The cGAS–STING Pathway in Dementia: An Emerging Mechanism of Neuroinflammation
Abstract
1. Introduction
2. Neuroinflammation in Dementia: A Shared Mechanism
2.1. Disease-Specific Mechanisms and Neuroinflammatory Pathways in Dementia
2.2. Genetic Drivers of Neuroinflammation in Dementia
3. cGAS-STING Pathway and Neuroinflammation in Dementia: A Potential Therapeutic Target
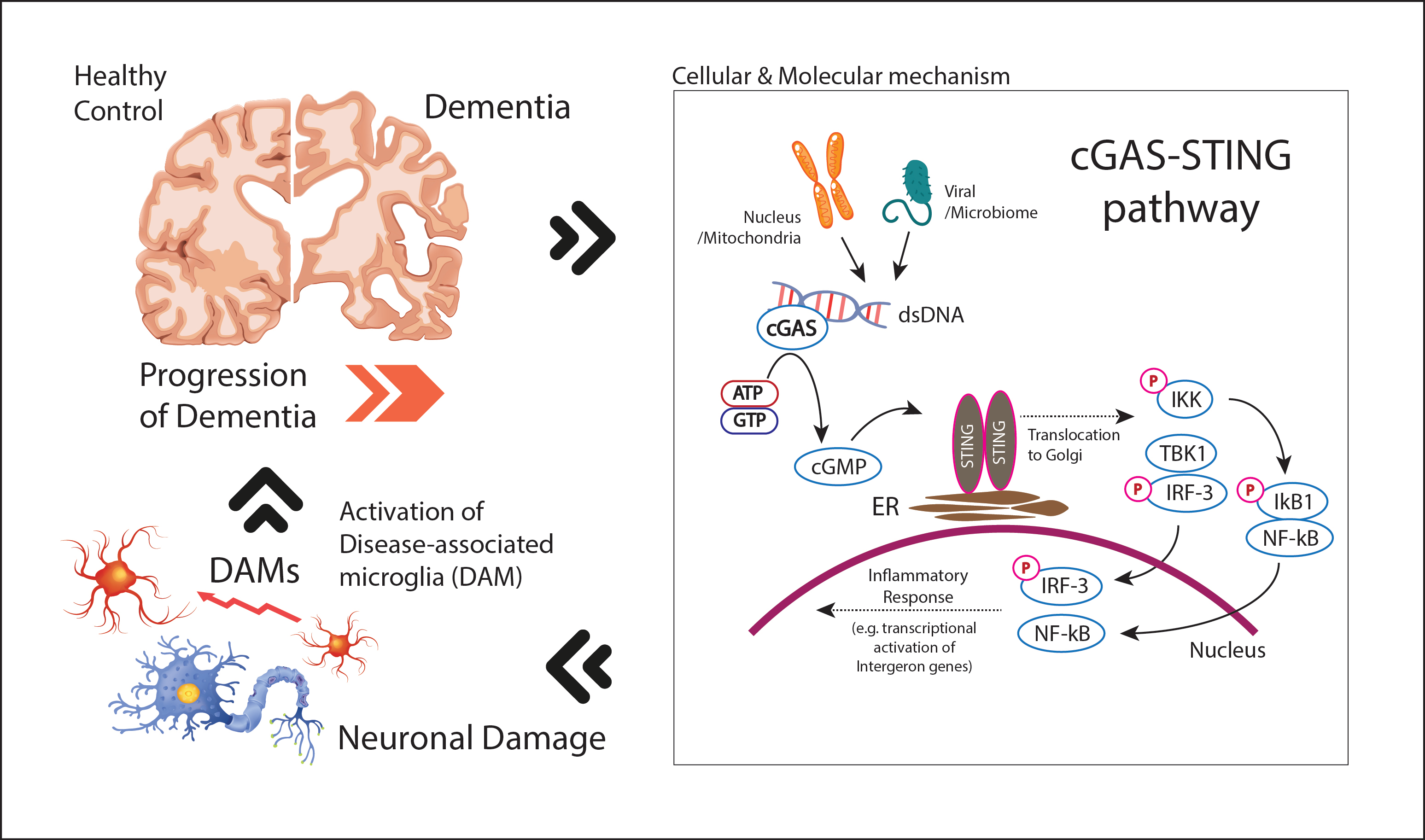
3.1. Molecular Mechanisms of cGAS-STING Pathway
3.2. cGAS-STING Activation in Dementia
4. Translational Insights: Therapeutic Targeting of cGAS-STING in Neurodegeneration
Therapeutic Modulation of cGAS-STING Pathway: Comparison Between Targeting cGAS vs. STING
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Spires-Jones, T.L.; Hyman, B.T. The Intersection of Amyloid Beta and Tau at Synapses in Alzheimer’s Disease. Neuron 2014, 82, 756–771. [Google Scholar] [CrossRef] [PubMed]
- Nichols, E.; Szoeke, C.; Vollset, S.E.; Abbasi, N.; Murray, C.J.L. Global, regional, and national burden of Alzheimer’s disease and other dementias, 1990–2016: A systematic analysis for the Global Burden of Disease Study 2016. Lancet Neurol. 2019, 18, 88–106. [Google Scholar] [CrossRef]
- Longhe, Z. 2020 Alzheimer’s disease facts and figures. Alzheimer’s Dement. 2020, 16, 391–460. [Google Scholar]
- Alzheimer’s Disease International. Dementia Statistics; Alzheimer’s Disease International: London, UK, 2024. [Google Scholar]
- Heneka, M.T.; Carson, M.J.; El Khoury, J.; Landreth, G.E.; Brosseron, F.; Feinstein, D.L.; Jacobs, A.H.; Wyss-Coray, T.; Vitorica, J.; Ransohoff, R.M.; et al. Neuroinflammation in Alzheimer’s disease. Lancet Neurol. 2015, 14, 388–405. [Google Scholar] [CrossRef]
- Tian, L.; Ma, L.; Kaarela, T.; Li, Z. Neuroimmune crosstalk in the central nervous system and its significance for neurological diseases. J. Neuroinflammation 2012, 9, 155. [Google Scholar] [CrossRef]
