Cellular Localization of gdnf in Adult Zebrafish Brain
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animal Care and Handling
2.2. Brain Dissection and Tissue Processing
2.3. In Situ Hybridization
2.4. Combined Fluorescence in Situ Hybridization and Immunohistochemistry
2.5. Microscopy
3. Results
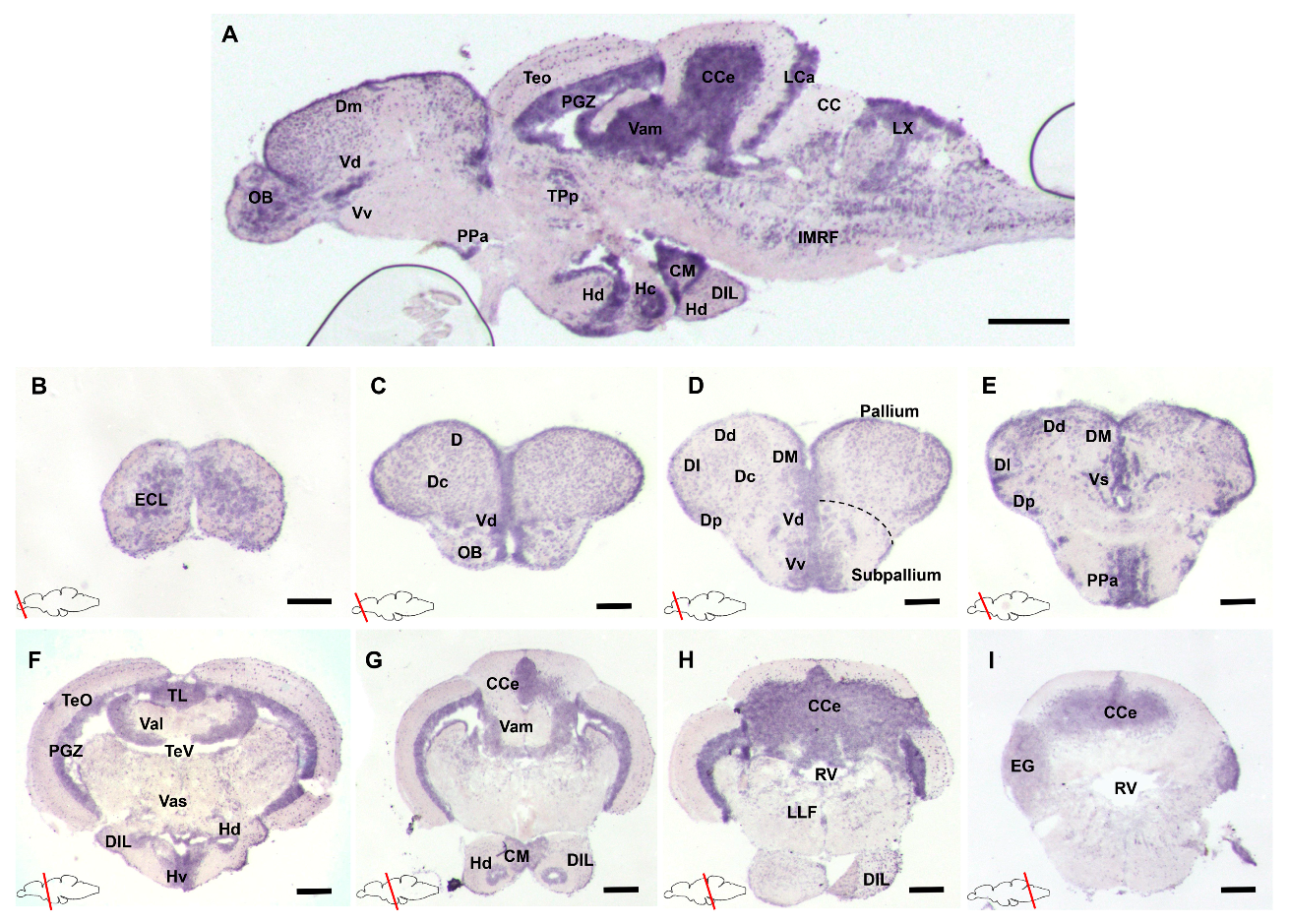
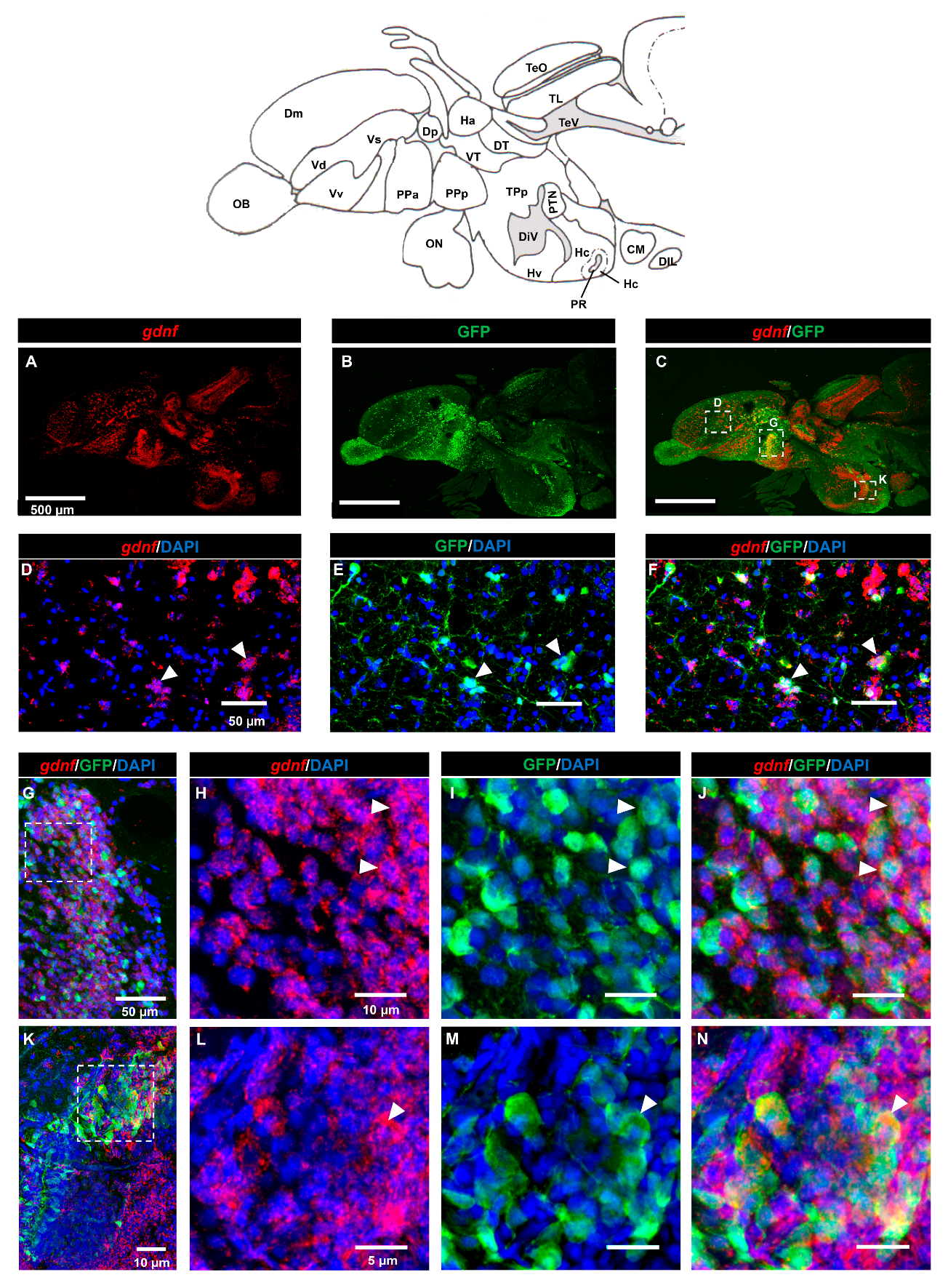
3.1. Characterization of gdnf Expression Patterns in the Adult Zebrafish Brain
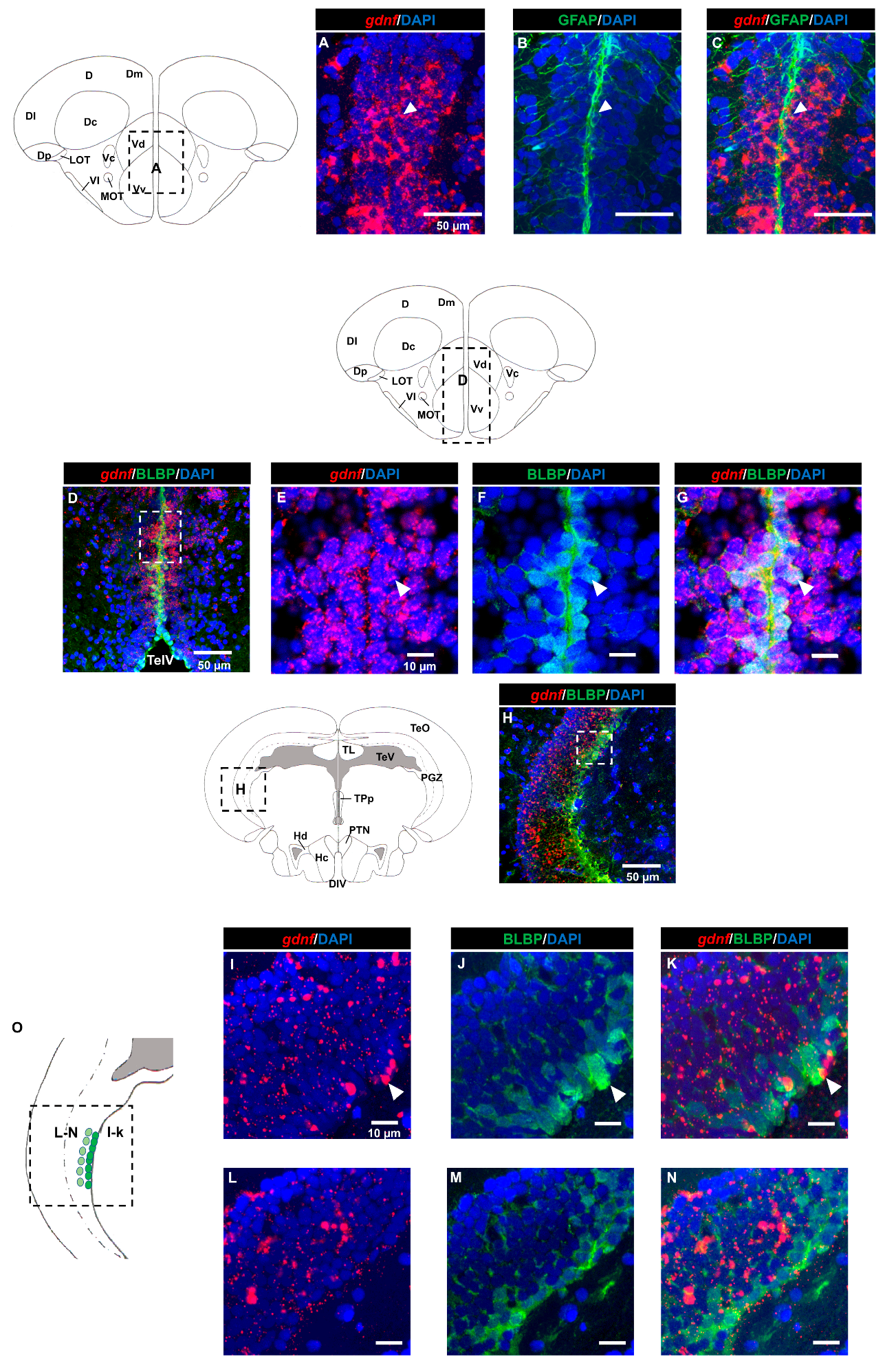
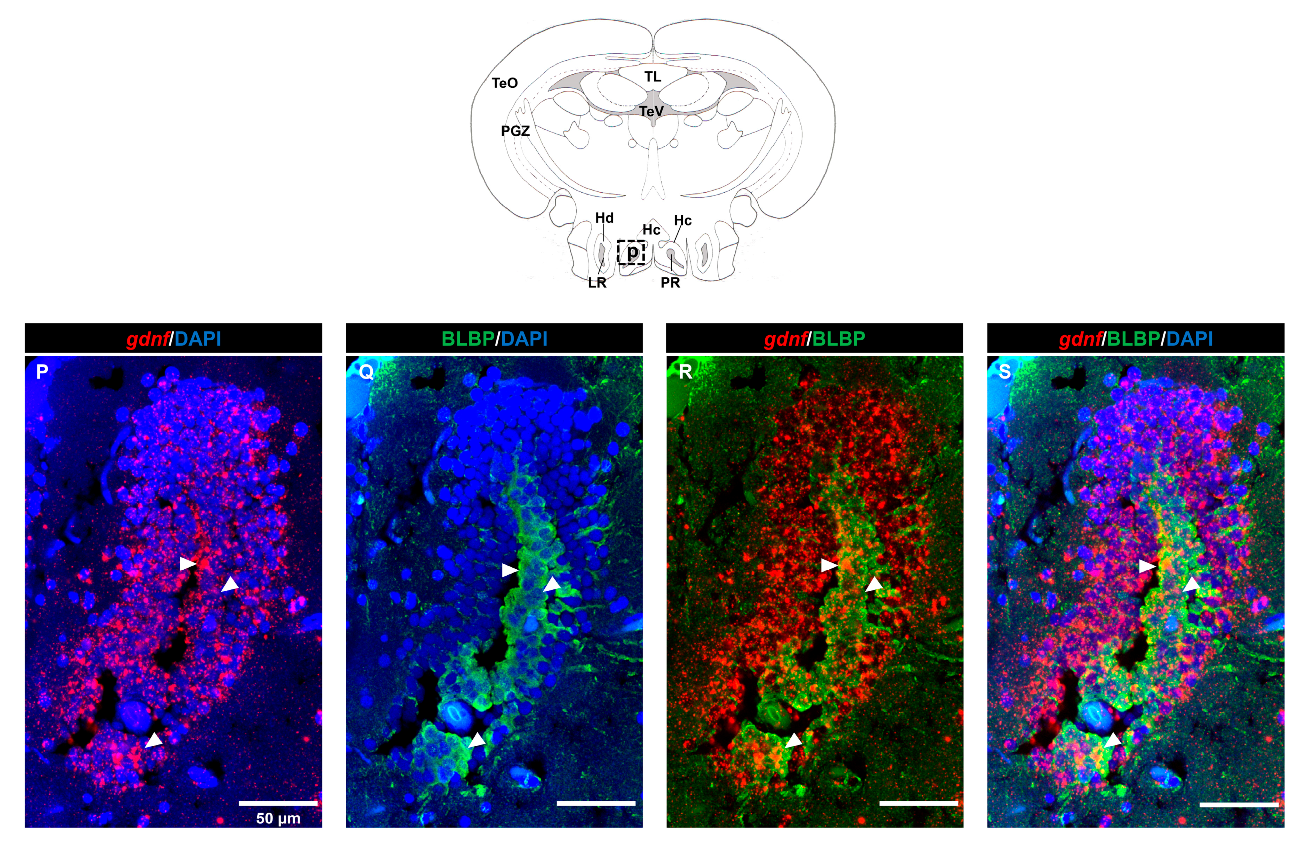
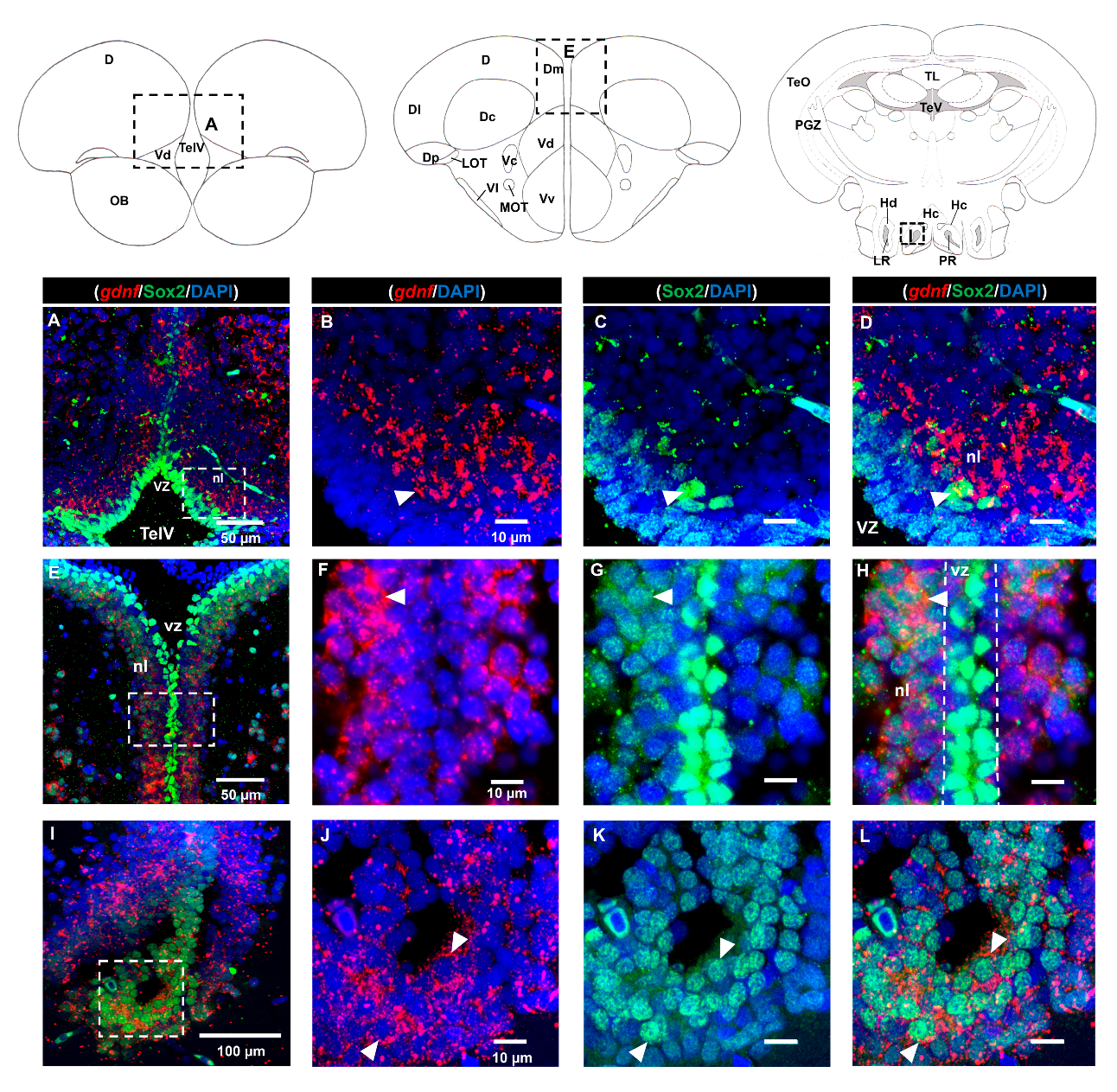
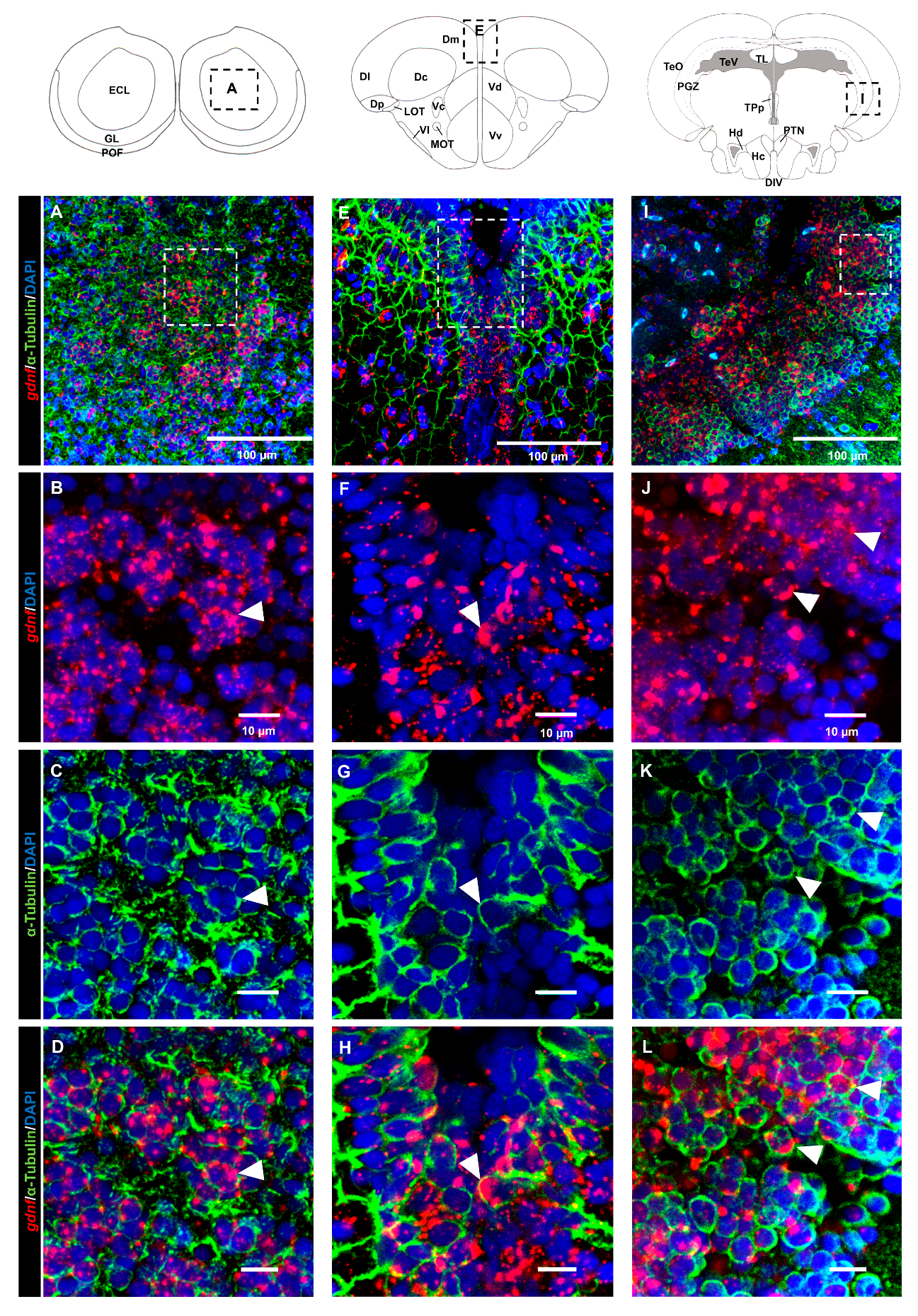
3.2. Characterization of gdnf Expression in Stem/Progenitor Cells
3.3. gdnf is Synthesized in Both Early Differentiated Neurons and Mature Neurons
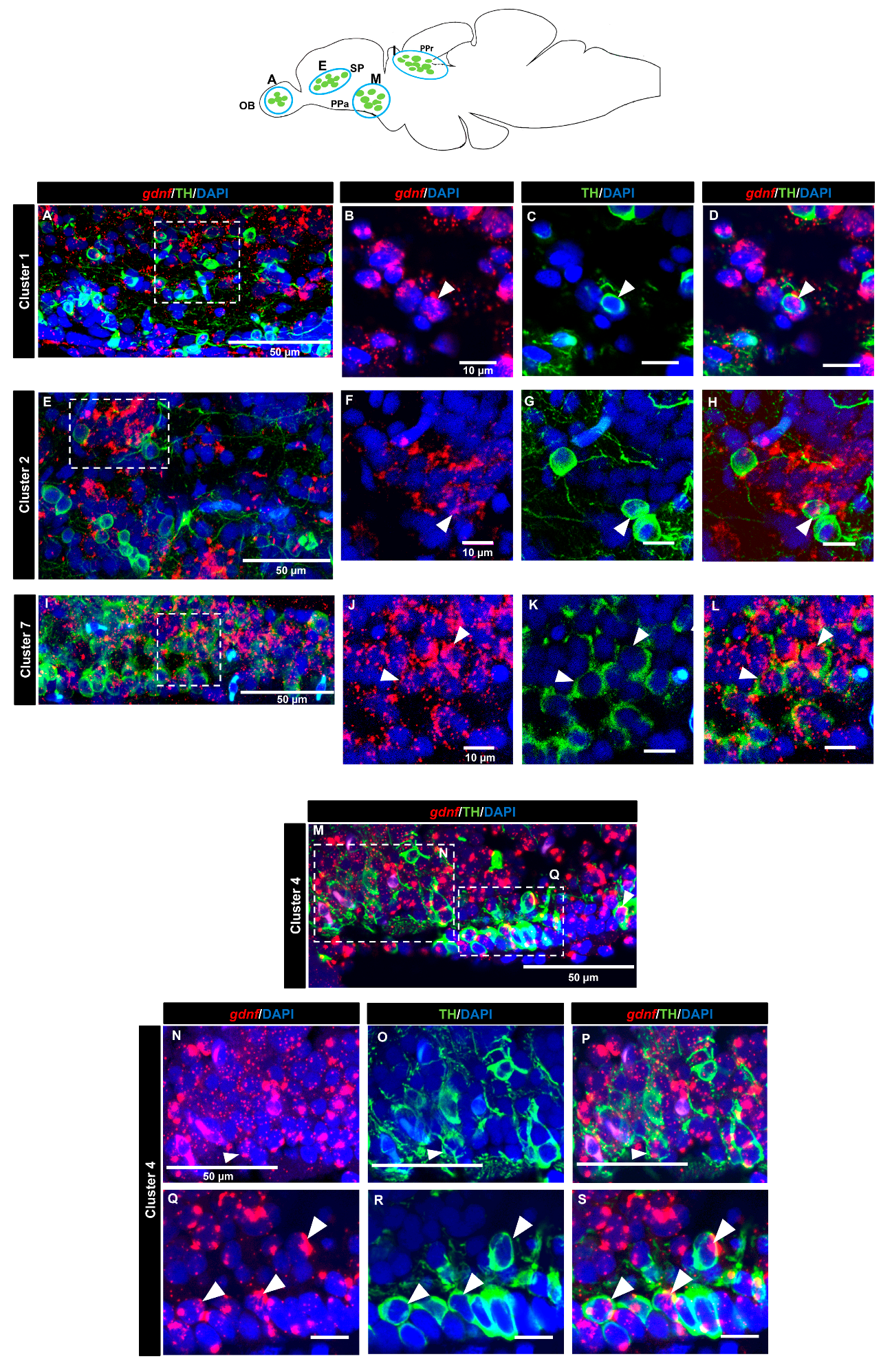
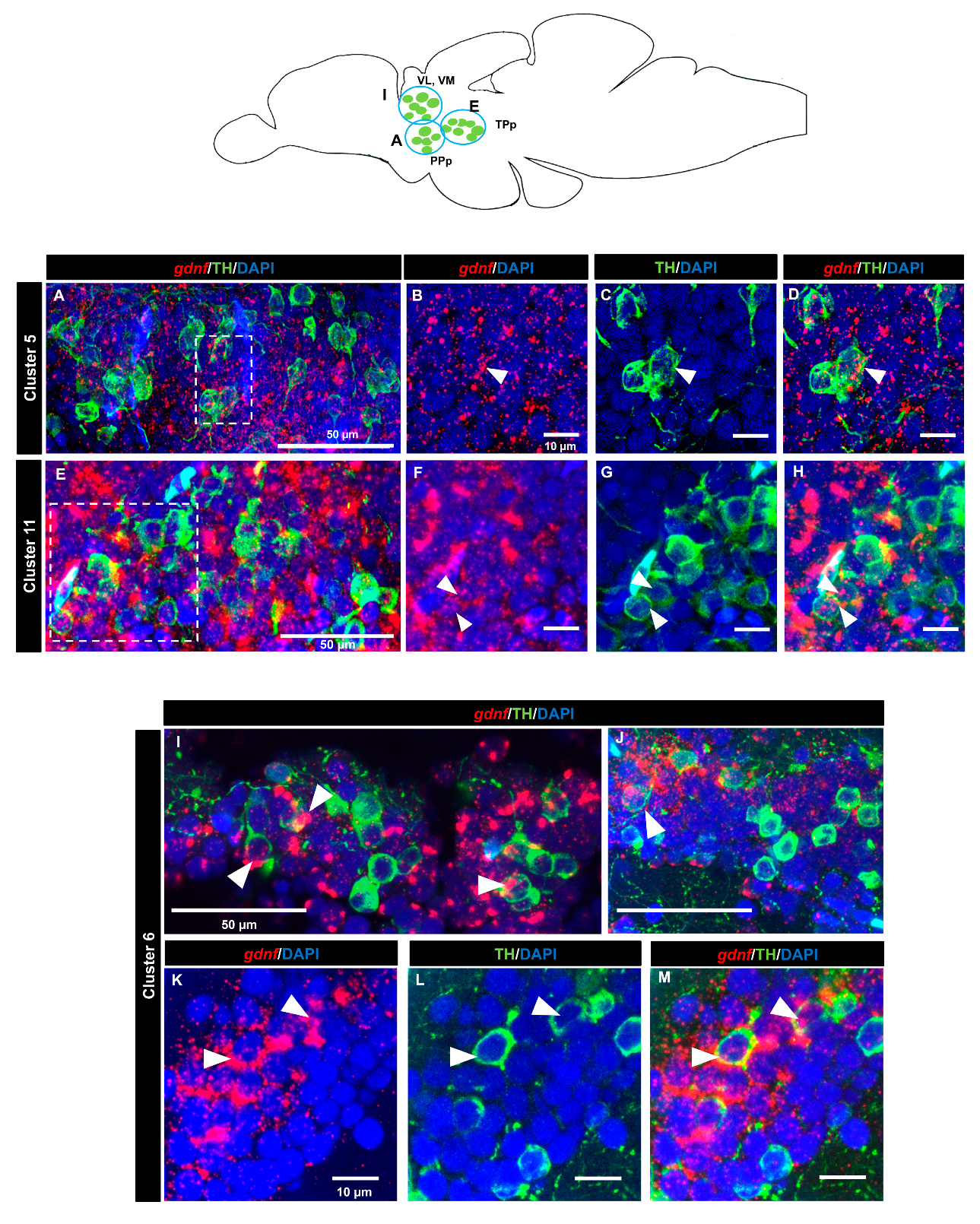
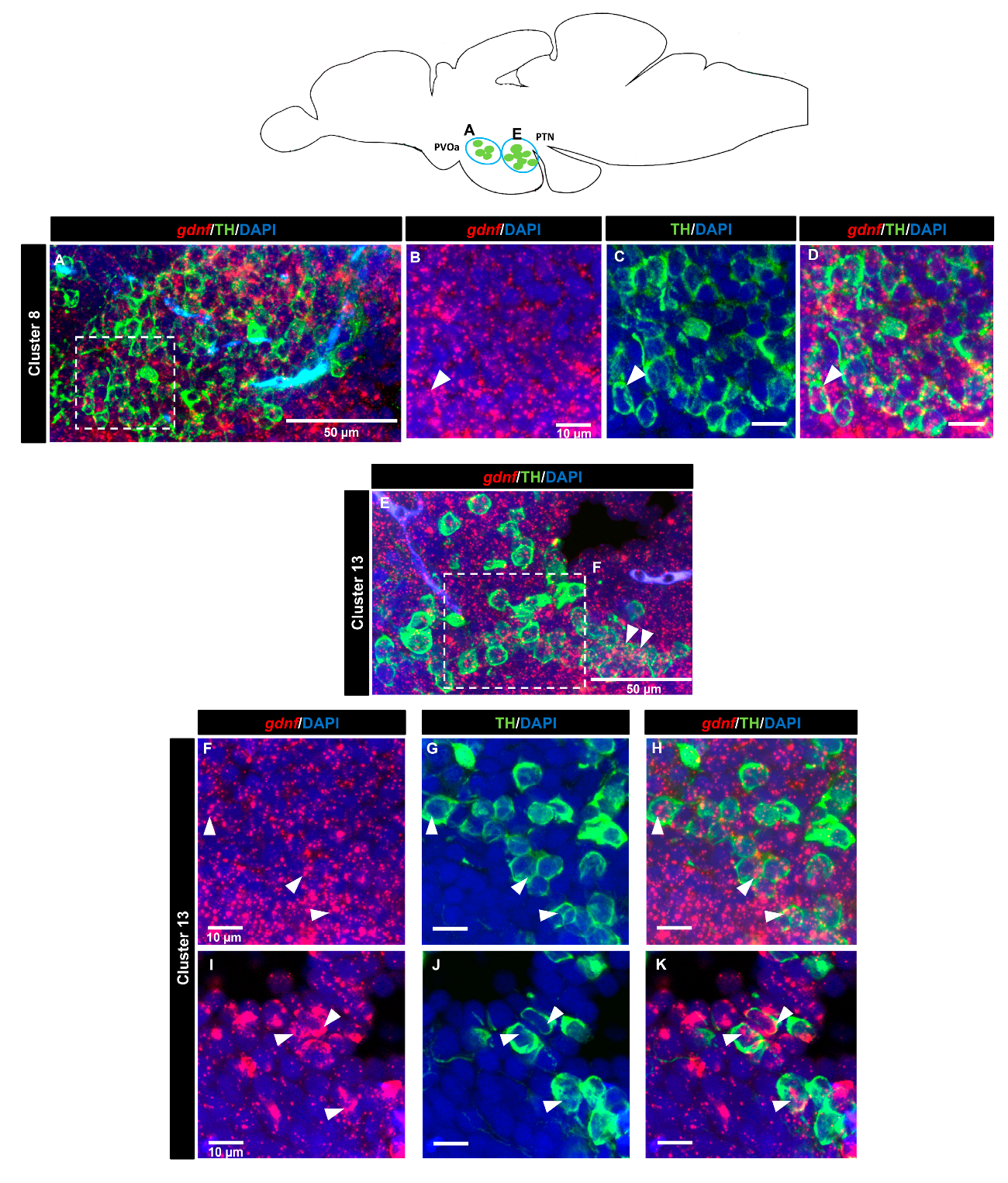
3.4. gdnf is Co-Expressed in the Subpopulation of Tyrosine Hydroxylase Immunoreactive Neurons
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Paratcha, G.; Ledda, F.; Ibáñez, C.F. The Neural Cell Adhesion Molecule NCAM Is an Alternative Signaling Receptor for GDNF Family Ligands. Cell 2003, 113, 867–879. [Google Scholar] [CrossRef] [Green Version]
- Ibáñez, C.F. Structure and Physiology of the RET Receptor Tyrosine Kinase. Cold Spring Harb. Perspect. Biol. 2013, 5, 009134. [Google Scholar] [CrossRef] [PubMed]
- Ibáñez, C.F.; Andressoo, J.-O. Biology of GDNF and Its Receptors—Relevance for Disorders of the Central Nervous System. Neurobiol. Dis. 2017, 97, 80–89. [Google Scholar] [CrossRef]
- Lin, L.F.; Doherty, D.H.; Lile, J.D.; Bektesh, S.; Collins, F. GDNF: A Glial Cell Line-Derived Neurotrophic Factor for Midbrain Dopaminergic Neurons. Science 1993, 260, 1130–1132. [Google Scholar] [CrossRef]
- Quintino, L.; Avallone, M.; Brännstrom, E.; Kavanagh, P.; Lockowandt, M.; Garcia Jareño, P.; Breger, L.S.; Lundberg, C. GDNF-Mediated Rescue of the Nigrostriatal System Depends on the Degree of Degeneration. Gene Ther. 2019, 26, 57–64. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.; Li, X.; Ge, G.; Liu, J.; Biju, K.C.; Laing, S.D.; Qian, Y.; Ballard, C.; He, Z.; Masliah, E.; et al. GDNF-Expressing Macrophages Mitigate Loss of Dopamine Neurons and Improve Parkinsonian Symptoms in MitoPark Mice. Sci. Rep. 2018, 8, 5460. [Google Scholar] [CrossRef]
- Gash, D.M.; Zhang, Z.; Ovadia, A.; Cass, W.A.; Yi, A.; Simmerman, L.; Russell, D.; Martin, D.; Lapchak, P.A.; Collins, F.; et al. Functional Recovery in Parkinsonian Monkeys Treated with GDNF. Nature 1996, 380, 252–255. [Google Scholar] [CrossRef]
- Oiwa, Y.; Nakai, K.; Itakura, T. Histological Effects of Intraputaminal Infusion of Glial Cell Line-Derived Neurotrophic Factor in Parkinson Disease Model Macaque Monkeys. Neurol. Med. Chir. 2006, 46, 267–276. [Google Scholar] [CrossRef] [Green Version]
- Enterría-Morales, D.; López-López, I.; López-Barneo, J.; d’Anglemont de Tassigny, X. Role of Glial Cell Line-Derived Neurotrophic Factor in the Maintenance of Adult Mesencephalic Catecholaminergic Neurons. Mov. Disord. 2020, 35, 565–576. [Google Scholar]
- Henderson, C.E.; Phillips, H.S.; Pollock, R.A.; Davies, A.M.; Lemeulle, C.; Armanini, M.; Simmons, L.; Moffet, B.; Vandlen, R.A.; to Simmons, L.; et al. GDNF: A Potent Survival Factor for Motoneurons Present in Peripheral Nerve and Muscle. Science 1994, 266, 1062–1064. [Google Scholar] [CrossRef]
- Moore, M.W.; Klein, R.D.; Fariñas, I.; Sauer, H.; Armanini, M.; Phillips, H.; Reichardt, L.F.; Ryan, A.M.; Carver-Moore, K.; Rosenthal, A. Renal and Neuronal Abnormalities in Mice Lacking GDNF. Nature 1996, 382, 76–79. [Google Scholar] [CrossRef] [PubMed]
- Koo, H.; Choi, B.H. Expression of Glial Cell Line-Derived Neurotrophic Factor (GDNF) in the Developing Human Fetal Brain. Int. J. Dev. Neurosci. 2001, 19, 549–558. [Google Scholar] [CrossRef]
- Hellmich, H.L.; Kos, L.; Cho, E.S.; Mahon, K.A.; Zimmer, A. Embryonic Expression of Glial Cell-Line Derived Neurotrophic Factor (GDNF) Suggests Multiple Developmental Roles in Neural Differentiation and Epithelial-Mesenchymal Interactions. Mech. Dev. 1996, 54, 95–105. [Google Scholar] [CrossRef]
- Cortés, D.; Carballo-Molina, O.A.; Castellanos-Montiel, M.J.; Velasco, I. The Non-Survival Effects of Glial Cell Line-Derived Neurotrophic Factor on Neural Cells. Front. Mol. Neurosci. 2017, 10, 258. [Google Scholar] [CrossRef] [PubMed]
- Pascual, A.; López-Barneo, J. Reply to “GDNF Is Not Required for Catecholaminergic Neuron Survival in Vivo”. Nat. Neurosci. 2015, 18, 322. [Google Scholar] [CrossRef]
- Kopra, J.; Vilenius, C.; Grealish, S.; Härma, M.-A.; Varendi, K.; Lindholm, J.; Castrén, E.; Võikar, V.; Björklund, A.; Piepponen, T.P.; et al. GDNF Is Not Required for Catecholaminergic Neuron Survival in Vivo. Nat. Neurosci. 2015, 18, 319. [Google Scholar] [CrossRef] [Green Version]
- Pascual, A.; Hidalgo-Figueroa, M.; Piruat, J.I.; Pintado, C.O.; Gómez-Díaz, R.; López-Barneo, J. Absolute Requirement of GDNF for Adult Catecholaminergic Neuron Survival. Nat. Neurosci. 2008, 11, 755. [Google Scholar] [CrossRef] [Green Version]
- Kumar, A.; Kopra, J.; Varendi, K.; Porokuokka, L.L.; Panhelainen, A.; Kuure, S.; Marshall, P.; Karalija, N.; Härma, M.-A.; Vilenius, C.; et al. GDNF Overexpression from the Native Locus Reveals Its Role in the Nigrostriatal Dopaminergic System Function. PLoS Genet. 2015, 11, 1005710. [Google Scholar] [CrossRef] [Green Version]
- Whone, A.; Luz, M.; Boca, M.; Woolley, M.; Mooney, L.; Dharia, S.; Broadfoot, J.; Cronin, D.; Schroers, C.; Barua, N.U.; et al. Randomized Trial of Intermittent Intraputamenal Glial Cell Line-Derived Neurotrophic Factor in Parkinson’s Disease. Brain 2019, 142, 512–525. [Google Scholar] [CrossRef] [Green Version]
- Pitchai, A.; Rajaretinam, R.K.; Freeman, J.L. Zebrafish as an Emerging Model for Bioassay-Guided Natural Product Drug Discovery for Neurological Disorders. Medicines 2019, 6, 61. [Google Scholar] [CrossRef] [Green Version]
- Friedrich, R.; Genoud, C.; Wanner, A. Analyzing the Structure and Function of Neuronal Circuits in Zebrafish. Front. Neural Circuits 2013, 7, 71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shepherd, I.T.; Beattie, C.E.; Raible, D.W. Functional Analysis of Zebrafish GDNF. Dev. Biol. 2001, 231, 420–435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schuster, K.; Dambly-Chaudière, C.; Ghysen, A. Glial Cell Line-Derived Neurotrophic Factor Defines the Path of Developing and Regenerating Axons in the Lateral Line System of Zebrafish. Proc. Natl. Acad. Sci. USA 2010, 107, 19531–19536. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, B.-K.; Yuan, R.-Y.; Lien, H.-W.; Hung, C.-C.; Hwang, P.-P.; Chen, R.P.-Y.; Chang, C.-C.; Liao, Y.-F.; Huang, C.-J. Multiple Signaling Factors and Drugs Alleviate Neuronal Death Induced by Expression of Human and Zebrafish Tau Proteins in Vivo. J. Biomed. Sci. 2016, 23, 25. [Google Scholar] [CrossRef] [Green Version]
- Zerucha, T.; Stühmer, T.; Hatch, G.; Park, B.K.; Long, Q.; Yu, G.; Gambarotta, A.; Schultz, J.R.; Rubenstein, J.L.R.; Ekker, M. A Highly Conserved Enhancer in the Dlx5/Dlx6Intergenic Region Is the Site of Cross-Regulatory Interactions BetweenDlx Genes in the Embryonic Forebrain. J. Neurosci. 2000, 20, 709–721. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cacialli, P.; Gueguen, M.-M.; Coumailleau, P.; D’Angelo, L.; Kah, O.; Lucini, C.; Pellegrini, E. BDNF Expression in Larval and Adult Zebrafish Brain: Distribution and Cell Identification. PLoS ONE 2016, 11, 1–22. [Google Scholar] [CrossRef] [Green Version]
- Wullimann, M.F.; Rupp, B.; Reichert, H. Neuroanatomy of the Zebrafish Brain. A Topological Atlas; Birkhauser: Basel, Switzerland, 1996. [Google Scholar]
- Hidalgo-Figueroa, M.; Bonilla, S.; Gutiérrez, F.; Pascual, A.; López-Barneo, J. GDNF Is Predominantly Expressed in the PV+ Neostriatal Interneuronal Ensemble in Normal Mouse and after Injury of the Nigrostriatal Pathway. J. Neurosci. 2012, 32, 864–872. [Google Scholar] [CrossRef]
- Lucini, C.; Maruccio, L.; Patruno, M.; Tafuri, S.; Staiano, N.; Mascarello, F.; Castaldo, L. Glial Cell Line-Derived Neurotrophic Factor Expression in the Brain of Adult Zebrafish (Danio Rerio). Histol. Histopathol. 2008, 23, 251–261. [Google Scholar]
- Pochon, N.A.-M.; Menoud, A.; Tseng, J.L.; Zurn, A.D.; Aebischer, P. Neuronal GDNF Expression in the Adult Rat Nervous System Identified By In Situ Hybridization. Eur. J. Neurosci. 1997, 9, 463–471. [Google Scholar] [CrossRef]
- Ortega-de San Luis, C.; Pascual, A. Simultaneous Detection of Both GDNF and GFRα1 Expression Patterns in the Mouse Central Nervous System. Front. Neuroanat. 2016, 10, 73. [Google Scholar] [CrossRef] [Green Version]
