; Divya, E.; Prabhas, K.S.; Syed Abudhagir, U. Performance Evaluation of Deep Learning Models for Alzheimer’s Disease Detection. In Proceedings of the 2024 10th International Conference on Communication and Signal Processing (ICCSP), Melmaruvathur, India, 12–14 April 2024; pp. 317–322. [Google Scholar] [CrossRef]
- Bharath, M.; Gowtham, S.; Vedanth, S.; Kodipalli, A.; Rao, T.; Rohini, B.R. Predicting Alzheimer’s Disease Progression through Machine Learning Algorithms. In Proceedings of the 2023 International Conference on Recent Advances in Science and Engineering Technology (ICRASET), B G Nagara, India, 23–24 November 2023; pp. 1–5. [Google Scholar] [CrossRef]
- Givian, H.; Calbimonte, J.P. Early diagnosis of Alzheimer’s disease and mild cognitive impairment using MRI analysis and machine learning algorithms. Discov. Appl. Sci. 2025, 7, 27. [Google Scholar] [CrossRef]
- Alahmed, H.; Al-Suhail, G. AlzONet: A deep learning optimized framework for multiclass Alzheimer’s disease diagnosis using MRI brain imaging. J. Supercomput. 2025, 81, 1234–1245. [Google Scholar] [CrossRef]
- Bortty, J.C.; Chakraborty, G.S.; Noman, I.R.; Batra, S.; Das, J.; Bishnu, K.K.; Tarafder, M.T.R.; Islam, A. A Novel Diagnostic Framework with an Optimized Ensemble of Vision Transformers and Convolutional Neural Networks for Enhanced Alzheimer’s Disease Detection in Medical Imaging. Diagnostics 2025, 15, 789. [Google Scholar] [CrossRef]
- Fujita, S.; Mori, S.; Onda, K.; Hanaoka, S.; Nomura, Y.; Nakao, T.; Yoshikawa, T.; Takao, H.; Hayashi, N.; Abe, O. Characterization of Brain Volume Changes in Aging Individuals With Normal Cognition Using Serial Magnetic Resonance Imaging. JAMA Netw. Open 2023, 6, e2318153. [Google Scholar] [CrossRef] [PubMed]
- Breijyeh, Z.; Karaman, R. Comprehensive Review on Alzheimer’s Disease: Causes and Treatment. Molecules 2020, 25, 5789. [Google Scholar] [CrossRef]
- Bai, W.; Chen, P.; Cai, H.; Zhang, Q.; Su, Z.; Cheung, T.; Jackson, T.; Sha, S.; Xiang, Y.T. Worldwide prevalence of mild cognitive impairment among community dwellers aged 50 years and older: A meta-analysis and systematic review of epidemiology studies. Age Ageing 2022, 51, afac173. [Google Scholar] [CrossRef]
- Zhang, Y.; Wu, K.M.; Yang, L.; Dong, Q.; Yu, J.T. Tauopathies: New perspectives and challenges. Mol. Neurodegener. 2022, 17, 28. [Google Scholar] [CrossRef] [PubMed]
- Bezprozvanny, I. Alzheimer’s disease—Where do we go from here? Biochem. Biophys. Res. Commun. 2022, 633, 72–76. [Google Scholar] [CrossRef]
- Rajeev, V.; Fann, D.Y.; Dinh, Q.N.; Kim, H.A.; De Silva, T.M.; Lai, M.K.; Chen, C.L.H.; Drummond, G.R.; Sobey, C.G.; Arumugam, T.V. Pathophysiology of blood brain barrier dysfunction during chronic cerebral hypoperfusion in vascular cognitive impairment. Theranostics 2022, 12, 1639–1658. [Google Scholar] [CrossRef]
- Xiong, Y.; Chen, X.; Zhao, X.; Fan, Y.; Zhang, Q.; Zhu, W. Altered regional homogeneity and functional brain networks in Type 2 diabetes with and without mild cognitive impairment. Sci. Rep. 2020, 10, 21254. [Google Scholar] [CrossRef]
