1. Introduction
Survey data from mobile applications can quickly generate large datasets suitable for deep learning. Recent literature demonstrates its potential in medical fields, in what is now referred to as ‘mobile health’ [
1,
2,
3,
4,
5].
A relevant example is the emergence of just-in-time adaptive interventions (JITAIs). These are mobile applications designed to provide users with personalized treatment, generally aimed at improving lifestyle, through surveys and sensors. JITAIs have been developed for, among others: gestational weight management [
6], insomnia [
7], anxiety in children [
8], sedentary behavior in the elderly [
9], diabetes self-management [
10], and smoking cessation [
11].
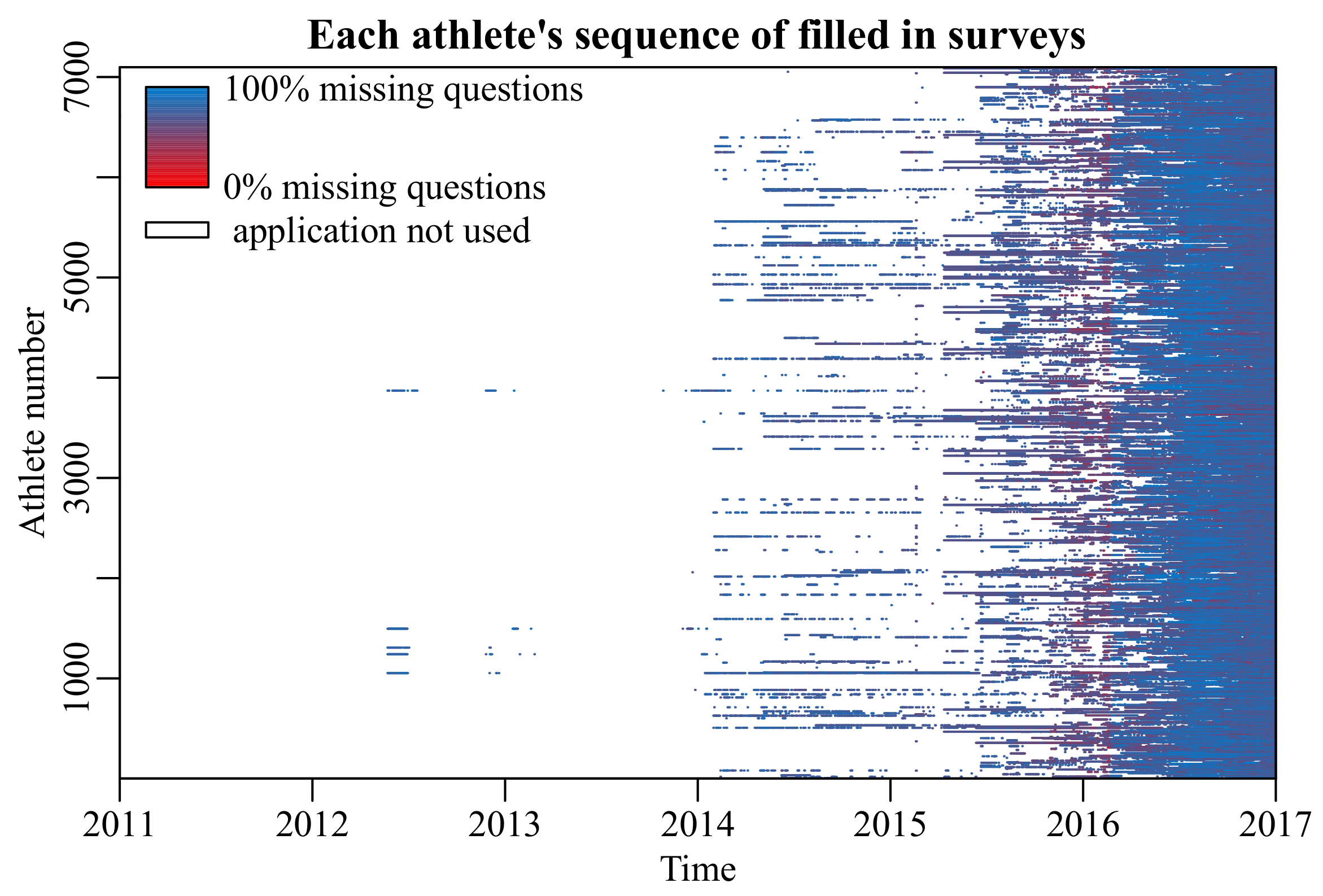
When surveys are included in the models that drive these applications, missing data are a persistent problem due to unanswered questions, or even entire missing surveys (
Figure 1) [
12,
13]. Developers could choose to make each question mandatory to avoid unanswered questions, but this would bias the results towards a group that is inclined to answer every question, or increase the number of entirely missing surveys. Furthermore, the missingness generating process must be taken into account (
Table 1). This raises the question of how to deal with missing data in longitudinal surveys.
