Deep Learning in the Biomedical Applications: Recent and Future Status
Abstract
1. Introduction
- Omics: genomics and proteomics are the study of the DNA/RNA sequencing and protein interactions and structure prediction.
- Bioimaging: the study of biological cell and tissue by analyzing the histopathology or immunohistochemically images.
- Medical imaging: the study of the human organs by analyzing magnetic resonance imaging (MRI), X-ray, etc.,
- Brain and body machine interface: the study of the brain and body decoding machine interface by analyzing different biosignals such as electroencephalogram (EEG), electroencephalogram (EEG), etc.
- Public and medical health management (PmHM): the study of big medical data to develop and enhance public health-care decisions for a humanity wellbeing.
2. Neural Network and Deep Learning
2.1. From Shallow to Deep Neural Networks
2.2. Most Popular Deep Neural Networks Architectures Used in Biomedical Applications
2.2.1. DNN for Non-Locally Correlated Input data
2.2.2. DNN for High Locally Correlated Data
3. Biomedical Applications
3.1. Omics
3.2. Bio and Medical Imaging
3.3. Brain and Body Machine Interfaces
3.4. Public and Medical Health Management
4. Omics
4.1. Around the Genome
4.1.1. Protein Binding Prediction
- DNA–RNA-binding proteins: The chemical interaction between the protein and a DNA or RNA strands is called the protein binding. Predicting these binding sites is very crucial in various biological activities such as gene regulation, transcription, DNA repair, and mutations [59] and it is essential for the interpretation of the genome. To model the binding sites, the position–frequency matrix is mainly used as a four input channels and the output is a binding score [108,109,110,111,112,113,114].
- Enhancer and promoter identification: Promoters and enhancers act via complex interactions across time and space in the nucleus to control when, where and at what magnitude genes are active [115]. Promoters and enhancers were early discoveries during the molecular characterization of genes [115]. Promoters specify and enable the positioning of RNA polymerase machinery at transcription initiation sites while enhancers are the short regions of DNA sequences bounded by a certain type of proteins, the transcription factors [115,116,117,118,119].
4.1.2. Gene Expression
- Alternative splicing: Alternative splicing is a process whereby the exons of a primary transcript may be connected in different ways during pre-mRNA splicing [120]. The objective is to build a model that can predict the outcome of AS from sequence information in different cellular contexts [120,121,122].
- Gene expression prediction: Gene expression refers to the process of producing a protein from sequence information encoded in DNA [123]. Predicting the gene expression from histone modification can be formulated as a binary classification where the outputs represent the gene expression level (high or low) [124,125,126].
4.1.3. Genomic Sequencing
- DNA sequencing: Learning the functional activity [127], quantifying the function [128] or identifying functional effects of noncoding variants [129] of DNA sequences from genOmics data are fundamental problems in Omics applications recently touched by the enthusiasm of deep learning. De novo identification of replication domains type using replication timing profiles is also a crucial application in the genomic sequencing [130].
- DNA methylation: DNA methylation is a process by which methyl groups are added to the DNA molecule and can change the activity of a DNA segment without changing the sequence. DNA methylation plays a crucial role in the establishment of tissue-specific gene expression and the regulation of key biological processes [131,132].
4.2. Around the Protein
4.2.1. Protein Structure Prediction (PSP)
- Protein loop modeling and disorder prediction: Biology and medicine have a long-standing interest in computational structure prediction and modeling of proteins. There are often missing regions or regions that need to be remodeled in protein structures. The process of predicting missing regions in a protein structure is called loop modeling [156,157,158]. Many proteins contain regions with unstable tertiary structure. These regions are called disorder regions [159].
4.2.2. Protein Interaction Prediction (PIP)
- Protein–Protein Interactions (PPIs): Protein–protein interactions (PPI) play critical roles in many cellular biological processes, such as signal transduction, immune response, and cellular organization. Analysis of protein–protein interactions is very important in the drug target detection and therapy design [160,161,162,163,164,165,166,167].
5. Brain and Body Machine Interface
5.1. Brain Decoding
5.1.1. Evoked Signals
- Steady state evoked potentials (SSEP): SSEP signals are brain signals that are generated when the subject perceives periodic external stimulus [174]. Table 4 gives an overview of papers using deep learning techniques for SSEP applications. The SSEP applications are divided into three main parts: human’s affective and emotion states classification, auditory evoked potential and visual evoked potential.