- Fang, E.F.; Xie, C.; Schenkel, J.A.; Wu, C.; Long, Q.; Cui, H.; Aman, Y.; Frank, J.; Liao, J.; Zou, H.; et al. A research agenda for ageing in China in the 21st century (2nd edition): Focusing on basic and translational research, long-term care, policy and social networks. Ageing Res. Rev. 2020, 64, 101174. [Google Scholar] [CrossRef]
- Sethi, M.; Rani, S.; Singh, A.; Mazón, J.L.V. A CAD System for Alzheimer’s Disease Classification Using Neuroimaging MRI 2D Slices. Comput. Math. Methods Med. 2022, 2022, 8680737. [Google Scholar] [CrossRef] [PubMed]
- Grueso, S.; Viejo-Sobera, R. Machine learning methods for predicting progression from mild cognitive impairment to Alzheimer’s disease dementia: A systematic review. Alzheimers Res. Ther. 2021, 13, 162. [Google Scholar] [CrossRef] [PubMed]
- Rajan, K.B.; Weuve, J.; Barnes, L.L.; McAninch, E.A.; Wilson, R.S.; Evans, D.A. Population estimate of people with clinical Alzheimer’s disease and mild cognitive impairment in the United States (2020–2060). Alzheimers Dement. 2021, 17, 1966–1975. [Google Scholar] [CrossRef]
- Mahendran, N.; Vincent, P.M.D.R.; Srinivasan, K.; Chang, C.-Y. Deep learning based ensemble model for classification of Alzheimer’s disease. Front. Biosci. 2022, 14, 27. [Google Scholar] [CrossRef]
- Gao, X.; Shi, F.; Shen, D.; Liu, M. Task-induced pyramid and attention GAN for multimodal brain image classification with application to Alzheimer’s disease. Front. Aging Neurosci. 2023, 15, 1242029. [Google Scholar] [CrossRef]
- Odusami, M.; Maskeliūnas, R.; Damaševičius, R. An Intelligent System for Early Recognition of Alzheimer’s Disease Using Neuroimaging. Sensors 2022, 22, 740. [Google Scholar] [CrossRef]
- Chen, Z.; Liu, Y.; Zhang, Y.; Zhu, J.; Li, Q.; Wu, X. Shared Manifold Regularized Joint Feature Selection for Joint Classification and Regression in Alzheimer’s Disease Diagnosis. IEEE Trans. Image Process. 2024, 33, 2730–2745. [Google Scholar] [CrossRef] [PubMed]



| Class | Original | Train | Validation | Augmented | Test | Total |
|---|---|---|---|---|---|---|
| Mild Demented | 321 | 289 | 32 | 867 | 49 | 370 |
| Moderate Demented | 591 | 532 | 59 | 1596 | 42 | 633 |
| Non-Demented | 316 | 285 | 31 | 855 | 22 | 338 |
| Severe Demented | 640 | 576 | 64 | 1728 | 47 | 687 |
| Very Mild Demented | 341 | 307 | 34 | 921 | 13 | 354 |
| Total | 2209 | 1989 | 220 | 5967 | 173 | 2382 |
| Class/Metric | ResNet50 | ViT | ResNet50 + ViT | Full Framework | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P | R | F1 | P | R | F1 | P | R | F1 | P | R | F1 | |
| Mild Demented | 1.00 | 0.96 | 0.98 | 0.98 | 0.96 | 0.97 | 0.98 | 0.92 | 0.95 | 1.00 | 0.98 | 0.99 |
| Moderate Demented | 1.00 | 0.95 | 0.98 | 1.00 | 0.95 | 0.98 | 1.00 | 0.95 | 0.98 | 1.00 | 1.00 | 1.00 |
| Non-Demented | 0.92 | 1.00 | 0.96 | 0.91 | 0.95 | 0.93 | 0.84 | 0.95 | 0.89 | 1.00 | 1.00 | 1.00 |