- Ransohoff, R.M. How neuroinflammation contributes to neurodegeneration. Science 2016, 353, 777–783. [Google Scholar] [CrossRef]
- Salter, M.W.; Stevens, B. Microglia emerge as central players in brain disease. Nat. Med. 2017, 23, 1018–1027. [Google Scholar] [CrossRef]
- Huang, Y.; Liu, B.; Sinha, S.C.; Amin, S.; Gan, L. Mechanism and therapeutic potential of targeting cGAS-STING signaling in neurological disorders. Mol. Neurodegener. 2023, 18, 79. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Yang, L.; Wang, J.; Wu, Y.; Li, Y.; Du, L.; Li, L.; Fang, Z.; Zhang, X. The cytosolic DNA-sensing cGAS-STING pathway in neurodegenerative diseases. CNS Neurosci. Ther. 2024, 30, e14671. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhang, B.; Duan, R.; Liu, Y. Mitochondrial DNA Leakage and cGas/STING Pathway in Microglia: Crosstalk Between Neuroinflammation and Neurodegeneration. Neuroscience 2024, 548, 1–8. [Google Scholar] [CrossRef]
- Sharma, O.; Kaur Grewal, A.; Khan, H.; Gurjeet Singh, T. Exploring the nexus of cGAS STING pathway in neurodegenerative terrain: A therapeutic odyssey. Int. Immunopharmacol. 2024, 142 Pt B, 113205. [Google Scholar] [CrossRef]
- Zhang, Y.; Zou, M.; Wu, H.; Zhu, J.; Jin, T. The cGAS-STING pathway drives neuroinflammation and neurodegeneration via cellular and molecular mechanisms in neurodegenerative diseases. Neurobiol. Dis. 2024, 202, 106710. [Google Scholar] [CrossRef]
- Paul, B.D.; Snyder, S.H.; Bohr, V.A. Signaling by cGAS-STING in Neurodegeneration, Neuroinflammation, and Aging. Trends Neurosci. 2021, 44, 83–96. [Google Scholar] [CrossRef]
- Alzheimer’s Association. “What Is Dementia?”. Available online: https://www.alz.org/alzheimers-dementia/what-is-dementia (accessed on 10 September 2025).