- Trupp, M.; Belluardo, N.; Funakoshi, H.; Ibáñez, C.F. Complementary and Overlapping Expression of Glial Cell Line-Derived Neurotrophic Factor (GDNF), c-Ret Proto-Oncogene, and GDNF Receptor-α Indicates Multiple Mechanisms of Trophic Actions in the Adult Rat CNS. J. Neurosci. 1997, 17, 3554–3567. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- März, M.; Chapouton, P.; Diotel, N.; Vaillant, C.; Hesl, B.; Takamiya, M.; Lam, C.S.; Kah, O.; Bally-Cuif, L.; Strähle, U. Heterogeneity in Progenitor Cell Subtypes in the Ventricular Zone of the Zebrafish Adult Telencephalon. Glia 2010, 58, 870–888. [Google Scholar] [CrossRef] [PubMed]
- Kizil, C.; Kaslin, J.; Kroehne, V.; Brand, M. Adult Neurogenesis and Brain Regeneration in Zebrafish. Dev. Neurobiol. 2012, 72, 429–461. [Google Scholar] [CrossRef] [PubMed]
- GermanÀ, A.; Montalbano, G.; Guerrera, M.C.; Amato, V.; LaurÀ, R.; Magnoli, D.; Campo, S.; Suarez-Fernandez, E.; Ciriaco, E.; Vega, J.A. Developmental Changes in the Expression of Sox2 in the Zebrafish Brain. Microsc. Res. Tech. 2011, 74, 347–354. [Google Scholar] [CrossRef]
- Ferri, A.L.M.; Cavallaro, M.; Braida, D.; Di Cristofano, A.; Canta, A.; Vezzani, A.; Ottolenghi, S.; Pandolfi, P.P.; Sala, M.; Debiasi, S.; et al. Sox2 Deficiency Causes Neurodegeneration and Impaired Neurogenesis in the Adult Mouse Brain. Dev. Dis. 2004, 131, 3805–3819. [Google Scholar] [CrossRef] [Green Version]
- Kaslin, J.; Ganz, J.; Geffarth, M.; Grandel, H.; Hans, S.; Brand, M. Stem Cells in the Adult Zebrafish Cerebellum: Initiation and Maintenance of a Novel Stem Cell Niche. J. Neurosci. 2009, 29, 6142–6153. [Google Scholar] [CrossRef] [Green Version]
- Duarte Azevedo, M.; Sander, S.; Tenenbaum, L. GDNF, A Neuron-Derived Factor Upregulated in Glial Cells during Disease. J. Clin. Med. 2020, 9, 456. [Google Scholar] [CrossRef] [Green Version]
- Bakshi, A.; Shimizu, S.; Keck, C.A.; Cho, S.; LeBold, D.G.; Morales, D.; Arenas, E.; Snyder, E.Y.; Watson, D.J.; McIntosh, T.K. Neural Progenitor Cells Engineered to Secrete GDNF Show Enhanced Survival, Neuronal Differentiation and Improve Cognitive Function Following Traumatic Brain Injury. Eur. J. Neurosci. 2006, 23, 2119–2134. [Google Scholar] [CrossRef]
- Gao, X.; Deng, L.; Wang, Y.; Yin, L.; Yang, C.; Du, J.; Yuan, Q. GDNF Enhances Therapeutic Efficiency of Neural Stem Cells-Based Therapy in Chronic Experimental Allergic Encephalomyelitis in Rat. Stem Cells Int. 2016, 2016, 1431349. [Google Scholar] [CrossRef] [Green Version]
- Deng, X.; Liang, Y.; Lu, H.; Yang, Z.; Liu, R.; Wang, J.; Song, X.; Long, J.; Li, Y.; Lei, D.; et al. Co-Transplantation of GDNF-Overexpressing Neural Stem Cells and Fetal Dopaminergic Neurons Mitigates Motor Symptoms in a Rat Model of Parkinson’s Disease. PLoS ONE 2013, 8, 1–9. [Google Scholar] [CrossRef]
- Appel, E.; Kolman, O.; Kazimirsky, G.; Blumberg, P.M.; Brodie, C. Regulation of GDNF Expression in Cultured Astrocytes by Inflammatory Stimuli. Neuroreport 1997, 8, 3309–3312. [Google Scholar] [CrossRef] [PubMed]
- Moretto, G.; Walker, D.G.; Lanteri, P.; Taioli, F.; Zaffagnini, S.; Xu, R.Y.; Rizzuto, N. Expression and Regulation of Glial-Cell-Line-Derived Neurotrophic Factor (GDNF) MRNA in Human Astrocytes in Vitro. Cell Tissue Res. 1996, 286, 257–262. [Google Scholar] [CrossRef] [PubMed]
