Domain Adaptation Using a Three-Way Decision Improves the Identification of Autism Patients from Multisite fMRI Data
Abstract
1. Introduction
- A three-way decision model based on triangular fuzzy similarity is proposed to reduce the cost loss of target domain data prediction. To the best of authors’ knowledge, it is the first time to combine the three-way decision model and the distribution adaptation method to reduce the distribution differences between domains. The proposed method extends the application of machine learning in the field of decision making.
- Our method utilizes the label information from the source domain and the structural information from the target domain at the same time, which not only reduces the distribution differences between domains but also further improves the recognition ability of the target domain data.
- Comprehensive experiments on the Autism Brain Imaging Data Exchange (ABIDE) dataset prove that our method is better than several state-of-the-art methods.
2. Related Work
2.1. Distribution Adaptation
2.2. Three-Way Decisions
2.3. Application of Machine Learning in Identification of ASD Patients
3. Preliminaries
4. Methods
4.1. Joint Distribution Adaptation
4.2. Three-Way Decision Model Based on Triangular Fuzzy Similarity
4.2.1. Information Difference Degree and Triangular Fuzzy Similarity
- (1)
- The greater the value of is, the greater the degree of information difference of objectunderand. When objecthas the same descriptionforand, the real part of the log function will have a denominator of 0, i.e.,. In this case, since, we can obtain that the final degree of information deviationis independent of the value of. For the reasonableness of the calculation, let.
- (2)
- For the convenience of the representation, we obtain the information difference matrix of object , which can be expressed as follows (Equation (16)):whererepresents the degree of information difference of objectunder attributesand.
- (1)
- Boundedness:.
- (2)
- Monotonicity: The degree of information difference ofaboutand increases monotonously as the difference increases.
- (3)
- Symmetry:.
- (1)
- According to Definition 4, and . When the description of under and appears in two extreme cases, namely, or , we can obtain , and the information difference reaches the maximum at this time, . □
- (1)
- .
- (2)
- if, andifandorand.
- (2)
- Since , when , i.e., , ,, we have , so . Similarly, since and , . When , . In this case, we can obtain and or and . □
4.2.2. Construction of the 3WD Model
| Algorithm 1 Three-way decision model based on the triangular fuzzy similarity |
| Input: target domain data , threshold , and . |
| Output: positive region object set , negative region object set , boundary region object set . |
| 1: BEGIN |
| 2: Calculate the degree of information difference of each object in the target domain under any two attributes according to Equation (15). |
| 3: Calculate the triangular fuzzy similarity between any two objects in the target domain using Equation (17). |
| 4: According to Equation (21), divide the target domain into three domains. |
| 5: END BEGIN |
4.3. Adaptation Via Iterative Refinement
| Algorithm 2 Our Proposed Model |
| Input: source domain data , target domain data , labels of source domain data, threshold , and |
| Output: as labels of target domain data |
| 1: BEGIN |
| 2: Initialize as Null |
| 3: while not converged do |
| 4: (1) Distribution adaptation in Equation (14) and let and |
| 5: (2) Assign using classifiers trained by |
| 6: (3) Obtain in Algorithm 1 |
| 7: (4) execute label propagation algorithm |
| 8: End while |
| 9: |
| 10: END BEGIN |
5. Experiments
5.1. Materials
5.1.1. Data Acquisition
5.1.2. Data Pre-Processing
5.2. Competing Methods
5.3. Experimental Setup
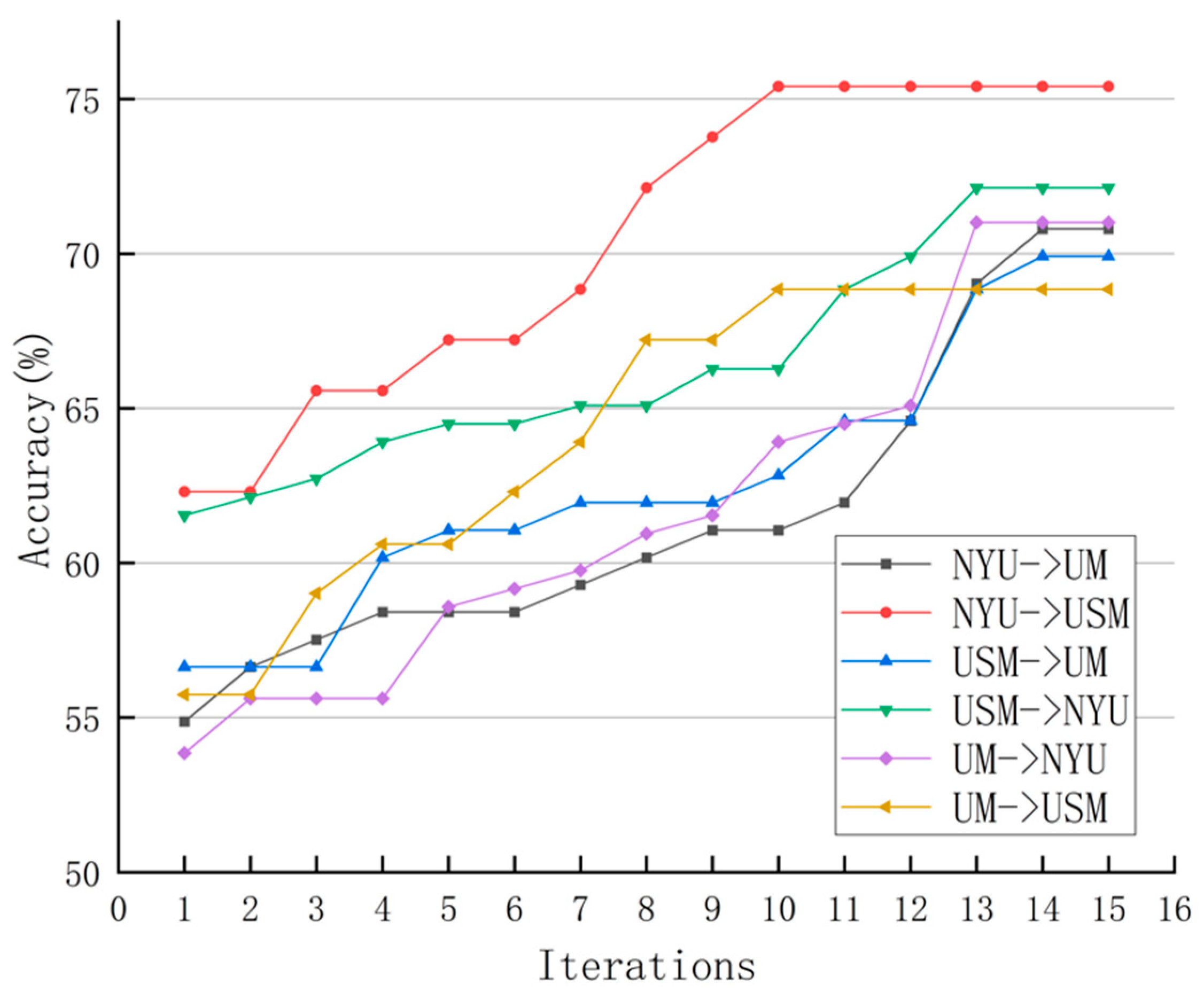
5.4. Results on ABIDE with Multisite fMRI Data
6. Discussion
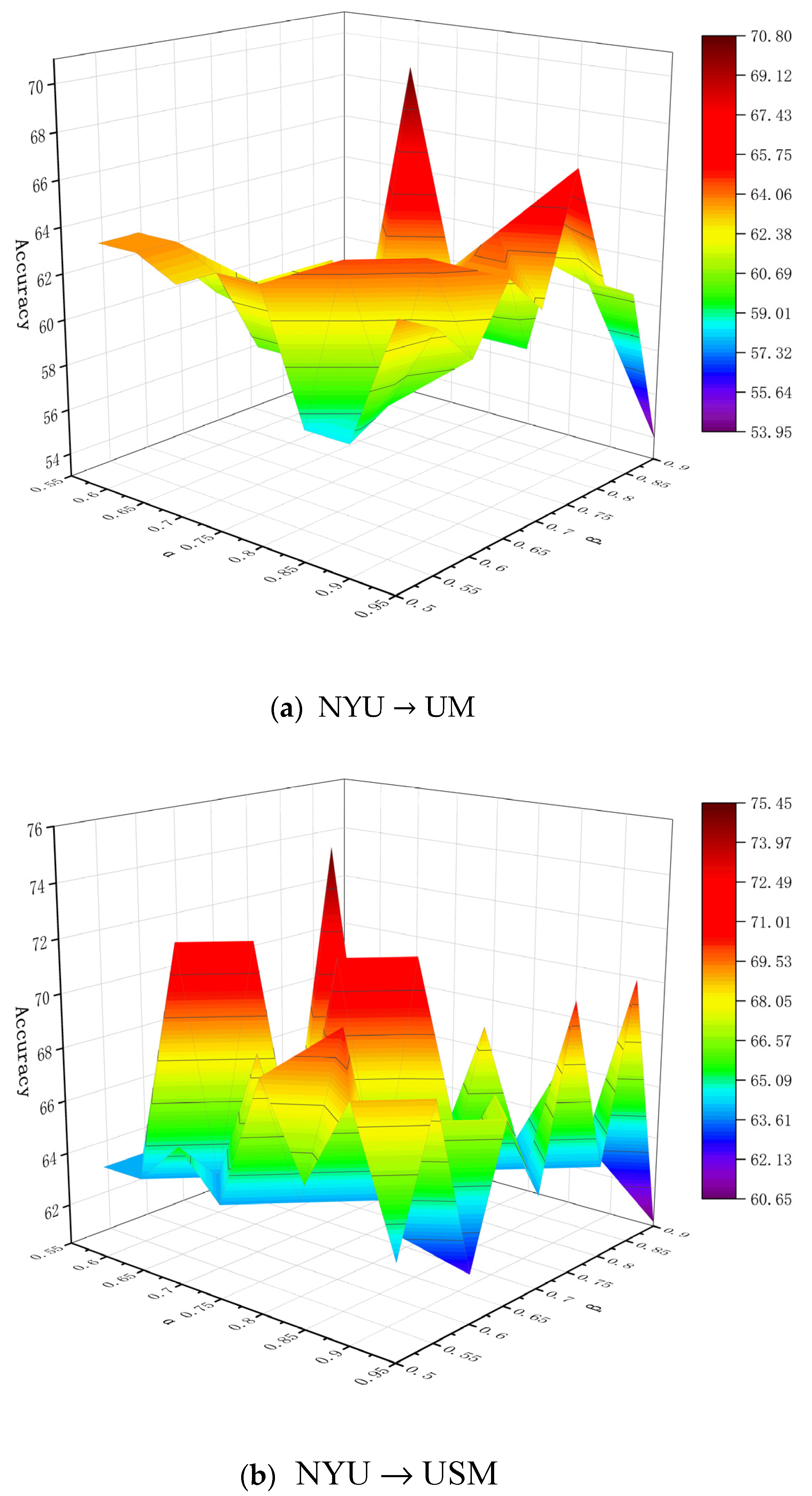
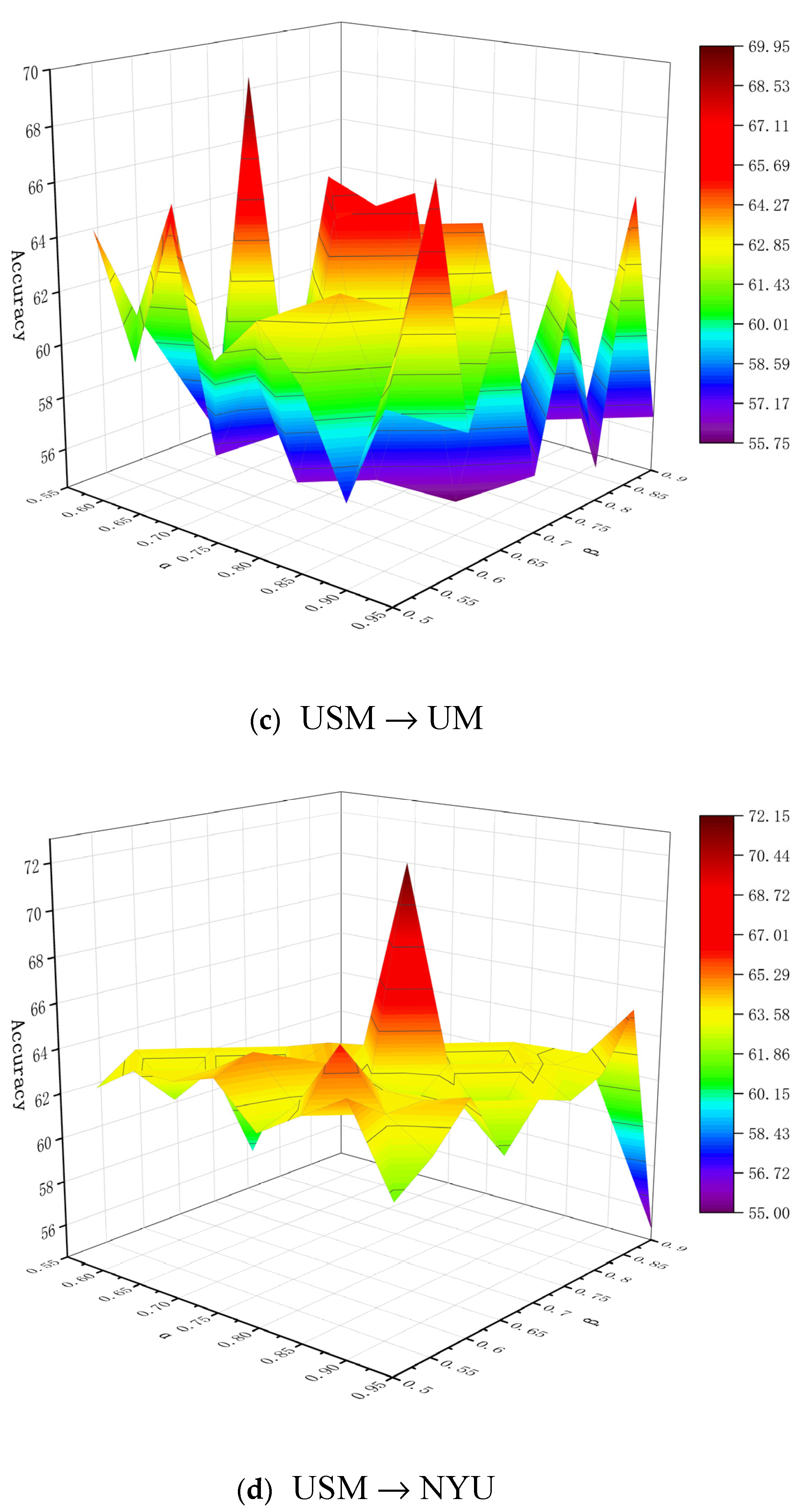
6.1. Parameter Analysis
6.2. Comparison with State-of-the-Art Methods
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Khan, N.A.; Waheeb, S.A.; Riaz, A.; Shang, X. A Three-Stage Teacher, Student Neural Networks and Sequential Feed Forward Selection-Based Feature Selection Approach for the Classification of Autism Spectrum Disorder. Brain Sci. 2020, 10, 754. [Google Scholar] [CrossRef] [PubMed]
- Kong, Y.; Gao, J.; Xu, Y.; Pan, Y.; Wang, J.; Liu, J. Classification of autism spectrum disorder by combining brain connectivity and deep neural network classifier. Neurocomputing 2019, 324, 63–68. [Google Scholar] [CrossRef]
- Zecavati, N.; Spence, S.J. Neurometabolic disorders and dysfunction in autism spectrum disorders. Curr. Neurol. Neurosci. 2009, 9, 129–136. [Google Scholar] [CrossRef] [PubMed]
- Amaral, D.G.; Schumann, C.M.; Nordahl, C.W. Neuroanatomy of autism. Trends Neurosci. 2008, 31, 137–145. [Google Scholar] [CrossRef] [PubMed]
- Khundrakpam, B.S.; Lewis, J.D.; Kostopoulos, P.; Carbonell, F.; Evans, A.C. Cortical Thickness Abnormalities in Autism Spectrum Disorders Through Late Childhood, Adolescence, and Adulthood: A Large-Scale MRI Study. Cereb. Cortex 2017, 27, 1721–1731. [Google Scholar] [CrossRef]
- Zablotsky, B.; Black, L.I.; Maenner, M.J.; Schieve, L.A.; Blumberg, S.J. Estimated Prevalence of Autism and Other Developmental Disabilities Following Questionnaire Changes in the 2014 National Health Interview Survey. Natl. Health Stat. Rep. 2015, 87, 1–20. [Google Scholar]
- Wang, M.; Zhang, D.; Huang, J.; Yap, P.; Shen, D.; Liu, M. Identifying Autism Spectrum Disorder With Multi-Site fMRI via Low-Rank Domain Adaptation. IEEE Trans. Med. Imaging 2020, 39, 644–655. [Google Scholar] [CrossRef]
- Fernell, E.; Eriksson, M.A.; Gillberg, C. Early diagnosis of autism and impact on prognosis: A narrative review. Clin. Epidemiol. 2013, 5, 33–43. [Google Scholar] [CrossRef] [PubMed]
- Mwangi, B.; Ebmeier, K.P.; Matthews, K.; Steele, J.D. Multi-centre diagnostic classification of individual structural neuroimaging scans from patients with major depressive disorder. Brain J. Neurol. 2012, 135, 1508–1521. [Google Scholar] [CrossRef]