- Jack, C.R., Jr.; Bennett, D.A.; Blennow, K.; Carrillo, M.C.; Dunn, B.; Haeberlein, S.B.; Holtzman, D.M.; Jagust, W.; Jessen, F.; Karlawish, J.; et al. NIA-AA Research Framework: Toward a biological definition of Alzheimer’s disease. Alzheimer’s Dement. 2018, 14, 535–562. [Google Scholar] [CrossRef]
- Moyse, E.; Krantic, S.; Djellouli, N.; Roger, S.; Angoulvant, D.; Debacq, C.; Leroy, V.; Fougere, B.; Aidoud, A. Neuroinflammation: A Possible Link Between Chronic Vascular Disorders and Neurodegenerative Diseases. Front. Aging Neurosci. 2022, 14, 827263. [Google Scholar] [CrossRef]
- Zhang, W.; Xiao, D.; Mao, Q.; Xia, H. Role of neuroinflammation in neurodegeneration development. Signal Transduct. Target. Ther. 2023, 8, 267. [Google Scholar] [CrossRef] [PubMed]
- Hardy, J.; Selkoe, D.J. The Amyloid Hypothesis of Alzheimer’s Disease: Progress and Problems on the Road to Therapeutics. Science 2002, 297, 353–356. [Google Scholar] [CrossRef] [PubMed]
- Heppner, F.L.; Ransohoff, R.M.; Becher, B. Immune attack: The role of inflammation in Alzheimer disease. Nat. Rev. Neurosci. 2015, 16, 358–372. [Google Scholar] [CrossRef]
- De Strooper, B.; Karran, E. The Cellular Phase of Alzheimer’s Disease. Cell 2016, 164, 603–615. [Google Scholar] [CrossRef]
- Venkat, P.; Chopp, M.; Chen, J. Models and mechanisms of vascular dementia. Exp. Neurol. 2015, 272, 97–108. [Google Scholar] [CrossRef] [PubMed]
- Tian, Z.; Ji, X.; Liu, J. Neuroinflammation in Vascular Cognitive Impairment and Dementia: Current Evidence, Advances, and Prospects. Int. J. Mol. Sci. 2022, 23, 6224. [Google Scholar] [CrossRef]
- Love, S.; Miners, J.S. Cerebrovascular disease in ageing and Alzheimer’s disease. Acta Neuropathol. 2016, 131, 645–658. [Google Scholar] [CrossRef]
- Gorelick, P.B.; Scuteri, A.; Black, S.E.; Decarli, C.; Greenberg, S.M.; Iadecola, C.; Launer, L.J.; Laurent, S.; Lopez, O.L.; Nyenhuis, D.; et al. Vascular contributions to cognitive impairment and dementia: A statement for healthcare professionals from the american heart association/american stroke association. Stroke 2011, 42, 2672–2713. [Google Scholar] [CrossRef]
- Kalaria, R.N. Neuropathological diagnosis of vascular cognitive impairment and vascular dementia with implications for Alzheimer’s disease. Acta Neuropathol. 2016, 131, 659–685. [Google Scholar]
- Loveland, P.M.; Yu, J.J.; Churilov, L.; Yassi, N.; Watson, R. Investigation of Inflammation in Lewy Body Dementia: A Systematic Scoping Review. Int. J. Mol. Sci. 2023, 24, 12116. [Google Scholar] [CrossRef] [PubMed]
- Surendranathan, A.; Su, L.; Mak, E.; Passamonti, L.; Hong, Y.T.; Arnold, R.; Vázquez Rodríguez, P.; Bevan-Jones, W.R.; Brain, S.A.E.; Fryer, T.D.; et al. Early microglial activation and peripheral inflammation in dementia with Lewy bodies. Brain 2018, 141, 3415–3427. [Google Scholar] [CrossRef]
- Vrillon, A.; Bousiges, O.; Götze, K.; Demuynck, C.; Muller, C.; Ravier, A.; Schorr, B.; Philippi, N.; Hourregue, C.; Cognat, E.; et al. Plasma biomarkers of amyloid, tau, axonal, and neuroinflammation pathologies in dementia with Lewy bodies. Alzheimer’s Res. Ther. 2024, 16, 146. [Google Scholar] [CrossRef]
- Bright, F.; Werry, E.L.; Dobson-Stone, C.; Piguet, O.; Ittner, L.M.; Halliday, G.M.; Hodges, J.R.; Kiernan, M.C.; Loy, C.T.; Kassiou, M.; et al. Neuroinflammation in frontotemporal dementia. Nat. Rev. Neurol. 2019, 15, 540–555. [Google Scholar] [CrossRef] [PubMed]
- Bevan-Jones, W.R.; Cope, T.E.; Jones, P.S.; Kaalund, S.S.; Passamonti, L.; Allinson, K.; Green, O.; Hong, Y.T.; Fryer, T.D.; Arnold, R.; et al. Neuroinflammation and protein aggregation co-localize across the frontotemporal dementia spectrum. Brain 2020, 143, 1010–1026. [Google Scholar]