A common solution to missing data in deep learning is to use indicators for missingness of the original variables [
15], such that the model can learn to associate each variables’ missingness with the outcome. However, this still leaves the question of how to impute the missing values in the original data. Moreover, including an indicator for each original variable doubles the input size, which may be problematic for data sets with a low number of observations relative to the number of features. Hence we propose a simple alternative for when the increase in variables is too high given the number of observations by Z-scoring, summing the missingness indicators, including the last recorded outcome in the input, and simplifying the padding of missing surveys.
The golden standard in handling missing data that are MCAR or MAR is through full conditional specification (FCS), which repeatedly imputes missing values of one variable, given the others [
16,
17]. Each imputation then trains a separate model, and the resulting models are pooled for inference or prediction. By drawing multiple imputations from the posterior, FCS can reflect the variance of the original variables. Furthermore, pre-existing relationships between variables can be preserved through passive imputation, resulting in greater precision [
18,
19].
A downside to FCS is that it cannot impute data that are MNAR without introducing bias. Moreover, the computational cost is greatly increased by drawing the imputations and separately training models on each of them. Recent literature suggests the required number of imputations depends on the amount of missingness and can even exceed 100, which would increase the computational cost by a hundred-fold from training the individual models alone [
20].
Gondara and Wang (2017) [
21] proposed training a denoising autoencoder on corrupted copies of a complete subset of the training data, minimizing the reconstruction error on the non-corrupt, non-missing subset. For purely predictive purposes, their method requires considerably less domain knowledge to use correctly compared to FCS, since the model automatically learns relationships between variables, rather than requiring careful consideration of each variable’s imputation method. Moreover, in their experiments, the autoencoder outperformed FCS both in terms of predictive accuracy and computational cost. However, their method relies on two strong assumptions: Firstly, there must be a large enough complete subset of the data to train such an autoencoder and secondly, this complete subset must accurately represent the incomplete data (i.e., missing data are MCAR).
Lipton et al., (2016) [
15] proposed simply including indicators for missingness of the original variables (Equation (
1)), followed by 0–1 normalization (Equation (
4)) and zero imputation. This method has the strong advantage that the model can learn direct relationships between missingness and the response, meaning that it should be able to model even MNAR. However, the number of variables can be increased by up to a two-fold, depending on how many columns contain missing values. For survey data largely consisting of partial entries, the individual missingness vectors may not bear enough information to justify the resulting increase in parameters. That is to say, the response variable might simply correlate with the number of questions an individual is inclined to answer on a given day.
Hence we propose a simple alternative for when the amount of missingness sufficiently correlates with the outcome by summing the missingness indicators (Equation (
2)), followed by Z-scoring (Equation (
3)) of the non-binary features, including the last recorded outcome in the input, and simplifying the padding of entirely missing surveys (
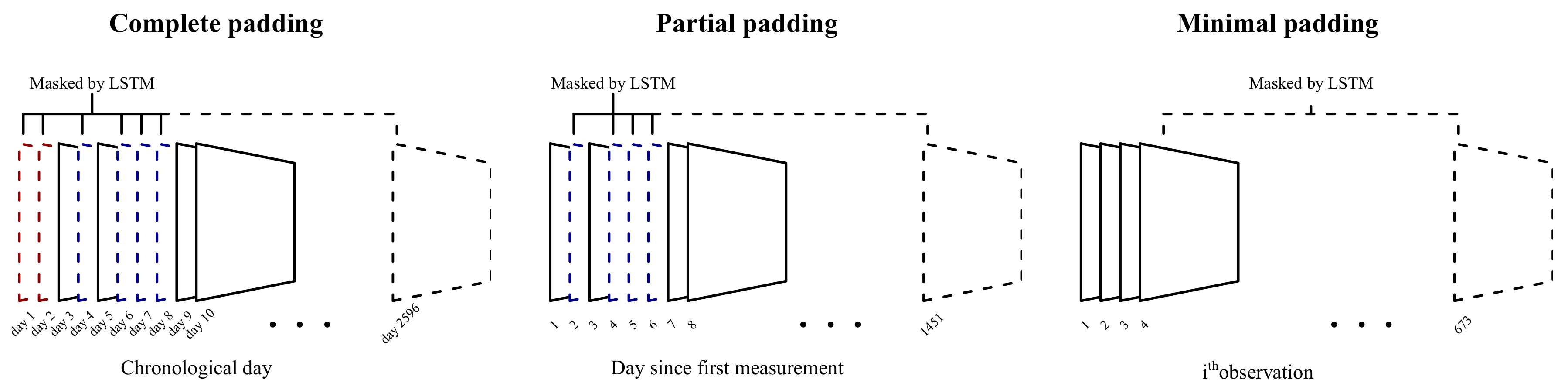
Figure 2).
2. Materials and Methods
This study has been approved by the ethical committee of Tokyo Institute of Technology under the approval number 2018069.
2.1. Description of Data
Repeated survey data spanning a 7-year period (2011–2017) was collected from a mobile application. This application was made available to 7098 athletes in Japan. Of these athletes, 5970 had at least one recorded outcome. These were randomly distributed into a train (80%), validation (10%), and test set (10%), each containing surveys from different athletes.
The survey consisted of 65 questions regarding the athletes’ physical activity, food intake, and biometric data (e.g., weight, body fat percentage) on a particular day. The outcome variable was the self-assessed condition on that day, ranging from 1 (very poor) to 10 (very good). Since the data are sparse and contain a considerable amount of missing data, the outcome variable was discretized to good condition (≥6) or poor condition (<6) to simplify the prediction problem to binary classification.
In total,
of survey answers on filled in surveys were missing. Questions which were answered less than
of the time were omitted, since these bear little information about the average user of the application. On average, participants only filled in the survey
of all days, leaving a large number of missing days (
Figure 1).
2.2. Imputation
For the FCS approach,
imputed data sets were generated, as recommended by Van Buuren (2016) [
16]. Although recent literature suggests a larger
m might be more precise [
20], the computation time already far exceeded other methods, taking upwards of
m times as long. Logistic regression was used to impute binary variables and predictive mean matching otherwise.
The missingness indicators approach was implemented in the same manner as described in Lipton et al. (2016) [
15], adding a matrix
of zeroes and ones corresponding to available and missing original data
, respectively as in Equation (
1), followed by zero imputation.
where
n is the number of observations and
p the number of questions on the survey. Categorical variables were restricted to two categories (yes/no), where missingness was imputed by the most sensible alternative. In our method, a missingness variable was added by summing the number of missing values in each row as in Equation (
2). Other combinations (mean, standard deviation) were also considered but did not appear to perform better in initial experimentation.
Studentized Z-scores were obtained from all non-binary variables using the non-missing training data as follows:
where
and
are the sample mean and standard deviation of non-missing observations from variable
j, respectively. Whereas 0-1 normalization as in Equation (
4) followed by zero imputation affects the mean (biased towards zero), Z-scoring as in Equation (
3) followed by zero imputation is effectively mean-imputation, preserving the original mean. This assigns a ’neutral’ value to missing values, whereas normalization followed by zero imputation assigns an extreme value. Categorical variables were treated as aforementioned.
2.3. Padding
Most participants filled in the survey sporadically, leaving large gaps in the sequences. Three padding strategies (
Figure 2) for these missing days were compared in terms of prediction error. The first involves simply padding all missing days as entries consisting entirely of zeroes (complete padding). These zeroes were then masked in the first layer of the model, allowing the LSTM to ignore these entries. This approach is very similar to that described in Che et al., (2018) [
22], except that their version was based on a gated recurrent unit [
23]. A quick comparison showed that an LSTM performed better on these data (results not included).
Since participants started using the application at different times, the second method ignores the missing days prior to a participant’s first entry (partial padding). In other words, the first day of entry was considered to be day 1. While this method cannot account for effects of a particular day (e.g., there might have been a national holiday), it reduces the total sequence length, which in turn reduces the model complexity and computational cost.
The third method simply ignores missing days and instead models the available data as an ordered sequence (minimal padding). While this method cannot account for the time between surveys, it further reduces the sequence length.
2.4. Lagged Response
Where available, the last recorded outcome was added to the input. For the first observation, this variable was set to zero instead, treating it in the same way as other missing values. In the results section, this is referred to as LR.
2.5. Experiments
All models consisted of a 3-layer LSTM network with 32, 24 and 16 nodes in the first, second, and third hidden layer, respectively [
24]. A combination of non-recurrent and recurrent dropout was used, as proposed by Semeniuta et al. (2016) [
25]. Non-recurrent dropout was set to
and recurrent dropout to
. Additional regularization and different layer types were considered, but did not appear to improve accuracy (results not included). Gradient updates were performed in batch sizes equal to the longest complete sequence (
) using adaptive moment estimation with hyperparameters equal to that of the original paper [
26]. Binary crossentropy was minimized on the validation set and overfitting was assessed by discrepancies between training and validation loss. Initial experimentation revealed that validation accuracy plateaued after around 2000 epochs. This number of epochs was used for each comparison in the results section, unless stated otherwise.