5.1.2. Spontaneous
- Motor and sensorimotor rhythms (MSR): Motor and sensorimotor rhythms are brain signals in relation with motor actions such as moving arms [174]. Most of the applications concerns the motor imagery tasks, which is the translation of the subject motor intention into control signals through motor imagery states (see Table 4).
5.1.3. Hybrid Signals
5.2. Diseases Diagnosis
6. Discussions and Outlooks
6.1. Overview of the Reviewed Papers
6.2. Overview of the Used Deep Neural Networks Architectures
- DNN: Deep neural architectures for non-locally correlated input data, which encompasses the deep multilayer perceptron, deep auto-encoders and the deep belief networks (see Section 2.2.1).
- CNN: Deep neural architectures for high locally correlated data.
6.3. Biomedical Data and Transfer Learning
6.4. Model Building
6.5. Emergent Architectures: The Generative Adversarial Networks
7. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- He, H.; Garcia, E.A. Learning from Imbalanced Data. IEEE Trans. Knowl. Data Eng. 2009, 21, 1263–1284. [Google Scholar] [CrossRef]
- Yu, H.; Yang, X.; Zheng, S.; Sun, C. Active Learning From Imbalanced Data: A Solution of Online Weighted Extreme Learning Machine. IEEE Trans. Neural Netw. Learn. Syst. 2018, 30, 1–16. [Google Scholar] [CrossRef]
- Ditzler, G.; Roveri, M.; Alippi, C.; Polikar, R. Learning in Nonstationary Environments: A Survey. IEEE Comput. Intell. Mag. 2015, 10, 12–25. [Google Scholar] [CrossRef]
- Alvarado-Díaz, W.; Lima, P.; Meneses-Claudio, B.; Roman-Gonzalez, A. Implementation of a brain-machine interface for controlling a wheelchair. In Proceedings of the 2017 CHILEAN Conference on Electrical, Electronics Engineering, Information and Communication Technologies (CHILECON), Pucon, Chile, 18–20 October 2017; pp. 1–6. [Google Scholar] [CrossRef]
- Mahmud, M.; Kaiser, M.; Hussain, A.; Vassanelli, S. Applications of Deep Learning and Reinforcement Learning to Biological Data. IEEE Trans. Neural Netw. Learn. Syst. 2018, 29, 2063–2079. [Google Scholar] [CrossRef]
- Cao, C.; Liu, F.; Tan, H.; Song, D.; Shu, W.; Li, W.; Zhou, Y.; Bo, X.; Xie, Z. Deep Learning and Its Applications in Biomedicine. Genom. Proteom. Bioinform. 2018, 16, 17–32. [Google Scholar] [CrossRef] [PubMed]
- Ravì, D.; Wong, C.; Deligianni, F.; Berthelot, M.; Andreu-Perez, J.; Lo, B.; Yang, G.Z. Deep Learning for Health Informatics. IEEE J. Biomed. Health Inform. 2017, 21, 4–21. [Google Scholar] [CrossRef]
- Jones, W.; Alasoo, K.; Fishman, D.; Parts, L. Computational biology: Deep learning. Emerg. Top. Life Sci. 2017, 1, 257–274. [Google Scholar] [CrossRef]
- Angermueller, C.; Pärnamaa, T.; Parts, L.; Stegle, O. Deep learning for computational biology. Mol. Syst. Biol. 2016, 12, 878. [Google Scholar] [CrossRef] [PubMed]
- Min, S.; Lee, B.; Yoon, S. Deep Learning in Bioinformatics. Brief. Bioinform. 2016, 18, 851–869. [Google Scholar] [CrossRef]
- Hubel, D.H.; Wiesel, T.N. Receptive fields, binocular interaction and functional architecture in the cat’s visual cortex. J. Physiol. 1962, 160, 106–154. [Google Scholar] [CrossRef] [PubMed]
- Hubel, D.H.; Wiesel, T.N. Receptive fields of single neurones in the cat’s striate cortex. J. Physiol. 1959, 148, 574–591. [Google Scholar] [CrossRef]
- Schmidhuber, J. Deep learning in neural networks: An overview. Neural Netw. 2015, 61, 85–117. [Google Scholar] [CrossRef]
- Hinton, G.E.; Osindero, S.; Teh, Y.W. A Fast Learning Algorithm for Deep Belief Nets. Neural Comput. 2006, 18, 1527–1554. [Google Scholar] [CrossRef]