- Plitt, M.; Barnes, K.A.; Martin, A. Functional connectivity classification of autism identifies highly predictive brain features but falls short of biomarker standards. NeuroImage Clin. 2015, 7, 359–366. [Google Scholar] [CrossRef]
- Shi, C.; Zhang, J.; Wu, X. An fMRI Feature Selection Method Based on a Minimum Spanning Tree for Identifying Patients with Autism. Symmetry 2020, 12, 1995. [Google Scholar] [CrossRef]
- Van den Heuvel, M.P.; Hulshoff Pol, H.E. Exploring the brain network: A review on resting-state fMRI functional connectivity. Eur. Neuropsychopharmacol. 2010, 20, 519–534. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Yoon, N.; Jang, S.; Lee, G.; Kim, B. Neuroimaging-Based Deep Learning in Autism Spectrum Disorder and Attention-Deficit/Hyperactivity Disorder. J. Child Adolesc. Psychiatry 2020, 31, 97–104. [Google Scholar] [CrossRef]
- Ktena, S.I.; Parisot, S.; Ferrante, E.; Rajchl, M.; Lee, M.; Glocker, B.; Rueckert, D. Metric learning with spectral graph convolutions on brain connectivity networks. Neuroimage 2018, 169, 431–442. [Google Scholar] [CrossRef] [PubMed]
- Abraham, A.; Milham, M.P.; Di Martino, A.; Craddock, R.C.; Samaras, D.; Thirion, B.; Varoquaux, G. Deriving reproducible biomarkers from multi-site resting-state data: An Autism-based example. Neuroimage 2017, 147, 736–745. [Google Scholar] [CrossRef] [PubMed]
- Heinsfeld, A.S.; Franco, A.R.; Craddock, R.C.; Buchweitz, A.; Meneguzzi, F. Identification of autism spectrum disorder using deep learning and the ABIDE dataset. NeuroImage Clin. 2017, 17, 16–23. [Google Scholar] [CrossRef]
- Nielsen, J.A.; Zielinski, B.A.; Fletcher, P.T.; Alexander, A.L.; Lange, N.; Bigler, E.D.; Lainhart, J.E.; Anderson, J.S. Multisite functional connectivity MRI classification of autism: ABIDE results. Front. Hum. Neurosci. 2013, 7. [Google Scholar] [CrossRef]
- Dai, W.; Yang, Q.; Xue, G.R.; Yu, Y. Boosting for transfer learning. In Proceedings of the 24th International Conference on Machine Learning, Corvalis, OR, USA, 20–24 June 2007; pp. 193–200. [Google Scholar]
- Ren, C.X.; Dai, D.Q.; Huang, K.K.; Lai, Z.R. Transfer learning of structured representation for face recognition. IEEE Trans. Image Process. 2014, 23, 5440–5454. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, J.; Huang, M.; Yu, H. Cross-position activity recognition with stratified transfer learning. Perv. Mob. Comput. 2019, 57, 1–13. [Google Scholar] [CrossRef]
- Yi, J.; Tao, J.; Wen, Z.; Bai, Y. Language-adversarial transfer learning for low-resource speech recognition. IEEE-ACM Trans. Audio Speech Lang. 2019, 27, 621–630. [Google Scholar] [CrossRef]
- Do, C.B.; Ng, A.Y. Transfer learning for text classification. Adv. Neural Inf. Process. Syst. 2005, 18, 299–306. [Google Scholar]
- Xu, Y.; Pan, S.J.; Xiong, H.; Wu, Q.; Luo, R.; Min, H.; Song, H. A Unified Framework for Metric Transfer Learning. IEEE Trans. Knowl. Data Eng. 2017, 29, 1158–1171. [Google Scholar] [CrossRef]
- Duan, L.; Tsang, I.W.; Xu, D.; Chua, T.S. Domain adaptation from multiple sources via auxiliary classifiers. In Proceedings of the 26th Annual International Conference on Machine Learning, Montreal, QC, Canada, 14–18 June 2009; pp. 289–296. [Google Scholar]
- Long, M.; Wang, J.; Ding, G.; Pan, S.J.; Yu, P.S. Adaptation Regularization: A General Framework for Transfer Learning. IEEE Trans. Knowl. Data Eng. 2014, 26, 1076–1089. [Google Scholar] [CrossRef]
- Fernando, B.; Habrard, A.; Sebban, M.; Tuytelaars, T. Unsupervised visual domain adaptation using subspace alignment. In Proceedings of the IEEE International Conference on Computer Vision, Sydney, Australia, 1–8 December 2013; pp. 2960–2967. [Google Scholar]
- Gong, B.; Shi, Y.; Sha, F.; Grauman, K. Geodesic flow kernel for unsupervised domain adaptation. In Proceedings of the 2012 IEEE Conference on Computer Vision and Pattern Recognition, Washington, DC, USA, 16–21 June 2012; pp. 2066–2073. [Google Scholar]
- Sun, B.; Feng, J.; Saenko, K. Return of frustratingly easy domain adaptation. In Proceedings of the AAAI Conference on Artificial Intelligence, Phoenix, AR, USA, 12–17 February 2016. [Google Scholar]
- Wang, J.; Chen, Y.; Yu, H.; Huang, M.; Yang, Q. Easy transfer learning by exploiting intra-domain structures. In Proceedings of the IEEE International Conference on Multimedia and Expo, Shanghai, China, 8–12 July 2019; pp. 1210–1215. [Google Scholar]
- Long, M.; Wang, J.; Ding, G.; Sun, J.; Yu, P.S. Transfer feature learning with joint distribution adaptation. In Proceedings of the IEEE International Conference on Computer Vision, Sydney, Australia, 1–8 December 2013; pp. 2200–2207. [Google Scholar]
- Wang, J.; Chen, Y.; Hao, S.; Feng, W.; Shen, Z. Balanced distribution adaptation for transfer learning. In Proceedings of the 2017 IEEE International Conference on Data Mining, New Orleans, LA, USA, 18–21 November 2017; pp. 1129–1134. [Google Scholar]
- Zhang, J.; Li, W.; Ogunbona, P. Joint geometrical and statistical alignment for visual domain adaptation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 1859–1867. [Google Scholar]
- Hou, C.; Tsai, Y.H.; Yeh, Y.; Wang, Y.F. Unsupervised Domain Adaptation with Label and Structural Consistency. IEEE T Image Process. 2016, 25, 5552–5562. [Google Scholar] [CrossRef]
- Tahmoresnezhad, J.; Hashemi, S. Visual domain adaptation via transfer feature learning. Knowl. Inf. Syst. 2017, 50, 586–605. [Google Scholar] [CrossRef]
- Zhang, Y.; Deng, B.; Jia, K.; Zhang, L. Label propagation with augmented anchors: A simple semi-supervised learning baseline for unsupervised domain adaptation. In Proceedings of the European Conference on Computer Vision, Glasgow, UK, 23–28 August 2020; pp. 781–797. [Google Scholar]
- Pan, S.J.; Tsang, I.W.; Kwok, J.T.; Yang, Q. Domain Adaptation via Transfer Component Analysis. IEEE Trans. Neural Netw. 2011, 22, 199–210. [Google Scholar] [CrossRef] [PubMed]
- Long, M.; Wang, J.; Ding, G.; Sun, J.; Yu, P.S. Transfer joint matching for unsupervised domain adaptation. In Proceedings of the IEEE conference on computer vision and pattern recognition, Columbus, OH, USA, 23–28 June 2014; pp. 1410–1417. [Google Scholar]
- Wang, J.; Chen, Y.; Hu, L.; Peng, X.; Philip, S.Y. Stratified transfer learning for cross-domain activity recognition. In Proceedings of the 2018 IEEE International Conference on Pervasive Computing and Communications, Athens, Greece, 19–23 March 2018; pp. 1–10. [Google Scholar]
- Yao, Y. Three-way decisions with probabilistic rough sets. Inf. Sci. 2010, 180, 341–353. [Google Scholar] [CrossRef]
- Chu, X.; Sun, B.; Huang, Q.; Zhang, Y. Preference degree-based multi-granularity sequential three-way group conflict decisions approach to the integration of TCM and Western medicine. Comput. Ind. Eng. 2020, 143. [Google Scholar] [CrossRef]
- Almasvandi, Z.; Vahidinia, A.; Heshmati, A.; Zangeneh, M.M.; Goicoechea, H.C.; Jalalvand, A.R. Coupling of digital image processing and three-way calibration to assist a paper-based sensor for determination of nitrite in food samples. RSC Adv. 2020, 10, 14422–14430. [Google Scholar] [CrossRef]
- Ren, F.; Wang, L. Sentiment analysis of text based on three-way decisions. J. Intell. Fuzzy Syst. 2017, 33, 245–254. [Google Scholar] [CrossRef]
- Yao, Y. Decision-theoretic rough set models. In International Conference on Rough Sets and Knowledge Technology; Springer: Berlin/Heidelberg, Germany, 14–16 May 2007; pp. 1–12. [Google Scholar]
- Zhang, H.; Yang, S. Three-way group decisions with interval-valued decision-theoretic rough sets based on aggregating inclusion measures. Int. J. Approx. Reason. 2019, 110, 31–45. [Google Scholar] [CrossRef]
- Liu, P.; Yang, H. Three-way decisions with intuitionistic uncertain linguistic decision-theoretic rough sets based on generalized Maclaurin symmetric mean operators. Int. J. Fuzzy Syst. 2020, 22, 653–667. [Google Scholar] [CrossRef]
- Agbodah, K. The determination of three-way decisions with decision-theoretic rough sets considering the loss function evaluated by multiple experts. Granul. Comput. 2019, 4, 285–297. [Google Scholar] [CrossRef]
- Liang, D.; Wang, M.; Xu, Z.; Liu, D. Risk appetite dual hesitant fuzzy three-way decisions with TODIM. Inf. Sci. 2020, 507, 585–605. [Google Scholar] [CrossRef]
- Qian, T.; Wei, L.; Qi, J. A theoretical study on the object (property) oriented concept lattices based on three-way decisions. Soft Comput. 2019, 23, 9477–9489. [Google Scholar] [CrossRef]
- Yang, C.; Zhang, Q.; Zhao, F. Hierarchical Three-Way Decisions with Intuitionistic Fuzzy Numbers in Multi-Granularity Spaces. IEEE Access 2019, 7, 24362–24375. [Google Scholar] [CrossRef]
- Yao, Y.; Deng, X. Sequential three-way decisions with probabilistic rough sets. In Proceedings of the IEEE 10th International Conference on Cognitive Informatics and Cognitive Computing, Banff, AB, Canada, 18–20 August 2011; pp. 120–125. [Google Scholar]
- Yang, X.; Li, T.; Liu, D.; Fujita, H. A temporal-spatial composite sequential approach of three-way granular computing. Inf. Sci. 2019, 486, 171–189. [Google Scholar] [CrossRef]
- Zhang, L.; Li, H.; Zhou, X.; Huang, B. Sequential three-way decision based on multi-granular autoencoder features. Inf. Sci. 2020, 507, 630–643. [Google Scholar] [CrossRef]
- Liu, D.; Ye, X. A matrix factorization based dynamic granularity recommendation with three-way decisions. Knowl. Based Syst. 2020, 191. [Google Scholar] [CrossRef]
- Yao, Y. Three-way granular computing, rough sets, and formal concept analysis. Int. J. Approx. Reason. 2020, 116, 106–125. [Google Scholar] [CrossRef]
- Lang, G.; Luo, J.; Yao, Y. Three-way conflict analysis: A unification of models based on rough sets and formal concept analysis. Knowl. Based Syst. 2020, 194. [Google Scholar] [CrossRef]
- Xin, X.; Song, J.; Peng, W. Intuitionistic Fuzzy Three-Way Decision Model Based on the Three-Way Granular Computing Method. Symmetry 2020, 12, 1068. [Google Scholar] [CrossRef]
- Yue, X.D.; Chen, Y.F.; Miao, D.Q.; Fujita, H. Fuzzy neighborhood covering for three-way classification. Inf. Sci. 2020, 507, 795–808. [Google Scholar] [CrossRef]
- Ma, Y.Y.; Zhang, H.R.; Xu, Y.Y.; Min, F.; Gao, L. Three-way recommendation integrating global and local information. J. Eng. 2018, 16, 1397–1401. [Google Scholar] [CrossRef]
- Yu, H.; Wang, X.; Wang, G.; Zeng, X. An active three-way clustering method via low-rank matrices for multi-view data. Inf. Sci. 2020, 507, 823–839. [Google Scholar] [CrossRef]
- Taneja, S.S. Re: Variability of the Positive Predictive Value of PI-RADS for Prostate MRI across 26 Centers: Experience of the Society of Abdominal Radiology Prostate Cancer Disease-Focused Panel. J. Urol. 2020, 204, 1380–1381. [Google Scholar] [CrossRef] [PubMed]
- Salama, S.; Khan, M.; Shanechi, A.; Levy, M.; Izbudak, I. MRI differences between MOG antibody disease and AQP4 NMOSD. Mult. Scler. 2020, 26, 1854–1865. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Lin, X.; Chen, X. Detecting Alzheimer’s disease Based on 4D fMRI: An exploration under deep learning framework. Neurocomputing 2020, 388, 280–287. [Google Scholar] [CrossRef]
- Riaz, A.; Asad, M.; Alonso, E.; Slabaugh, G. DeepFMRI: End-to-end deep learning for functional connectivity and classification of ADHD using fMRI. J. Neurosci. Meth. 2020, 335. [Google Scholar] [CrossRef]
- Diciotti, S.; Orsolini, S.; Salvadori, E.; Giorgio, A.; Toschi, N.; Ciulli, S.; Ginestroni, A.; Poggesi, A.; De Stefano, N.; Pantoni, L.; et al. Resting state fMRI regional homogeneity correlates with cognition measures in subcortical vascular cognitive impairment. J. Neurol. Sci. 2017, 373, 1–6. [Google Scholar] [CrossRef]
- Lu, H.; Liu, S.; Wei, H.; Tu, J. Multi-kernel fuzzy clustering based on auto-encoder for fMRI functional network. Exp. Syst. Appl. 2020, 159. [Google Scholar] [CrossRef]
- Leming, M.; Górriz, J.M.; Suckling, J. Ensemble Deep Learning on Large, Mixed-Site fMRI Datasets in Autism and Other Tasks. Int. J. Neural Syst. 2020, 30. [Google Scholar] [CrossRef]
- Benabdallah, F.Z.; Maliani, A.D.E.; Lotfi, D.; Hassouni, M.E. Analysis of the Over-Connectivity in Autistic Brains Using the Maximum Spanning Tree: Application on the Multi-Site and Heterogeneous ABIDE Dataset. In Proceedings of the 2020 8th International Conference on Wireless Networks and Mobile Communications (WINCOM), Reims, France, 27–29 October 2020; pp. 1–7. [Google Scholar]
- Eslami, T.; Mirjalili, V.; Fong, A.; Laird, A.R.; Saeed, F. ASD-DiagNet: A Hybrid Learning Approach for Detection of Autism Spectrum Disorder Using fMRI Data. Front. Neuroinf. 2019, 13. [Google Scholar] [CrossRef] [PubMed]
- Bi, X.; Wang, Y.; Shu, Q.; Sun, Q.; Xu, Q. Classification of Autism Spectrum Disorder Using Random Support Vector Machine Cluster. Front. Genet. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Rakić, M.; Cabezas, M.; Kushibar, K.; Oliver, A.; Lladó, X. Improving the detection of autism spectrum disorder by combining structural and functional MRI information. NeuroImage Clin. 2020, 25. [Google Scholar] [CrossRef] [PubMed]
- Borgwardt, K.M.; Gretton, A.; Rasch, M.J.; Kriegel, H.; Schölkopf, B.; Smola, A.J. Integrating structured biological data by Kernel Maximum Mean Discrepancy. Bioinformatics 2006, 22, e49–e57. [Google Scholar] [CrossRef]
- Steinwart, I. On the influence of the kernel on the consistency of support vector machines. J. Mach. Learn. Res. 2001, 2, 67–93. [Google Scholar]
- Xu, Z.S. Study on method for triangular fuzzy number-based multi-attribute decision making with preference information on alternatives. Syst. Eng. Electron. 2002, 24, 9–12. [Google Scholar]
- Yao, Y. The superiority of three-way decisions in probabilistic rough set models. Inf. Sci. 2011, 180, 1080–1096. [Google Scholar] [CrossRef]
- Yao, Y. An outline of a theory of three-way decisions. In Proceedings of the International Conference on Rough Sets and Current Trends in Computing, Chengdu, China, 17–20 August 2012; pp. 1–17. [Google Scholar]
- Chao-Gan, Y.; Yu-Feng, Z. DPARSF: A MATLAB Toolbox for “Pipeline” Data Analysis of Resting-State fMRI. Front. Syst. Neurosci. 2010, 4. [Google Scholar] [CrossRef]
- Tzourio-Mazoyer, N.; Landeau, B.; Papathanassiou, D.; Crivello, F.; Etard, O.; Delcroix, N.; Mazoyer, B.; Joliot, M. Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. Neuroimage 2002, 15, 273–289. [Google Scholar] [CrossRef] [PubMed]
- Niu, K.; Guo, J.; Pan, Y.; Gao, X.; Peng, X.; Li, N.; Li, H. Multichannel deep attention neural networks for the classification of autism spectrum disorder using neuroimaging and personal characteristic data. Complexity 2020. [Google Scholar] [CrossRef]

| Action | Cost Function | |
|---|---|---|
| Site | ASD | Normal Control | ||
|---|---|---|---|---|
| Age (m ± std) | Gender (M/F) | Age (m ± std) | Gender (M/F) | |
| NYU | 14.92 7.04 | 64/9 | 15.75 6.23 | 70/36 |
| USM | 24.59 8.46 | 38/0 | 22.33 7.69 | 23/0 |
| UM | 13.85 2.29 | 39/9 | 15.03 3.64 | 49/16 |
| Task | Method | ACC (%) | SEN (%) | SPE (%) | BAC (%) | PPV (%) | NPV (%) |
|---|---|---|---|---|---|---|---|
| NYU→UM | Baseline | 54.87 | 49.23 | 62.5 | 55.87 | 64 | 47.62 |
| TCA | 62.83 | 58.46 | 68.75 | 63.61 | 71.69 | 55.00 | |
| JDA | 64.50 | 66.67 | 61.64 | 64.16 | 69.57 | 58.44 | |
| DALSC | 64.60 | 56.92 | 75.00 | 65.96 | 75.51 | 56.25 | |
| Ours | 70.80 | 72.31 | 68.75 | 70.53 | 75.81 | 64.71 | |
| NYU→USM | Baseline | 67.21 | 78.26 | 60.53 | 69.39 | 54.55 | 82.14 |
| TCA | 68.85 | 82.61 | 60.53 | 71.57 | 55.88 | 85.19 | |
| JDA | 70.49 | 86.96 | 60.53 | 73.74 | 57.14 | 88.46 | |
| DALSC | 72.13 | 73.91 | 71.05 | 72.48 | 60.71 | 81.81 | |
| Ours | 75.41 | 91.30 | 65.79 | 78.55 | 61.76 | 92.59 | |
| USM→UM | Baseline | 57.52 | 35.38 | 87.50 | 61.44 | 79.31 | 50.00 |
| TCA | 58.41 | 38.46 | 85.42 | 61.94 | 78.13 | 50.62 | |
| JDA | 61.06 | 61.54 | 60.42 | 60.98 | 67.80 | 53.70 | |
| DALSC | 64.60 | 73.85 | 52.08 | 62.96 | 67.61 | 59.52 | |
| Ours | 69.91 | 76.92 | 60.42 | 68.67 | 72.46 | 65.91 | |
| USM→NYU | Baseline | 53.25 | 35.42 | 76.71 | 56.06 | 66.67 | 47.46 |
| TCA | 57.39 | 40.63 | 79.45 | 60.04 | 72.22 | 50.43 | |
| JDA | 60.36 | 64.58 | 54.79 | 59.69 | 65.26 | 54.05 | |
| DALSC | 63.91 | 65.63 | 61.64 | 63.63 | 69.23 | 57.69 | |
| Ours | 72.13 | 78.26 | 68.42 | 73.34 | 60.00 | 83.87 | |
| UM→NYU | Baseline | 58.58 | 83.33 | 26.03 | 54.68 | 59.70 | 54.29 |
| TCA | 61.54 | 82.29 | 34.25 | 58.27 | 62.20 | 59.50 | |
| JDA | 63.31 | 82.29 | 38.35 | 60.32 | 63.71 | 62.22 | |
| DALSC | 64.49 | 92.70 | 27.39 | 60.05 | 62.68 | 74.07 | |
| Ours | 71.01 | 90.63 | 45.21 | 67.92 | 68.50 | 78.57 | |
| UM→USM | Baseline | 54.09 | 78.26 | 39.47 | 58.87 | 43.90 | 75.00 |
| TCA | 60.66 | 73.91 | 52.63 | 63.27 | 48.57 | 76.92 | |
| JDA | 60.66 | 78.26 | 50.00 | 64.13 | 48.65 | 79.17 | |
| DALSC | 57.38 | 73.91 | 47.37 | 60.64 | 45.95 | 75.00 | |
| Ours | 68.85 | 82.61 | 60.53 | 71.57 | 55.88 | 85.19 |
| Method | Feature Type | Feature Dimension | Classifier | ACC (%) |
|---|---|---|---|---|
| sGCN + Hing Loss [14] | HOA | 111 × 111 | K-Nearest Neighbor (KNN) | 60.50 |
| sGCN + Global Loss [14] | HOA | 111 × 111 | KNN | 63.50 |
| sGCN + Constrained Variance Loss [14] | HOA | 111 × 111 | KNN | 68.00 |
| FCA [17] | GMR | 7266 × 7266 | t-test | 63.00 |
| DAE [16] | CC200 Atlas | 19,900 | Softmax Regression | 66.00 |
| DANN [78] | AAL | 6670 | Deep neural network | 70.90 |
| Ours | AAL | 4005 | SVM | 72.13/71.01 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shi, C.; Xin, X.; Zhang, J. Domain Adaptation Using a Three-Way Decision Improves the Identification of Autism Patients from Multisite fMRI Data. Brain Sci. 2021, 11, 603. https://doi.org/10.3390/brainsci11050603
Shi C, Xin X, Zhang J. Domain Adaptation Using a Three-Way Decision Improves the Identification of Autism Patients from Multisite fMRI Data. Brain Sciences. 2021; 11(5):603. https://doi.org/10.3390/brainsci11050603
Chicago/Turabian StyleShi, Chunlei, Xianwei Xin, and Jiacai Zhang. 2021. "Domain Adaptation Using a Three-Way Decision Improves the Identification of Autism Patients from Multisite fMRI Data" Brain Sciences 11, no. 5: 603. https://doi.org/10.3390/brainsci11050603
APA StyleShi, C., Xin, X., & Zhang, J. (2021). Domain Adaptation Using a Three-Way Decision Improves the Identification of Autism Patients from Multisite fMRI Data. Brain Sciences, 11(5), 603. https://doi.org/10.3390/brainsci11050603