All experiments were conducted using the Windows version of the RStudio interface to Keras Tensorflow, with GPU acceleration enabled, on the system summarized in
Table 2. ROC statistics were calculated using the ROCR package [
27].
4. Discussion
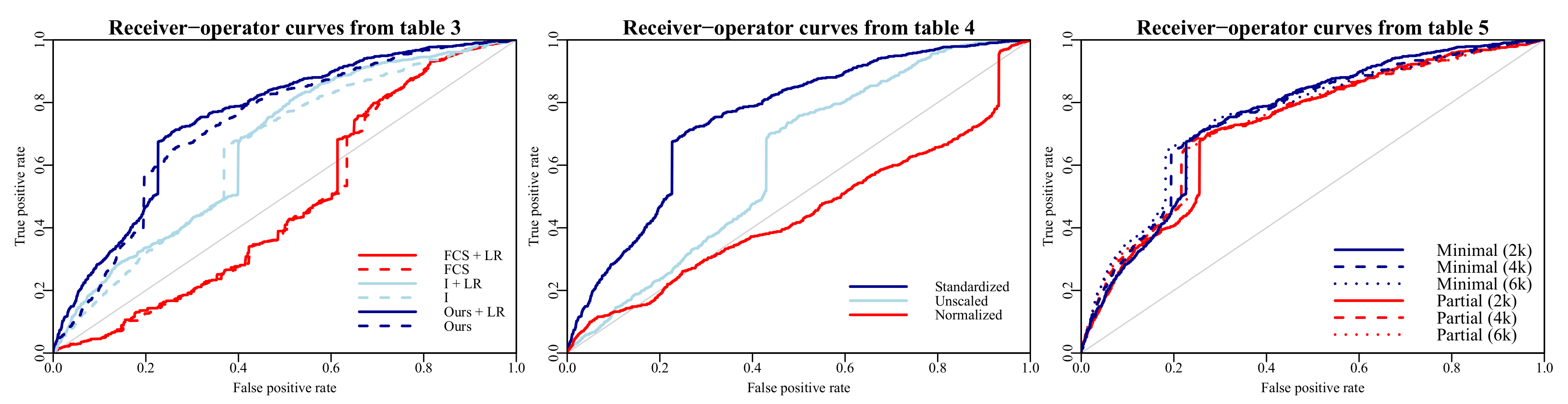
The FCS approach with
imputations underperformed all other methods, both in terms of prediction accuracy and computation time (increased by a factor
m). The most likely explanation for its subpar accuracy is that the missing data were MNAR, and FCS is not suitable for MNAR. This assertion is supported by the increased performance observed when directly specifying missingness through indicators (
Table 3). Another contributing factor might be that the amount of missingness (
on filled in surveys;
in total) is simply too large, causing issues when imputing missing data and then ‘reusing’ the data for the predictive model.
While FCS might have benefitted from a considerably larger number of imputations, it was already the most computationally costly method. Since model averaging generally tends to increase performance, the additional computation time—if available—might perhaps be better spent on e.g., bagging other methods, rather than drawing additional imputations with FCS.
Interestingly, correctly padding and masking the missing days did not improve accuracy, nor did the forward filling strategy proposed in Lipton et al. (2016) [
15]. The former could either be attributed to a small effect of time in between survey entries on the outcome, or inability of the model to learn this effect using the number of observations at hand (
) in conjunction with the high amount of missingness. A limitation of this comparison is that we did not further investigate how different regularization techniques might have affected the performance of the padding strategies. The forward filling strategy might be better justifiable for clinical data, where measurements are taken at intervals after which they are expected to change.
While ignoring the missing days resulted in the greatest performance (
Table 6), reasonable performance could also be achieved by masking and padding the missing days in between measurements. This method might achieve higher accuracy on problems where the time in between observations is of greater relevance to the outcome.
These results demonstrate that with appropriate adjustments, a deep learning algorithm can even learn reasonably from a sparse sequence of gappy data. Various simplifications of previously suggested methods greatly reduced computational time, while simultaneously outperforming more costly methods in terms of prediction accuracy. The greatest accuracy was achieved on these data, by including a LR variable and a count of the missing variables, followed by Z-scoring and zero imputation.
Importantly, our method does not require a subset of the data which is complete, nor does it require the missing data to be MCAR or MAR. It must be noted, however, that our method might not perform well if the amount of missingness is not as informative about the outcome as it is in these survey data. Future work can demonstrate whether this approach also performs well on other time series with various forms of missingness.