- Hinton, G.E.; Salakhutdinov, R.R. Reducing the Dimensionality of Data with Neural Networks. Science 2006, 313, 504–507. [Google Scholar] [CrossRef]
- Bengio, Y.; Lamblin, P.; Popovici, D.; Larochelle, H. Greedy Layer-Wise Training of Deep Networks. In Advances in Neural Information Processing Systems (NIPS 06); Schölkopf, B., Platt, J., Hoffman, T., Eds.; MIT Press: Cambridge, MA, USA, 2007; pp. 153–160. [Google Scholar]
- Rumelhart, D.E.; Hinton, G.E.; Williams, R.J. Learning representations by back-propagating errors. Nature 1986, 323, 533–536. [Google Scholar] [CrossRef]
- Choi, B.; Lee, J.H.; Kim, D.H. Solving local minima problem with large number of hidden nodes on two-layered feed-forward artificial neural networks. Neurocomputing 2008, 71, 3640–3643. [Google Scholar] [CrossRef]
- Behera, L.; Kumar, S.; Patnaik, A. On Adaptive Learning Rate That Guarantees Convergence in Feedforward Networks. IEEE Trans. Neural Netw. 2006, 17, 1116–1125. [Google Scholar] [CrossRef]
- Duffner, S.; Garcia, C. An Online Backpropagation Algorithm with Validation Error-Based Adaptive Learning Rate. In Artificial Neural Networks—ICANN 2007: 17th International Conference, Porto, Portugal, 9–13 September 2007, Proceedings, Part I; de Sá, J.M., Alexandre, L.A., Duch, W., Mandic, D., Eds.; Springer: Berlin/Heidelberg, Germany, 2007; pp. 249–258. [Google Scholar]
- Zeraatkar, E.; Soltani, M.; Karimaghaee, P. A fast convergence algorithm for BPNN based on optimal control theory based learning rate. In Proceedings of the 2nd International Conference on Control, Instrumentation and Automation, Shiraz, Iran, 27–29 December 2011; pp. 292–297. [Google Scholar] [CrossRef]
- Zhang, R.; Xu, Z.B.; Huang, G.B.; Wang, D. Global Convergence of Online BP Training With Dynamic Learning Rate. IEEE Trans. Neural Netw. Learn. Syst. 2012, 23, 330–341. [Google Scholar] [CrossRef]
- Zeiler, M.D. ADADELTA: An Adaptive Learning Rate Method. arXiv, 2012; arXiv:1212.5701. [Google Scholar]
- Shrestha, S.B.; Song, Q. Adaptive learning rate of SpikeProp based on weight convergence analysis. Neural Netw. 2015, 63, 185–198. [Google Scholar] [CrossRef]
- Asha, C.; Narasimhadhan, A. Adaptive Learning Rate for Visual Tracking Using Correlation Filters. Procedia Comput. Sci. 2016, 89, 614–622. [Google Scholar] [CrossRef]
- Narayanan, S.J.; Bhatt, R.B.; Perumal, B. Improving the Accuracy of Fuzzy Decision Tree by Direct Back Propagation with Adaptive Learning Rate and Momentum Factor for User Localization. Procedia Comput. Sci. 2016, 89, 506–513. [Google Scholar] [CrossRef][Green Version]
- Gorunescu, F.; Belciug, S. Boosting backpropagation algorithm by stimulus-sampling: Application in computer-aided medical diagnosis. J. Biomed. Inform. 2016, 63, 74–81. [Google Scholar] [CrossRef]
- Glorot, X.; Bordes, A.; Bengio, Y. Deep Sparse Rectifier Neural Networks. In Proceedings of the Fourteenth International Conference on Artificial Intelligence and Statistics (AISTATS-11), Fort Lauderdale, FL, USA, 11–13 April 2011; Volume 15, pp. 315–323. [Google Scholar]
- Chandra, B.; Sharma, R.K. Deep learning with adaptive learning rate using laplacian score. Expert Syst. Appl. 2016, 63, 1–7. [Google Scholar] [CrossRef]
- Zhang, N.; Ding, S.; Zhang, J.; Xue, Y. An overview on Restricted Boltzmann Machines. Neurocomputing 2018, 275, 1186–1199. [Google Scholar] [CrossRef]