- Zhang, J. Mapping neuroinflammation in frontotemporal dementia with molecular PET imaging. J. Neuroinflammation 2015, 12, 108. [Google Scholar] [CrossRef]
- Mathur, V.; Burai, R.; Vest, R.T.; Bonanno, L.N.; Lehallier, B.; Zardeneta, M.E.; Mistry, K.N.; Do, D.; Marsh, S.E.; Abud, E.M.; et al. Activation of the STING-Dependent Type I Interferon Response Reduces Microglial Reactivity and Neuroinflammation. Neuron 2017, 96, 1290–1302.e6. [Google Scholar] [CrossRef]
- Bachiller, S.; Jiménez-Ferrer, I.; Paulus, A.; Yang, Y.; Swanberg, M.; Deierborg, T.; Boza-Serrano, A. Microglia in Neurological Diseases: A Road Map to Brain-Disease Dependent-Inflammatory Response. Front. Cell. Neurosci. 2018, 12, 488. [Google Scholar] [CrossRef] [PubMed]
- Daniilidou, M.; Holleman, J.; Hagman, G.; Kåreholt, I.; Aspö, M.; Brinkmalm, A.; Zetterberg, H.; Blennow, K.; Solomon, A.; Kivipelto, M.; et al. Neuroinflammation, cerebrovascular dysfunction and diurnal cortisol biomarkers in a memory clinic cohort: Findings from the Co-STAR study. Transl. Psychiatry 2024, 14, 364. [Google Scholar] [CrossRef] [PubMed]
- Ewers, M.; Biechele, G.; Suárez-Calvet, M.; Sacher, C.; Blume, T.; Morenas-Rodriguez, E.; Deming, Y.; Piccio, L.; Cruchaga, C.; Kleinberger, G.; et al. Higher CSF sTREM2 and microglia activation are associated with slower rates of beta-amyloid accumulation. EMBO Mol. Med. 2020, 12, e12308. [Google Scholar] [CrossRef]
- Nabizadeh, F.; Seyedmirzaei, H.; Karami, S. Neuroimaging biomarkers and CSF sTREM2 levels in Alzheimer’s disease: A longitudinal study. Sci. Rep. 2024, 14, 15318. [Google Scholar] [CrossRef]
- Pan, J.; Hu, J.; Meng, D.; Chen, L.; Wei, X. Neuroinflammation in dementia: A meta-analysis of PET imaging studies. Medicine 2024, 103, e38086. [Google Scholar] [CrossRef]
- Gouilly, D.; Saint-Aubert, L.; Ribeiro, M.J.; Salabert, A.S.; Tauber, C.; Péran, P.; Arlicot, N.; Pariente, J.; Payoux, P. Neuroinflammation PET imaging of the translocator protein (TSPO) in Alzheimer’s disease: An update. Eur. J. Neurosci. 2022, 55, 1322–1343. [Google Scholar] [CrossRef]
- Kunkle, B.W.; Grenier-Boley, B.; Sims, R.; Bis, J.C.; Damotte, V.; Naj, A.C.; Boland, A.; Vronskaya, M.V.; van der Lee, S.J.; Amlie-Wolf, A.; et al. Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Aβ, tau, immunity and lipid processing. Nat. Genet. 2019, 51, 414–430. [Google Scholar] [CrossRef]
- Chia, R.; Sabir, M.S.; Bandres-Ciga, S.; Saez-Atienzar, S.; Reynolds, R.H.; Gustavsson, E.; Walton, R.L.; Ahmed, S.; Viollet, C.; Ding, J.; et al. Genome sequencing analysis identifies new loci associated with Lewy body dementia and provides insights into its genetic architecture. Nat. Genet. 2021, 53, 294–303. [Google Scholar] [CrossRef] [PubMed]
- Yin, Z.; Rosenzweig, N.; Kleemann, K.L.; Zhang, X.; Brandão, W.; Margeta, M.A.; Schroeder, C.; Sivanathan, K.N.; Silveira, S.; Gauthier, C.; et al. APOE4 impairs the microglial response in Alzheimer’s disease by inducing TGFβ-mediated checkpoints. Nat. Immunol. 2023, 24, 1839–1853. [Google Scholar] [CrossRef]
- Lee, S.; Devanney, N.A.; Golden, L.R.; Smith, C.T.; Schwartz, J.L.; Walsh, A.E.; Clarke, H.A.; Goulding, D.S.; Allenger, E.J.; Morillo-Segovia, G.; et al. APOE modulates microglial immunometabolism in response to age, amyloid pathology, and inflammatory challenge. Cell Rep. 2023, 42, 112196. [Google Scholar] [CrossRef]
- Ulland, T.K.; Colonna, M. TREM2—A key player in microglial biology and Alzheimer disease. Nat. Rev. Neurol. 2018, 14, 667–675. [Google Scholar] [CrossRef]
- Siddiqui, S.S.; Matar, R.; Merheb, M.; Hodeify, R.; Vazhappilly, C.G.; Marton, J.; Shamsuddin, S.A.; Al Zouabi, H. Siglecs in Brain Function and Neurological Disorders. Cells 2019, 8, 1125. [Google Scholar] [CrossRef]
- Griciuc, A.; Serrano-Pozo, A.; Parrado, A.R.; Lesinski, A.N.; Asselin, C.N.; Mullin, K.; Hooli, B.; Choi, S.H.; Hyman, B.T.; Tanzi, R.E. Alzheimer’s disease risk gene CD33 inhibits microglial uptake of amyloid beta. Neuron 2013, 78, 631–643. [Google Scholar] [CrossRef] [PubMed]
- Dib, S.; Pahnke, J.; Gosselet, F. Role of ABCA7 in Human Health and in Alzheimer’s Disease. Int. J. Mol. Sci. 2021, 22, 4603. [Google Scholar] [CrossRef]
- Naj, A.C.; Jun, G.; Beecham, G.W.; Wang, L.S.; Vardarajan, B.N.; Buros, J.; Gallins, P.J.; Buxbaum, J.D.; Jarvik, G.P.; Crane, P.K.; et al. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer’s disease. Nat. Genet. 2011, 43, 436–441. [Google Scholar] [CrossRef]
- Leyns, C.E.G.; Ulrich, J.D.; Finn, M.B.; Stewart, F.R.; Koscal, L.J.; Remolina Serrano, J.; Robinson, G.O.; Anderson, E.; Colonna, M.; Holtzman, D.M. TREM2 deficiency attenuates neuroinflammation and protects against neurodegeneration in a mouse model of tauopathy. Proc. Natl. Acad. Sci. USA 2017, 114, 11524–11529. [Google Scholar] [CrossRef]
- Baker, M.; Mackenzie, I.R.; Pickering-Brown, S.M.; Gass, J.; Rademakers, R.; Lindholm, C.; Snowden, J.; Adamson, J.; Sadovnick, A.D.; Rollinson, S. Mutations in progranulin cause tau-negative frontotemporal dementia linked to chromosome 17. Nature 2006, 442, 916–919. [Google Scholar] [CrossRef]
- O’Rourke, J.G.; Bogdanik, L.; Yáñez, A.; Lall, D.; Wolf, A.J.; Muhammad, A.K.; Ho, R.; Carmona, S.; Vit, J.P.; Zarrow, J.; et al. C9orf72 is required for proper macrophage and microglial function in mice. Science 2016, 351, 1324–1329. [Google Scholar] [CrossRef]
- Jonsson, T.; Stefansson, H.; Steinberg, S.; Jonsdottir, I.; Jonsson, P.V.; Snaedal, J.; Bjornsson, S.; Huttenlocher, J.; Levey, A.I.; Lah, J.J.; et al. Variant of TREM2 associated with the risk of Alzheimer’s disease. N. Engl. J. Med. 2013, 368, 107–116. [Google Scholar] [CrossRef] [PubMed]
- Guerreiro, R.; Wojtas, A.; Bras, J.; Carrasquillo, M.; Rogaeva, E.; Majounie, E.; Cruchaga, C.; Sassi, C.; Kauwe, J.S.; Younkin, S.; et al. TREM2 variants in Alzheimer’s disease. N. Engl. J. Med. 2013, 368, 117–127. [Google Scholar] [CrossRef]
- Shi, Y.; Yamada, K.; Liddelow, S.A.; Smith, S.T.; Zhao, L.; Luo, W.; Tsai, R.M.; Spina, S.; Grinberg, L.T.; Rojas, J.C.; et al. ApoE4 markedly exacerbates tau-mediated neurodegeneration in a mouse model of tauopathy. Nature 2017, 549, 523–527. [Google Scholar] [CrossRef] [PubMed]
- Sierksma, A.; Lu, A.; Mancuso, R.; Fattorelli, N.; Thrupp, N.; Salta, E.; Zoco, J.; Blum, D.; Buée, L.; De Strooper, B.; et al. Novel Alzheimer risk genes determine the microglia response to amyloid-β but not to TAU pathology. EMBO Mol. Med. 2020, 12, e10606. [Google Scholar] [CrossRef]
- Mishra, S.; Knupp, A.; Young, J.E.; Jayadev, S. Depletion of the AD risk gene SORL1 causes endo-lysosomal dysfunction in human microglia. Alzheimer’s Dement. 2022, 18, e068943. [Google Scholar] [CrossRef]
- Feng, T.; Mai, S.; Roscoe, J.M.; Sheng, R.R.; Ullah, M.; Zhang, J.; Katz, I.I.; Yu, H.; Xiong, W.; Hu, F. Loss of TMEM106B and PGRN leads to severe lysosomal abnormalities and neurodegeneration in mice. EMBO Rep. 2020, 21, e50219. [Google Scholar] [CrossRef]
- Ledo, J.H.; Liebmann, T.; Zhang, R.; Chang, J.C.; Azevedo, E.P.; Wong, E.; Silva, H.M.; Troyanskaya, O.G.; Bustos, V.; Greengard, P. Presenilin 1 phosphorylation regulates amyloid-β degradation by microglia. Mol. Psychiatry 2021, 26, 5620–5635. [Google Scholar] [CrossRef]
- Karch, C.M.; Goate, A.M. Alzheimer’s disease risk genes and mechanisms of disease pathogenesis. Biol. Psychiatry 2015, 77, 43–51. [Google Scholar] [CrossRef]
- Chapuis, J.; Hansmannel, F.; Gistelinck, M.; Mounier, A.; Van Cauwenberghe, C.; Kolen, K.V.; Geller, F.; Sottejeau, Y.; Harold, D.; Dourlen, P.; et al. Increased expression of BIN1 mediates Alzheimer genetic risk by modulating tau pathology. Mol. Psychiatry 2013, 18, 1225–1234. [Google Scholar] [CrossRef]
- Zhao, P.; Xu, Y.; Jiang, L.L.; Fan, X.; Ku, Z.; Li, L.; Liu, X.; Deng, M.; Arase, H.; Zhu, J.J.; et al. LILRB2-mediated TREM2 signaling inhibition suppresses microglia functions. Mol. Neurodegener. 2022, 17, 44. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhao, Y. Progress on the roles of MEF2C in neuropsychiatric diseases. Mol. Brain 2022, 15, 8. [Google Scholar] [CrossRef]
- Pasinelli, P.; Brown, R.H. Molecular biology of amyotrophic lateral sclerosis: Insights from genetics. Nat. Rev. Neurosci. 2006, 7, 710–723. [Google Scholar] [CrossRef] [PubMed]
- Ablasser, A.; Chen, Z.J. cGAS in action: Expanding roles in immunity and inflammation. Science 2019, 363, eaat8657. [Google Scholar]
- He, S.; Li, X.; Mittra, N.; Bhattacharjee, A.; Wang, H.; Song, S.; Zhao, S.; Liu, F.; Han, X. Microglial cGAS Deletion Preserves Intercellular Communication and Alleviates Amyloid-β-Induced Pathogenesis of Alzheimer’s Disease. Adv. Sci. 2025, 12, e2410910. [Google Scholar] [CrossRef]
- Thanos, J.M.; Campbell, O.C.; Cowan, M.N.; Bruch, K.R.; Moore, K.A.; Ennerfelt, H.E.; Natale, N.R.; Mangalmurti, A.; Kerur, N.; Lukens, J.R. STING deletion protects against amyloid β-induced Alzheimer’s disease pathogenesis. Alzheimer’s Dement. 2025, 21, e70305. [Google Scholar]
- Hinkle, J.T.; Patel, J.; Panicker, N.; Karuppagounder, S.S.; Biswas, D.; Belingon, B.; Chen, R.; Brahmachari, S.; Pletnikova, O.; Troncoso, J.C.; et al. STING mediates neurodegeneration and neuroinflammation in nigrostriatal α-synucleinopathy. Proc. Natl. Acad. Sci. USA 2022, 119, e2118819119. [Google Scholar] [CrossRef] [PubMed]
- Bright, F.; Chan, G.; van Hummel, A.; Ittner, L.M.; Ke, Y.D. TDP-43 and Inflammation: Implications for Amyotrophic Lateral Sclerosis and Frontotemporal Dementia. Int. J. Mol. Sci. 2021, 22, 7781. [Google Scholar] [CrossRef] [PubMed]
- Jauhari, A.; Baranov, S.V.; Suofu, Y.; Kim, J.; Singh, T.; Yablonska, S.; Li, F.; Wang, X.; Oberly, P.; Minnigh, M.B.; et al. Melatonin inhibits cytosolic mitochondrial DNA-induced neuroinflammatory signaling in accelerated aging and neurodegeneration. J. Clin. Investig. 2020, 130, 3124–3136. [Google Scholar] [CrossRef] [PubMed]
- Ferecskó, A.S.; Smallwood, M.J.; Moore, A.; Liddle, C.; Newcombe, J.; Holley, J.; Whatmore, J.; Gutowski, N.J.; Eggleton, P. STING-Triggered CNS Inflammation in Human Neurodegenerative Diseases. Biomedicines 2023, 11, 1375. [Google Scholar] [CrossRef]
- Marques, C.; Held, A.; Dorfman, K.; Sung, J.; Song, C.; Kavuturu, A.S.; Aguilar, C.; Russo, T.; Oakley, D.H.; Albers, M.W.; et al. Neuronal STING activation in amyotrophic lateral sclerosis and frontotemporal dementia. Acta Neuropathol. 2024, 147, 56. [Google Scholar] [CrossRef]
- Govindarajulu, M.; Ramesh, S.; Beasley, M.; Lynn, G.; Wallace, C.; Labeau, S.; Pathak, S.; Nadar, R.; Moore, T.; Dhanasekaran, M. Role of cGAS-Sting Signaling in Alzheimer’s Disease. Int. J. Mol. Sci. 2023, 24, 8151. [Google Scholar]
- Bryant, J.D.; Lei, Y.; VanPortfliet, J.J.; Winters, A.D.; West, A.P. Assessing Mitochondrial DNA Release into the Cytosol and Subsequent Activation of Innate Immune-related Pathways in Mammalian Cells. Curr. Protoc. 2022, 2, e372. [Google Scholar] [CrossRef]
- Picca, A.; Calvani, R.; Coelho-Junior, H.J.; Landi, F.; Bernabei, R.; Marzetti, E. Mitochondrial Dysfunction, Oxidative Stress, and Neuroinflammation: Intertwined Roads to Neurodegeneration. Antioxidants 2020, 9, 647. [Google Scholar] [CrossRef] [PubMed]
- Frozza, R.L.; Lourenco, M.V.; De Felice, F.G. Challenges for Alzheimer’s Disease Therapy: Insights from Novel Mechanisms Beyond Memory Defects. Front. Neurosci. 2018, 12, 37. [Google Scholar] [CrossRef] [PubMed]
- Decout, A.; Katz, J.D.; Venkatraman, S.; Ablasser, A. The cGAS–STING pathway as a therapeutic target in inflammatory diseases. Nat. Rev. Immunol. 2021, 21, 548–569. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Cai, L.; Yao, J.; Li, C.; Wang, X. Agonists and Inhibitors of the cGAS-STING Pathway. Molecules 2024, 29, 3121. [Google Scholar] [CrossRef]
- Zhang, H.; He, Z.; Yin, C.; Yang, S.; Li, J.; Lin, H.; Hu, G.; Wu, A.; Qin, D.; Hu, G.; et al. STING-mediated neuroinflammation: A therapeutic target in neurodegenerative diseases. Front. Aging Neurosci. 2025, 17, 1659216. [Google Scholar] [CrossRef] [PubMed]
| Gene Symbols | Full Name | Chromosomal Location | Role in Microglia Activation | Associated Dementia | References |
|---|---|---|---|---|---|
| Genes governing microglial activation and phagocytosis | |||||
| TREM2 | Triggering Receptor Expressed on Myeloid cells 2 | 6p21.1 | Impair microglial activation, reduce phagocytosis of Aβ and damaged cells. | AD, FTD | [52,53] |
| APOE ε4 | Apolipoprotein E (Allele ε4) | 19q13.32 | impairs microglial Aβ clearance; detrimental TREM2 signaling pathway driving microglia from a resting state to a protective | AD | [54] |
| CD33 | Cluster of Differentiation 33 | 19q13.41 | Inhibitory signal that suppresses microglial phagocytosis of Aβ | AD | [45,46] |
| PLCG2 | Phospholipase C Gamma 2 | 16q23.3 | downstream effector of TREM2, promote microglial activation and phagocytosis, improving immune response in AD. | AD | [55] |
| MAPT | Microtubule-associated Protein Tau | 17q21.32 | Promotes tau pathology, induce reactive microglial state | FTD | [49] |
| Genes involved in protein and debris clearance within the microglia | |||||
| GRN | Granulin (progranulin) | 17q21.31 | Lysosomal dysfunction, excessive microglial activation, impaired microglia debris clearance | FTD | [50] |
| SORL1 | Sortilin Related Receptor 1 | 11q24.2 | lysosome dysfunction of microglia, malfunction in processing and trafficking of amyloid precursor protein and Aβ | AD | [56] |
| TMEM106B | Transmembrane Protein 106B | 7p21.3 | lysosomal dysfunction in microglia, excessive microglial activation, impaired waste clearance. | FTD | [57] |
| ABCA7 | ATP Binding Cassette Subfamily A Member 7 | 19p13.3 | abnormal cholesterol efflux and lipid metabolism, impaired microglial phagocytosis and inflammatory response | AD | [47] |
| PS1 (PSEN1) | Presenilin 1 | 14q24.2 | Cause massive Aβ burden leading secondary microglial toxicity, impaired lysosomal calcium signaling and autophagy leading abnormal brain homeostasis | AD | [58] |
| SNCA | Synuclein Alpha | 4q22.1 | Lipid peroxidation, loss of antioxidant defense, microglial phagocytic exhaustion | LBD | [41] |
| Genes affecting complement and inflammatory signaling that microglia use to label and clear damaged cells | |||||
| CR1 | Complement Receptor type 1 | 1q32.2 | Variants in CR1 impair complement regulation and enhance microglial activation via the complement cascade. | AD | [59] |
| MS4A | Membrane Spanning 4 domain, subfamily A | 11q12.2 | Altered membrane protein interaction, abnormal microglial inflammatory signaling | AD | [40,48] |
| BIN1 | Bridging Integrator 1 | 2q14.3 | The second most significant AD risk gene, highly expressed in microglia, Abnormal microglial phagocytosis and endocytosis of Aβ | AD | [60] |
| LILRB2 | Leukocyte Immunoglobulin-Like Receptor B2 | 19q13.42 | co-expressed with TREM2 in microglia, inhibitory receptor, suppresses microglial phagocytosis of Aβ | AD | [61] |
| Genes associated with a non-cell-autonomous role contributing to neurotoxicity | |||||
| C9orf72 | Chromosome 9 open reading frame 72 | 9p21.2 | dysregulated microglial activation, lysosomal storage defects, toxic inflammatory activation | FTD, ALS | [51] |
| MEF2C | Myocyte Enhancer Factor 2C | 5q14.3 | A transcriptional factor, modulating inflammatory environment in microglia | AD | [62] |
| SOD1 | Superoxide Dismutase 1 | 21q22.11 | increases oxidative stress, enhanced toxic microglial activation | ALS | [63] |
| Component | Cellular Location/Target | Function in Activation | Primary Downstream Effect |
|---|---|---|---|
| cGAS | Cytosol /Nucleus | Recognizes cytosolic dsDNA (DAMPs/PAMPs); dimerizes/catalyzes cGAMP synthesis | cGAMP production |
| cGAMP | Cytosol | Second messenger; binds and activates STING protein | STING conformational change and trafficking (from ER to Golgi) |
| STING | Endoplasmic Reticulum | Activated by cGAMP; recruits and activates TBK1 | Recruitment and phosphorylation of TBK1 |
| TBK1 | Cytosol /Golgi | Kinase; Phosphorylates STING, IRF3, and IKK | Activation of IRF3/NF-κB pathways |
| IRF3 | Cytosol /Nucleus | Transcription Factor; drives transcription of inflammatory genes | Transcription of Type I IFNs |
| Compound Name/Class | Target | Mechanism of Action (MOA) | Key Preclinical Use/Disease Model | References |
|---|---|---|---|---|
| RU.521, RU.365 (Tricyclic benzofluoropyrimidine compounds) | cGAS | Inhibit cGAS catalytic activity by competitive binding to DNA-binding domain; blocks dsDNA-cGAS interaction and cGAMP synthesis | subarachnoid hemorrhage(SAH)/macrophages from autoimmune mice Trex1−/− mice, human iPSC-derived Huntington’s disease (HD) neurons, ed in iPSC-derived ALS neurons/cGAS inhibition, Neuroprotection, reduced cognitive dysfunction, microglial inflammation | [69,76,78] |
| A151 | cGAS | Inhibitor that competes with dsDNA for cGAS binding; Synthetic oligonucleotide antagonist | Acute brain trauma/mouse model of ischemic stroke; reduced neuronal loss, immune cell infiltration and infarct volume | [14,76] |
| TDI-6570 | cGAS | Brain-permeable cGAS inhibitor | AD/P301S tauopathy mouse model/protect impaired cognitive and synaptic function | [9] |
| H-151 | STING | Covalent binding to Cys91; blocks palmitoylation/clustering (irreversible) | AD, ALS, FTD/5xFAD mice, TDP-43 mutant mice, iPSC-derived ALS neurons/reduced phospho-IRF3, improved cognitive function in aged mice | [71,72,76] |
| ,SN-011 | STING | Potent small molecule inhibitor | Aicardi–Goutières syndrome (AGS)/Trex1-deficient model/Mitigating neurological impairment) | [78] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Min, Y.; Lee, Y.-S.; Lee, J.; Keum, D.-Y.; Gwag, J.-Y.; Jeon, S.-M.; Jo, H.; Kang, S.-U. The cGAS–STING Pathway in Dementia: An Emerging Mechanism of Neuroinflammation. Brain Sci. 2025, 15, 1241. https://doi.org/10.3390/brainsci15111241
Min Y, Lee Y-S, Lee J, Keum D-Y, Gwag J-Y, Jeon S-M, Jo H, Kang S-U. The cGAS–STING Pathway in Dementia: An Emerging Mechanism of Neuroinflammation. Brain Sciences. 2025; 15(11):1241. https://doi.org/10.3390/brainsci15111241
Chicago/Turabian StyleMin, Young, Yoon-Seob Lee, Juwon Lee, Da-Young Keum, Joo-Young Gwag, Sung-Min Jeon, Heejin Jo, and Sung-Ung Kang. 2025. "The cGAS–STING Pathway in Dementia: An Emerging Mechanism of Neuroinflammation" Brain Sciences 15, no. 11: 1241. https://doi.org/10.3390/brainsci15111241
APA StyleMin, Y., Lee, Y.-S., Lee, J., Keum, D.-Y., Gwag, J.-Y., Jeon, S.-M., Jo, H., & Kang, S.-U. (2025). The cGAS–STING Pathway in Dementia: An Emerging Mechanism of Neuroinflammation. Brain Sciences, 15(11), 1241. https://doi.org/10.3390/brainsci15111241