| Severe Demented | 0.96 | 1.00 | 0.98 | 0.96 | 1.00 | 0.98 | 0.96 | 1.00 | 0.98 | 1.00 | 1.00 | 1.00 |
| Very Mild Demented | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| Macro Precision | 0.98 | 0.97 | 0.96 | 1.00 | ||||||||
| Macro Recall | 0.98 | 0.97 | 0.97 | 0.998 | ||||||||
| Macro F1-Score | 0.98 | 0.97 | 0.96 | 0.998 | ||||||||
| Weighted Precision | 0.98 | 0.97 | 0.96 | 1.00 | ||||||||
| Weighted Recall | 0.98 | 0.97 | 0.96 | 0.994 | ||||||||
| Weighted F1-Score | 0.98 | 0.97 | 0.96 | 0.996 | ||||||||
| Overall Accuracy | 97.69% | 97.11% | 95.95% | 99.42% | ||||||||
| Class/Metric | KNN | Random Forest | Proposed Framework | ||||||
|---|---|---|---|---|---|---|---|---|---|
| P | R | F1 | P | R | F1 | P | R | F1 | |
| Mild Demented | 0.98 | 0.84 | 0.90 | 1.00 | 0.94 | 0.97 | 1.00 | 0.98 | 0.99 |
| Moderate Demented | 0.95 | 0.95 | 0.95 | 1.00 | 0.98 | 0.99 | 1.00 | 1.00 | 1.00 |
| Non-Demented | 0.78 | 0.95 | 0.86 | 0.88 | 1.00 | 0.94 | 1.00 | 1.00 | 1.00 |
| Severe Demented | 0.96 | 1.00 | 0.98 | 0.98 | 0.98 | 0.98 | 1.00 | 1.00 | 1.00 |
| Very Mild Demented | 1.00 | 1.00 | 1.00 | 0.93 | 1.00 | 0.96 | 1.00 | 1.00 | 1.00 |
| Macro Precision | 0.93 | 0.96 | 1.00 | ||||||
| Macro Recall | 0.95 | 0.98 | 0.998 | ||||||
| Macro F1-Score | 0.94 | 0.97 | 0.998 | ||||||
| Weighted Precision | 0.94 | 0.97 | 1.00 | ||||||
| Weighted Recall | 0.94 | 0.97 | 0.994 | ||||||
| Weighted F1-Score | 0.94 | 0.97 | 0.996 | ||||||
| Overall Accuracy | 93.64% | 97.11% | 99.42% | ||||||
| Author(s) | Model | Classes | Dataset | Accuracy (%) |
|---|---|---|---|---|
| Pradhan et al. [9] | DenseNet169 | MD, VMD, MOD, ND, SD | AD5C | 92.85 |
| Mahendran et al. [66] | CNN Ensemble | MD, VMD, MOD, ND, SD | AD5C | 95.2 |
| Gao et al. [67] | CNN-Transformer | MD, VMD, MOD, ND, SD | AD5C | 96.8 |
| Zia-ur-Rehman et al. [13] | DenseNet-201 | MD, VMD, MOD, ND, SD | AD5C | 98.24 |
| Proposed Framework | ResNet50+ViT | MD, VMD, MOD, ND, SD | AD5C | 99.42 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Muhammad, A.; Jin, Q.; Elwasila, O.; Gulzar, Y. Hybrid Deep Learning Architecture with Adaptive Feature Fusion for Multi-Stage Alzheimer’s Disease Classification. Brain Sci. 2025, 15, 612. https://doi.org/10.3390/brainsci15060612
Muhammad A, Jin Q, Elwasila O, Gulzar Y. Hybrid Deep Learning Architecture with Adaptive Feature Fusion for Multi-Stage Alzheimer’s Disease Classification. Brain Sciences. 2025; 15(6):612. https://doi.org/10.3390/brainsci15060612
Chicago/Turabian StyleMuhammad, Ahmad, Qi Jin, Osman Elwasila, and Yonis Gulzar. 2025. "Hybrid Deep Learning Architecture with Adaptive Feature Fusion for Multi-Stage Alzheimer’s Disease Classification" Brain Sciences 15, no. 6: 612. https://doi.org/10.3390/brainsci15060612
APA StyleMuhammad, A., Jin, Q., Elwasila, O., & Gulzar, Y. (2025). Hybrid Deep Learning Architecture with Adaptive Feature Fusion for Multi-Stage Alzheimer’s Disease Classification. Brain Sciences, 15(6), 612. https://doi.org/10.3390/brainsci15060612