- Nicole, O.; Ali, C.; Docagne, F.; Plawinski, L.; MacKenzie, E.T.; Vivien, D.; Buisson, A. Neuroprotection Mediated by Glial Cell Line-Derived Neurotrophic Factor: Involvement of a Reduction of NMDA-Induced Calcium Influx by the Mitogen-Activated Protein Kinase Pathway. J. Neurosci. 2001, 21, 3024–3033. [Google Scholar] [CrossRef] [PubMed]
- Ubhi, K.; Rockenstein, E.; Mante, M.; Inglis, C.; Adame, A.; Patrick, C.; Whitney, K.; Masliah, E. Neurodegeneration in a Transgenic Mouse Model of Multiple System Atrophy Is Associated with Altered Expression of Oligodendroglial-Derived Neurotrophic Factors. J. Neurosci. 2010, 30, 6236–6246. [Google Scholar] [CrossRef] [Green Version]
- März, M.; Schmidt, R.; Rastegar, S.; Strähle, U. Regenerative Response Following Stab Injury in the Adult Zebrafish Telencephalon. Dev. Dyn. 2011, 240, 2221–2231. [Google Scholar] [CrossRef]
- Kroehne, V.; Freudenreich, D.; Hans, S.; Kaslin, J.; Brand, M. Regeneration of the Adult Zebrafish Brain from Neurogenic Radial Glia-Type Progenitors. Development 2011, 138, 4831–4841. [Google Scholar] [CrossRef] [Green Version]
- Bresjanac, M.; Antauer, G. Reactive Astrocytes of the Quinolinic Acid-Lesioned Rat Striatum Express GFRα1 as Well as GDNF in Vivo. Exp. Neurol. 2000, 164, 53–59. [Google Scholar] [CrossRef]
- Chapouton, P.; Skupien, P.; Hesl, B.; Coolen, M.; Moore, J.C.; Madelaine, R.; Kremmer, E.; Faus-Kessler, T.; Blader, P.; Lawson, N.D.; et al. Notch Activity Levels Control the Balance between Quiescence and Recruitment of Adult Neural Stem Cells. J. Neurosci. 2010, 30, 7961–7974. [Google Scholar] [CrossRef] [Green Version]
- de Oliveira-Carlos, V.; Ganz, J.; Hans, S.; Kaslin, J.; Brand, M. Notch Receptor Expression in Neurogenic Regions of the Adult Zebrafish Brain. PLoS ONE 2013, 8, 73384. [Google Scholar] [CrossRef] [Green Version]
- Khazaei, M.; Ahuja, C.S.; Nakashima, H.; Nagoshi, N.; Li, L.; Wang, J.; Chio, J.; Badner, A.; Seligman, D.; Ichise, A.; et al. GDNF Rescues the Fate of Neural Progenitor Grafts by Attenuating Notch Signals in the Injured Spinal Cord in Rodents. Sci. Transl. Med. 2020, 12, 3538. [Google Scholar] [CrossRef]
- Ito, Y.; Tanaka, H.; Okamoto, H.; Ohshima, T. Characterization of Neural Stem Cells and Their Progeny in the Adult Zebrafish Optic Tectum. Dev. Biol. 2010, 342, 26–38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shimizu, Y.; Ueda, Y.; Ohshima, T. Wnt Signaling Regulates Proliferation and Differentiation of Radial Glia in Regenerative Processes after Stab Injury in the Optic Tectum of Adult Zebrafish. Glia 2018, 66, 1382–1394. [Google Scholar] [CrossRef] [PubMed]
- Lindsey, B.W.; Aitken, G.E.; Tang, J.K.; Khabooshan, M.; Douek, A.M.; Vandestadt, C.; Kaslin, J. Midbrain Tectal Stem Cells Display Diverse Regenerative Capacities in Zebrafish. Sci. Rep. 2019, 9, 4420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- MacDonald, R.B.; Debiais-Thibaud, M.; Talbot, J.C.; Ekker, M. The Relationship between Dlx and Gad1 Expression Indicates Highly Conserved Genetic Pathways in the Zebrafish Forebrain. Dev. Dyn. 2010, 239, 2298–2306. [Google Scholar] [CrossRef] [Green Version]
- Yu, M.; Xi, Y.; Pollack, J.; Debiais-Thibaud, M.; MacDonald, R.B.; Ekker, M. Activity of Dlx5a/Dlx6a Regulatory Elements during Zebrafish GABAergic Neuron Development. Int. J. Dev. Neurosci. 2011, 29, 681–691. [Google Scholar] [CrossRef]
- Solek, C.M.; Feng, S.; Perin, S.; Weinschutz Mendes, H.; Ekker, M. Lineage Tracing of Dlx1a/2a and Dlx5a/6a Expressing Cells in the Developing Zebrafish Brain. Dev. Biol. 2017, 427, 131–147. [Google Scholar] [CrossRef]
- d’Anglemont de Tassigny, X.; Pascual, A.; López-Barneo, J. GDNF-Based Therapies, GDNF-Producing Interneurons, and Trophic Support of the Dopaminergic Nigrostriatal Pathway. Implications for Parkinson’s Disease. Front. Neuroanat. 2015, 9, 10. [Google Scholar]
- Adolf, B.; Chapouton, P.; Lam, C.S.; Topp, S.; Tannhäuser, B.; Strähle, U.; Götz, M.; Bally-Cuif, L. Conserved and Acquired Features of Adult Neurogenesis in the Zebrafish Telencephalon. Dev. Biol. 2006, 295, 278–293. [Google Scholar] [CrossRef] [Green Version]
- Blandini, F.; Cova, L.; Armentero, M.-T.; Zennaro, E.; Levandis, G.; Bossolasco, P.; Calzarossa, C.; Mellone, M.; Giuseppe, B.; Deliliers, G.L.; et al. Transplantation of Undifferentiated Human Mesenchymal Stem Cells Protects against 6-Hydroxydopamine Neurotoxicity in the Rat. Cell Transplant. 2010, 19, 203–218. [Google Scholar] [CrossRef]
- Chauhan, N.B.; Siegel, G.J.; Lee, J.M. Depletion of Glial Cell Line-Derived Neurotrophic Factor in Substantia Nigra Neurons of Parkinson’s Disease Brain. J. Chem. Neuroanat. 2001, 21, 277–288. [Google Scholar] [CrossRef]
- Tay, T.L.; Ronneberger, O.; Ryu, S.; Nitschke, R.; Driever, W. Comprehensive Catecholaminergic Projectome Analysis Reveals Single-Neuron Integration of Zebrafish Ascending and Descending Dopaminergic Systems. Nat Commun 2011, 2, 171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaslin, J.; Panula, P. Comparative Anatomy of the Histaminergic and Other Aminergic Systems in Zebrafish (Danio Rerio). J. Comp. Neurol. 2001, 440, 342–377. [Google Scholar] [CrossRef] [PubMed]
- Rink, E.; Wullimann, M.F. The Teleostean (Zebrafish) Dopaminergic System Ascending to the Subpallium (Striatum) Is Located in the Basal Diencephalon (Posterior Tuberculum). Brain Res. 2001, 889, 316–330. [Google Scholar] [CrossRef]
- Sallinen, V.; Torkko, V.; Sundvik, M.; Reenilä, I.; Khrustalyov, D.; Kaslin, J.; Panula, P. MPTP and MPP+ Target Specific Aminergic Cell Populations in Larval Zebrafish. J. Neurochem. 2009, 108, 719–731. [Google Scholar] [CrossRef]
- Panula, P.; Chen, Y.-C.; Priyadarshini, M.; Kudo, H.; Semenova, S.; Sundvik, M.; Sallinen, V. The Comparative Neuroanatomy and Neurochemistry of Zebrafish CNS Systems of Relevance to Human Neuropsychiatric Diseases. Neurobiol. Dis. 2010, 40, 46–57. [Google Scholar] [CrossRef]
- Barroso-Chinea, P.; Cruz-Muros, I.; Aymerich, M.S.; Rodríguez-Díaz, M.; Afonso-Oramas, D.; Lanciego, J.L.; González-Hernández, T. Striatal Expression of GDNF and Differential Vulnerability of Midbrain Dopaminergic Cells. Eur. J. Neurosci. 2005, 21, 1815–1827. [Google Scholar] [CrossRef] [Green Version]
- Bretaud, S.; Lee, S.; Guo, S. Sensitivity of Zebrafish to Environmental Toxins Implicated in Parkinson’s Disease. Neurotoxicol. Teratol. 2004, 26, 857–864. [Google Scholar] [CrossRef]
- Lam, C.S.; Korzh, V.; Strahle, U. Zebrafish Embryos Are Susceptible to the Dopaminergic Neurotoxin MPTP. Eur. J. Neurosci. 2005, 21, 1758–1762. [Google Scholar] [CrossRef]
- McKinley, E.T.; Baranowski, T.C.; Blavo, D.O.; Cato, C.; Doan, T.N.; Rubinstein, A.L. Neuroprotection of MPTP-Induced Toxicity in Zebrafish Dopaminergic Neurons. Brain Res. Mol. Brain Res. 2005, 141, 128–137. [Google Scholar] [CrossRef]
- Stednitz, S.J.; Freshner, B.; Shelton, S.; Shen, T.; Black, D.; Gahtan, E. Selective Toxicity of L-DOPA to Dopamine Transporter-Expressing Neurons and Locomotor Behavior in Zebrafish Larvae. Neurotoxicol. Teratol. 2015, 52, 51–56. [Google Scholar] [CrossRef] [Green Version]
- Wen, L.; Wei, W.; Gu, W.; Huang, P.; Ren, X.; Zhang, Z.; Zhu, Z.; Lin, S.; Zhang, B. Visualization of Monoaminergic Neurons and Neurotoxicity of MPTP in Live Transgenic Zebrafish. Dev. Biol. 2008, 314, 84–92. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kalyn, M.; Hua, K.; Mohd Noor, S.; Wong, C.E.D.; Ekker, M. Comprehensive Analysis of Neurotoxin-Induced Ablation of Dopaminergic Neurons in Zebrafish Larvae. Biomedicines 2020, 8, 1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, T.; Zondervan-van der Linde, H.; Severijnen, L.-A.; Oostra, B.A.; Willemsen, R.; Bonifati, V. Dopaminergic Neuronal Loss and Dopamine-Dependent Locomotor Defects in Fbxo7-Deficient Zebrafish. PLoS ONE 2012, 7, 48911. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sheng, D.; Qu, D.; Kwok, K.H.H.; Ng, S.S.; Lim, A.Y.M.; Aw, S.S.; Lee, C.W.H.; Sung, W.K.; Tan, E.K.; Lufkin, T.; et al. Deletion of the WD40 Domain of LRRK2 in Zebrafish Causes Parkinsonism-Like Loss of Neurons and Locomotive Defect. PLoS Genet. 2010, 6, 1000914. [Google Scholar] [CrossRef] [Green Version]
- Soman, S.; Keatinge, M.; Moein, M.; Da Costa, M.; Mortiboys, H.; Skupin, A.; Sugunan, S.; Bazala, M.; Kuznicki, J.; Bandmann, O. Inhibition of the Mitochondrial Calcium Uniporter Rescues Dopaminergic Neurons in Pink1(-/-) Zebrafish. Eur. J. Neurosci. 2017, 45, 528–535. [Google Scholar] [CrossRef]
- Anichtchik, O.; Diekmann, H.; Fleming, A.; Roach, A.; Goldsmith, P.; Rubinsztein, D.C. Loss of PINK1 Function Affects Development and Results in Neurodegeneration in Zebrafish. J. Neurosci. 2008, 28, 8199–8207. [Google Scholar] [CrossRef]
- Young, A.; Assey, K.S.; Sturkie, C.D.; West, F.D.; Machacek, D.W.; Stice, S.L. Glial Cell Line-Derived Neurotrophic Factor Enhances in Vitro Differentiation of Mid-/Hindbrain Neural Progenitor Cells to Dopaminergic-like Neurons. J. Neurosci. Res. 2010, 88, 3222–3232. [Google Scholar] [CrossRef]
- Jo, J.; Xiao, Y.; Sun, A.X.; Cukuroglu, E.; Tran, H.-D.; Göke, J.; Tan, Z.Y.; Saw, T.Y.; Tan, C.-P.; Lokman, H.; et al. Midbrain-like Organoids from Human Pluripotent Stem Cells Contain Functional Dopaminergic and Neuromelanin-Producing Neurons. Cell Stem Cell 2016, 19, 248–257. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Liu, X.; Zhang, Y.; Luan, Z.; Yang, Y.; Wang, Z.; Zhang, C. Human Neural Stem Cells with GDNF Site-Specific Integration at AAVS1 by Using AAV Vectors Retained Their Stemness. Neurochem. Res. 2018, 43, 930–937. [Google Scholar] [CrossRef]
- Wang, S.; Zou, C.; Fu, L.; Wang, B.; An, J.; Song, G.; Wu, J.; Tang, X.; Li, M.; Zhang, J.; et al. Autologous IPSC-Derived Dopamine Neuron Transplantation in a Nonhuman Primate Parkinson’s Disease Model. Cell Discov. 2015, 1, 15012. [Google Scholar] [CrossRef] [Green Version]

| Host | Target | Dilution | Catalog |
|---|---|---|---|
| Mouse | HuC/HuD | 1:200 | A-2127 (Invitrogen) |
| Rabbit | Sox2 | 1:200 | Ab97959 (Abcam) |
| Rabbit | Tyrosine hydroxylase (TH) | 1:200 | AB152 (Merck Millipore) |
| Mouse | Green fluorescence protein (GFP) | 1:200 | 632381 (Clontech) |
| Rabbit | Brain lipid-binding protein (BLBP) | 1:200 | ABN14 (Merck Millipore) |
| Rabbit | Glial fibrillary acidic protein (GFAP) | 1:200 | Z0334 (Dako) |
| Mouse | Acetylated α-tubulins | 1:200 | T7451 (Sigma-Aldrich) |
| Brain Areas | TH+ Groups | Expression Level | |
|---|---|---|---|
| Telencephalon | Olfactory bulb (OB) | Cluster 1 | + |
| Subpallium (SP) | Cluster 2 | + | |
| Diencephalon | Preoptic area | Cluster 3 | ND |
| Cluster 4 | +++ | ||
| Prethalamic | Cluster 5 | + | |
| Cluster 6 | ++ | ||
| Cluster 11 | ++ | ||
| Periventricular pretectal nucleus (PPr) | Cluster 7 | +++ | |
| Anterior part of paraventricular organ (PVOa) | Cluster 8 | ++ | |
| Intermediate part of paraventricular organ (PVOi) | Cluster 9 | ND | |
| posterior part of paraventricular organ (PVOp) | Cluster 10 | + | |
| Posterior tuberculum (TP) | Cluster 12 | ND | |
| Posterior tuberal nucleus (PTN) | Cluster 13 | ++ |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wong, C.E.D.; Hua, K.; Monis, S.; Norazit, A.; Noor, S.M.; Ekker, M. Cellular Localization of gdnf in Adult Zebrafish Brain. Brain Sci. 2020, 10, 286. https://doi.org/10.3390/brainsci10050286
Wong CED, Hua K, Monis S, Norazit A, Noor SM, Ekker M. Cellular Localization of gdnf in Adult Zebrafish Brain. Brain Sciences. 2020; 10(5):286. https://doi.org/10.3390/brainsci10050286
Chicago/Turabian StyleWong, Chee Ern David, Khang Hua, Simon Monis, Anwar Norazit, Suzita Mohd Noor, and Marc Ekker. 2020. "Cellular Localization of gdnf in Adult Zebrafish Brain" Brain Sciences 10, no. 5: 286. https://doi.org/10.3390/brainsci10050286
APA StyleWong, C. E. D., Hua, K., Monis, S., Norazit, A., Noor, S. M., & Ekker, M. (2020). Cellular Localization of gdnf in Adult Zebrafish Brain. Brain Sciences, 10(5), 286. https://doi.org/10.3390/brainsci10050286