- Hosseini-Asl, E.; Zurada, J.M.; Nasraoui, O. Deep Learning of Part-Based Representation of Data Using Sparse Autoencoders With Nonnegativity Constraints. IEEE Trans. Neural Netw. Learn. Syst. 2016, 27, 2486–2498. [Google Scholar] [CrossRef]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Srivastava, N.; Hinton, G.; Krizhevsky, A.; Sutskever, I.; Salakhutdinov, R. Dropout: A Simple Way to Prevent Neural Networks from Overfitting. J. Mach. Learn. Res. 2014, 15, 1929–1958. [Google Scholar]
- Dahl, G.E.; Sainath, T.N.; Hinton, G.E. Improving deep neural networks for LVCSR using rectified linear units and dropout. In Proceedings of the 2013 IEEE International Conference on Acoustics, Speech and Signal Processing, Vancouver, BC, Canada, 26–31 May 2013; pp. 8609–8613. [Google Scholar] [CrossRef]
- Goodfellow, I.J.; Warde-farley, D.; Mirza, M.; Courville, A.; Bengio, Y. Maxout networks. arXiv, 2013; arXiv:1302.4389. [Google Scholar]
- Hastie, T.; Tibshirani, R.; Friedman, J. The Elements of Statistical Learning; Springer: New York, NY, USA, 2009. [Google Scholar] [CrossRef]
- Yao, X. A review of evolutionary artificial neural networks. Int. J. Intell. Syst. 1993, 8, 539–567. [Google Scholar] [CrossRef]
- Pérez-Sánchez, B.; Fontenla-Romero, O.; Guijarro-Berdiñas, B. A review of adaptive online learning for artificial neural networks. Artif. Intell. Rev. 2016, 49, 281–299. [Google Scholar] [CrossRef]
- Kalaiselvi, T.; Sriramakrishnan, P.; Somasundaram, K. Survey of using GPU CUDA programming model in medical image analysis. Inform. Med. Unlocked 2017, 9, 133–144. [Google Scholar] [CrossRef]
- Smistad, E.; Falch, T.L.; Bozorgi, M.; Elster, A.C.; Lindseth, F. Medical image segmentation on GPU—A comprehensive review. Med. Image Anal. 2015, 20, 1–18. [Google Scholar] [CrossRef]
- Eklund, A.; Dufort, P.; Forsberg, D.; LaConte, S.M. Medical image processing on the GPU—Past, present and future. Med. Image Anal. 2013, 17, 1073–1094. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet Classification with Deep Convolutional Neural Networks. In Proceedings of the 25th International Conference on Neural Information Processing Systems, Lake Tahoe, NV, USA, 3–6 December 2012; Curran Associates Inc.: Lake Tahoe, NV, USA, 2012; pp. 1097–1105. [Google Scholar]
- Fischer, A.; Igel, C. Training restricted Boltzmann machines: An introduction. Pattern Recognit. 2014, 47, 25–39. [Google Scholar] [CrossRef]
- Hopfield, J.J. Neural networks and physical systems with emergent collective computational abilities. Proc. Natl. Acad. Sci. USA 1982, 79, 2554–2558. [Google Scholar] [CrossRef]
- Sheri, A.M.; Rafique, A.; Pedrycz, W.; Jeon, M. Contrastive divergence for memristor-based restricted Boltzmann machine. Eng. Appl. Artif. Intell. 2015, 37, 336–342. [Google Scholar] [CrossRef]
- Sankaran, A.; Goswami, G.; Vatsa, M.; Singh, R.; Majumdar, A. Class sparsity signature based Restricted Boltzmann Machine. Pattern Recognit. 2017, 61, 674–685. [Google Scholar] [CrossRef]
- Längkvist, M.; Karlsson, L.; Loutfi, A. A review of unsupervised feature learning and deep learning for time-series modeling. Pattern Recognit. Lett. 2014, 42, 11–24. [Google Scholar] [CrossRef]
- Hinton, G.E. Deep belief networks. Scholarpedia 2009, 4, 5947. [Google Scholar] [CrossRef]
- Lecun, Y.; Bottou, L.; Bengio, Y.; Haffner, P. Gradient-based learning applied to document recognition. Proc. IEEE 1998, 86, 2278–2324. [Google Scholar] [CrossRef]
- Matsugu, M.; Mori, K.; Mitari, Y.; Kaneda, