The Role of Superoxide Dismutase 1 in Amyotrophic Lateral Sclerosis: Identification of Signaling Pathways, Regulators, Molecular Interaction Networks, and Biological Functions through Bioinformatics
Abstract
1. Introduction
2. Materials and Methods
2.1. IPA Toxicity Analysis of SOD1
2.2. IPA Exploration of the Pathway/Pathway Molecules between SOD1 and ALS
2.3. Gene Ontology Analysis of SOD1-ALS Pathway Genes
2.4. Construction of Molecular Interaction Network of SOD1-ALS Pathway Molecules
3. Results
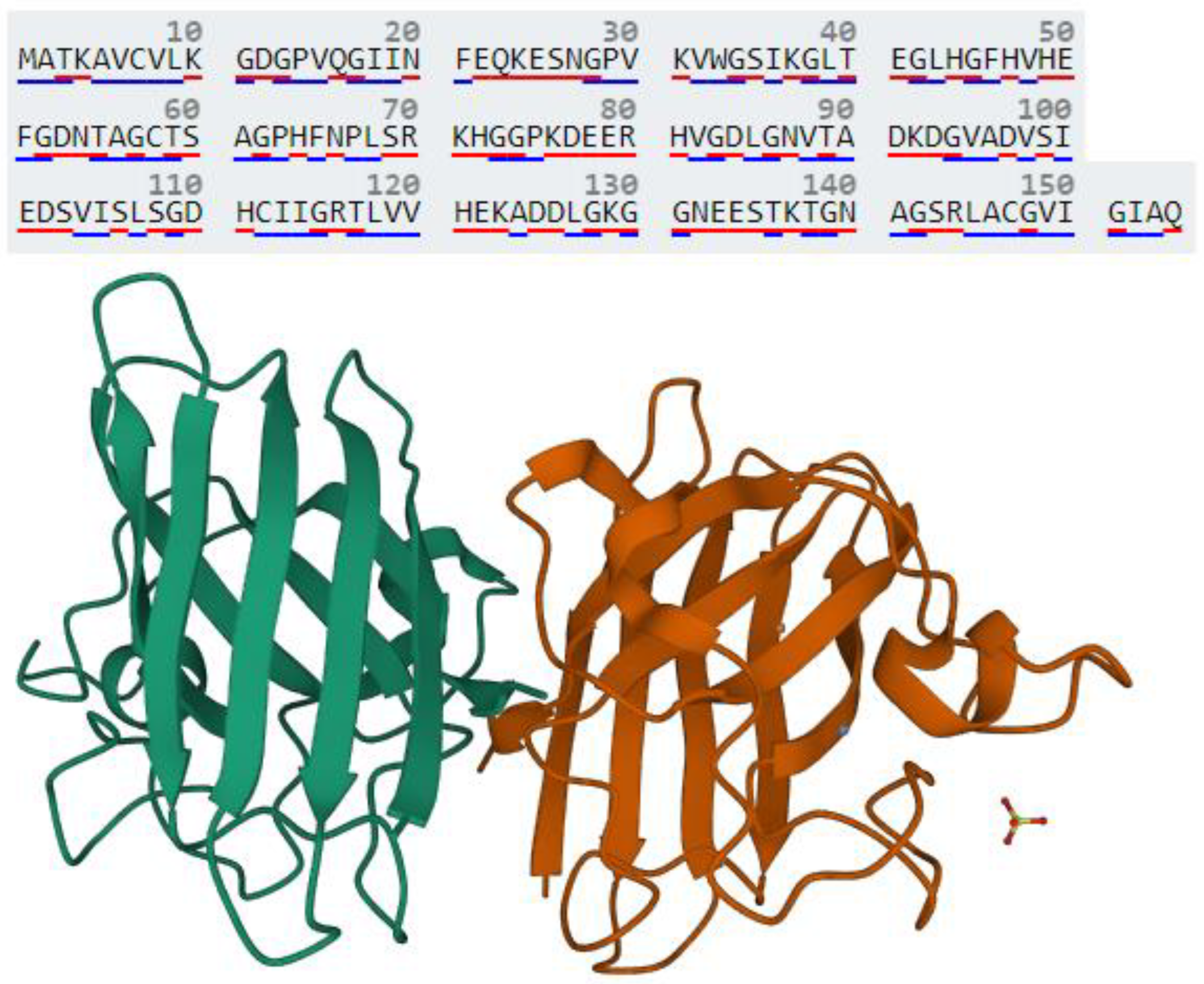
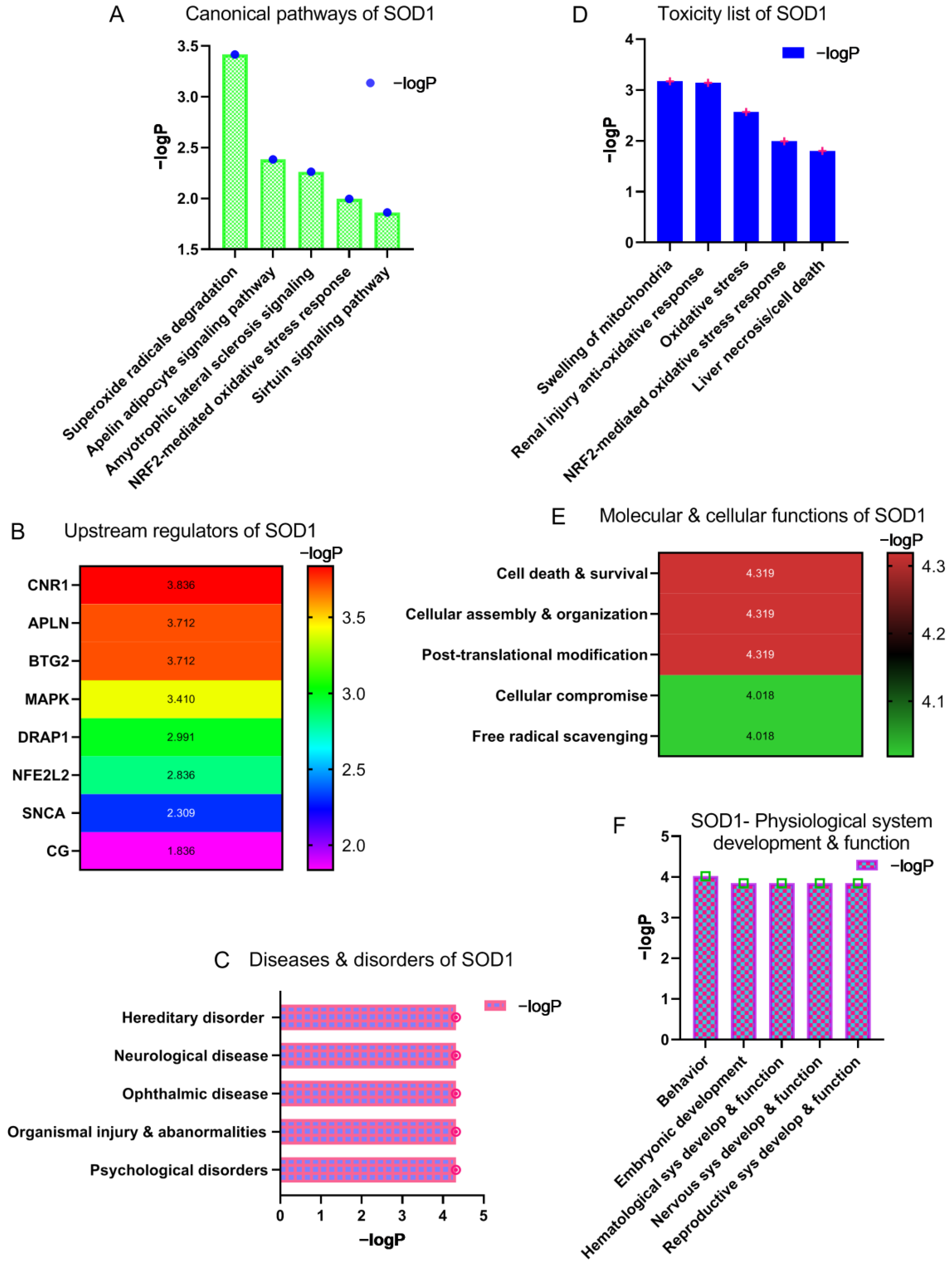
3.1. Identification of Canonical Pathways, Regulatory Molecules, Biological Functions and Toxicity Outcome of SOD1
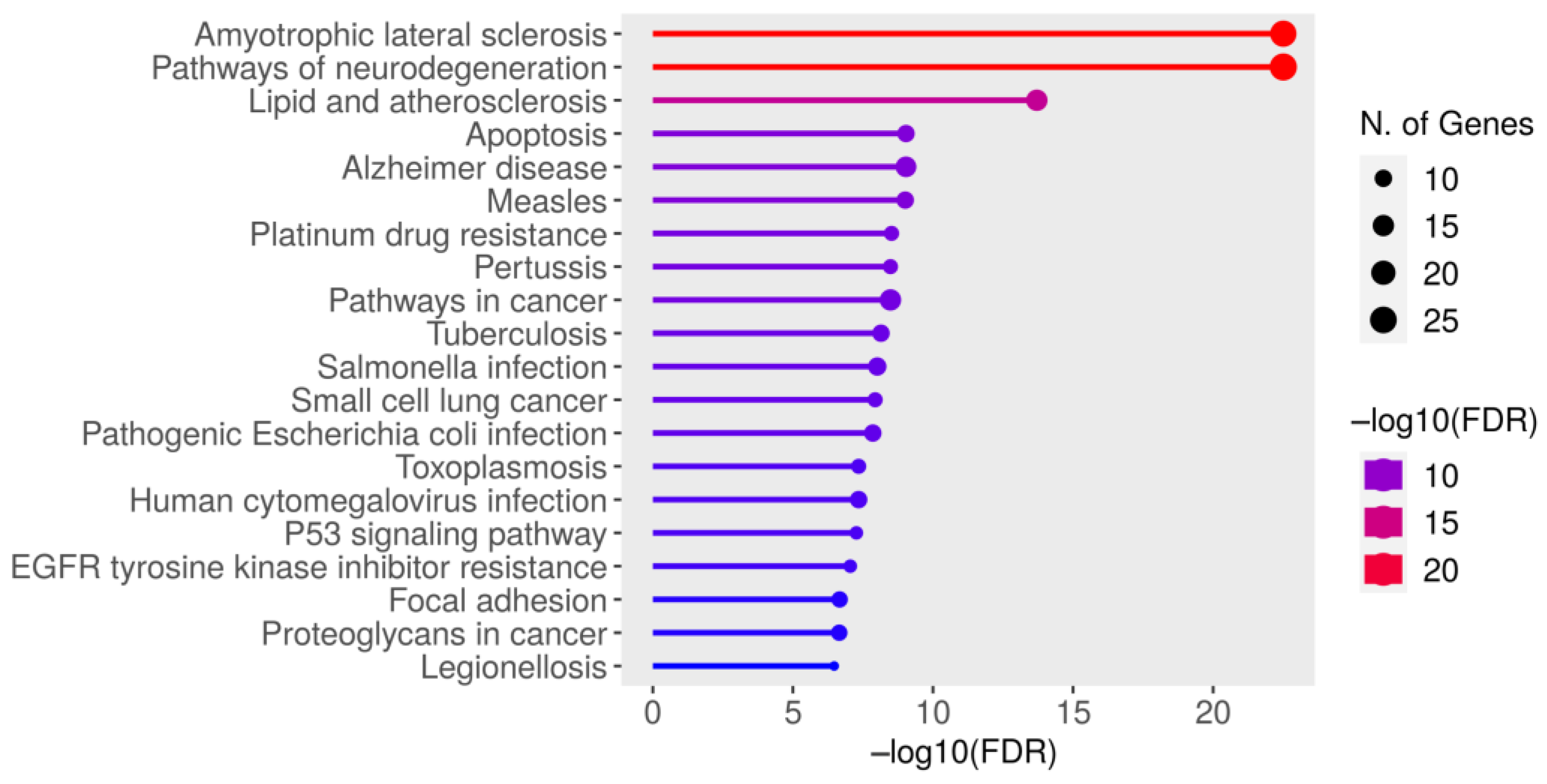
3.2. Identification of SOD1-ALS Pathway Molecules
3.3. GO Analysis of SOD1-ALS Pathway Genes
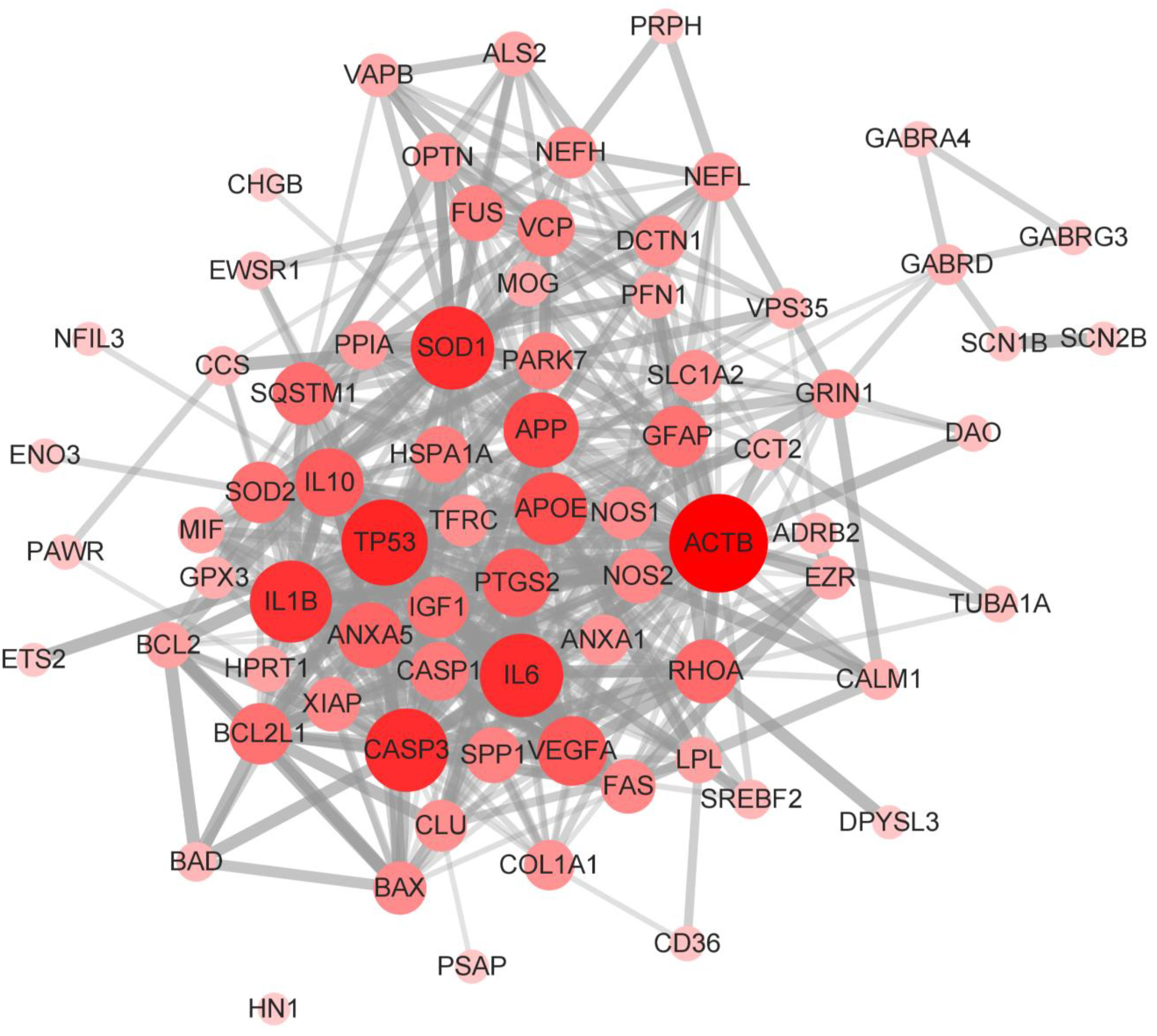
3.4. Construction and Analysis of Interaction Network of SOD1-ALS Pathway Molecules
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- McCord, J.M.; Fridovich, I. Superoxide Dismutase. An Enzymic Function for Erythrocuprein (Hemocuprein). J. Biol. Chem. 1969, 244, 6049–6055. [Google Scholar] [CrossRef] [PubMed]
- Sturtz, L.A.; Diekert, K.; Jensen, L.T.; Lill, R.; Culotta, V.C. A Fraction of Yeast Cu,Zn-Superoxide Dismutase and its Metallochaperone, CCS, Localize to the Intermembrane Space of Mitochondria. A Physiological Role for SOD1 in Guarding Against Mitochondrial Oxidative Damage. J. Biol. Chem. 2001, 276, 38084–38089. [Google Scholar] [CrossRef]
- Mesecke, N.; Terziyska, N.; Kozany, C.; Baumann, F.; Neupert, W.; Hell, K.; Herrmann, J.M. A Disulfide Relay System in the Intermembrane Space of Mitochondria that Mediates Protein Import. Cell 2005, 121, 1059–1069. [Google Scholar] [CrossRef] [PubMed]
- Field, L.S.; Furukawa, Y.; O’Halloran, T.V.; Culotta, V.C. Factors Controlling the Uptake of Yeast Copper/Zinc Superoxide Dismutase into Mitochondria. J. Biol. Chem. 2003, 278, 28052–28059. [Google Scholar] [CrossRef] [PubMed]
- Montllor-Albalate, C.; Kim, H.; Thompson, A.E.; Jonke, A.P.; Torres, M.P.; Reddi, A.R. SOD1 Integrates Oxygen Availability to Redox Regulate NADPH Production and the Thiol Redoxome. Proc. Natl. Acad. Sci. USA 2022, 119, e2023328119. [Google Scholar] [CrossRef] [PubMed]
- Tsang, C.K.; Liu, Y.; Thomas, J.; Zhang, Y.; Zheng, X.F.S. Superoxide Dismutase 1 Acts as a Nuclear Transcription Factor to Regulate Oxidative Stress Resistance. Nat. Commun. 2014, 5, 3446. [Google Scholar] [CrossRef]
- Rosen, D.R.; Siddique, T.; Patterson, D.; Figlewicz, D.A.; Sapp, P.; Hentati, A.; Donaldson, D.; Goto, J.; O’Regan, J.P.; Deng, H.X.; et al. Mutations in Cu/Zn Superoxide Dismutase Gene are Associated with Familial Amyotrophic Lateral Sclerosis. Nature 1993, 362, 59–62. [Google Scholar] [CrossRef]
- Mulder, D.W.; Kurland, L.T.; Offord, K.P.; Beard, C.M. Familial Adult Motor Neuron Disease: Amyotrophic Lateral Sclerosis. Neurology 1986, 36, 511–517. [Google Scholar] [CrossRef]
- Bruijn, L.I.; Miller, T.M.; Cleveland, D.W. Unraveling the Mechanisms Involved in Motor Neuron Degeneration in ALS. Annu. Rev. Neurosci. 2004, 27, 723–749. [Google Scholar] [CrossRef]
- ALSoD—Amyotrophic Lateral Sclerosis Online Database. Available online: https://alsod.ac.uk/output/gene.php/SOD1 (accessed on 10 November 2022).
- Berdyński, M.; Miszta, P.; Safranow, K.; Andersen, P.M.; Morita, M.; Filipek, S.; Żekanowski, C.; Kuźma-Kozakiewicz, M. SOD1 Mutations Associated with Amyotrophic Lateral Sclerosis Analysis of Variant Severity. Sci. Rep. 2022, 12, 103. [Google Scholar] [CrossRef]
- Rodriguez, J.A.; Shaw, B.F.; Durazo, A.; Sohn, S.H.; Doucette, P.A.; Nersissian, A.M.; Faull, K.F.; Eggers, D.K.; Tiwari, A.; Hayward, L.J.; et al. Destabilization of Apoprotein is Insufficient to Explain Cu,Zn-Superoxide Dismutase-linked ALS Pathogenesis. Proc. Natl. Acad. Sci. USA 2005, 102, 10516–10521. [Google Scholar] [CrossRef]
- Banci, L.; Bertini, I.; Boca, M.; Calderone, V.; Cantini, F.; Girotto, S.; Vieru, M. Structural and Dynamic Aspects Related to Oligomerization of Apo SOD1 and its Mutants. Proc. Natl. Acad. Sci. USA 2009, 106, 6980–6985. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Prakash, A.; Pandey, P.; Lynn, A.M.; Hassan, M.I. TFE-induced local unfolding and fibrillation of SOD1: Bridging the experiment and simulation studies. Biochem. J. 2018, 475, 1701–1719. [Google Scholar] [CrossRef]
- Prakash, A.; Kumar, V.; Pandey, P.; Bharti, D.R.; Vishwakarma, P.; Singh, R.; Hassan, M.I.; Lynn, A.M. Solvent sensitivity of protein aggregation in Cu, Zn superoxide dismutase: A molecular dynamics simulation study. J. Biomol. Struct. Dyn. 2018, 36, 2605–2617. [Google Scholar] [CrossRef] [PubMed]
- DiDonato, M.; Craig, L.; Huff, M.E.; Thayer, M.M.; Cardoso, R.M.F.; Kassmann, C.J.; Lo, T.P.; Bruns, C.K.; Powers, E.T.; Kelly, J.W.; et al. ALS Mutants of Human Superoxide Dismutase Form Fibrous Aggregates via Framework Destabilization. J. Mol. Biol. 2003, 332, 601–615. [Google Scholar] [CrossRef]
- Stathopulos, P.B.; Rumfeldt, J.A.O.; Scholz, G.A.; Irani, R.A.; Frey, H.E.; Hallewell, R.A.; Lepock, J.R.; Meiering, E.M. Cu/Zn Superoxide Dismutase Mutants Associated with Amyotrophic Lateral Sclerosis Show Enhanced Formation of Aggregates In Vitro. Proc. Natl. Acad. Sci. USA 2003, 100, 7021–7026. [Google Scholar] [CrossRef]
- Trist, B.G.; Hilton, J.B.; Hare, D.J.; Crouch, P.J.; Double, K.L. Superoxide Dismutase 1 in Health and Disease: How a Frontline Antioxidant Becomes Neurotoxic. Angew. Chem. Int. Ed. Engl. 2021, 60, 9215–9246. [Google Scholar] [CrossRef] [PubMed]
- Peggion, C.; Scalcon, V.; Massimino, M.L.; Nies, K.; Lopreiato, R.; Rigobello, M.P.; Bertoli, A. SOD1 in ALS: Taking Sock in Pathogenic Mechanisms and the Role of Glial and Muscle Cells. Antioxidants 2022, 11, 614. [Google Scholar] [CrossRef]
- NCBI (2022). National Center for Biotechnology (NCBI). Available online: https://www.ncbi.nlm.nih.gov/gene (accessed on 30 September 2022).
- IPA (2021). Ingenuity Pathway Analysis (IPA), Qiagen. Available online: https://digitalinsights.qiagen.com/products-overview/discovery-insights-portfolio/analysis-and-visualization/qiagen-ipa/ (accessed on 30 September 2022).
- Suthar, S.K.; Alam, M.M.; Lee, J.; Monga, J.; Joseph, A.; Lee, S.-Y. Bioinformatic Analyses of Canonical Pathways of TSPOAP1 and its Roles in Human Diseases. Front. Mol. Biosci. 2021, 8, 667947. [Google Scholar] [CrossRef]
- Suthar, S.K.; Lee, S.-Y. Ingenuity Pathway Analysis of α-Synuclein Predicts Potential Signaling Pathways, Network Molecules, Biological Functions, and its Role in Neurological Diseases. Front. Mol. Neurosci. 2022, 15, 1029682. [Google Scholar] [CrossRef]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A Graphical Gene-set Enrichment Tool for Animals and Plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P.; et al. STRING v10: Protein-Protein Interaction Networks, Integrated Over the Tree of Life. Nucleic Acids Res. 2015, 43, D447–D452. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]

| Description (Term) | Gene Counts | p-Value | |
|---|---|---|---|
| BP | Response to chemical (GO:0042221) | 55 | 7.84 × 10−23 |
| Regulation of biological quality (GO:0065008) | 53 | 3.34 × 10−22 | |
| Response to organic substance (GO:0010033) | 47 | 8.62 × 10−22 | |
| Positive regulation of transport (GO:0051050) | 29 | 4.89 × 10−20 | |
| Signaling (GO:0023052) | 56 | 1.02 × 10−19 | |
| MF | Identical protein binding (GO:0042802) | 38 | 2.88 × 10−20 |
| Protein binding (GO:0005515) | 62 | 4.54 × 10−19 | |
| Protein domain specific binding (GO:0019904) | 18 | 8.00 × 10−11 | |
| Enzyme binding (GO:0019899) | 29 | 2.83 × 10−10 | |
| Signaling receptor binding (GO:0005102) | 23 | 4.65 × 10−09 | |
| CC | Vesicle (GO:0031982) | 43 | 7.79 × 10−14 |
| Extracellular space (GO:0005615) | 37 | 5.07 × 10−12 | |
| Extracellular region (GO:0005576) | 41 | 2.86 × 10−11 | |
| Cell junction (GO:0030054) | 28 | 2.63 × 10−10 | |
| Synapse (GO:0045202) | 23 | 2.30 × 10−10 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Suthar, S.K.; Lee, S.-Y. The Role of Superoxide Dismutase 1 in Amyotrophic Lateral Sclerosis: Identification of Signaling Pathways, Regulators, Molecular Interaction Networks, and Biological Functions through Bioinformatics. Brain Sci. 2023, 13, 151. https://doi.org/10.3390/brainsci13010151
Suthar SK, Lee S-Y. The Role of Superoxide Dismutase 1 in Amyotrophic Lateral Sclerosis: Identification of Signaling Pathways, Regulators, Molecular Interaction Networks, and Biological Functions through Bioinformatics. Brain Sciences. 2023; 13(1):151. https://doi.org/10.3390/brainsci13010151
Chicago/Turabian StyleSuthar, Sharad Kumar, and Sang-Yoon Lee. 2023. "The Role of Superoxide Dismutase 1 in Amyotrophic Lateral Sclerosis: Identification of Signaling Pathways, Regulators, Molecular Interaction Networks, and Biological Functions through Bioinformatics" Brain Sciences 13, no. 1: 151. https://doi.org/10.3390/brainsci13010151
APA StyleSuthar, S. K., & Lee, S.-Y. (2023). The Role of Superoxide Dismutase 1 in Amyotrophic Lateral Sclerosis: Identification of Signaling Pathways, Regulators, Molecular Interaction Networks, and Biological Functions through Bioinformatics. Brain Sciences, 13(1), 151. https://doi.org/10.3390/brainsci13010151







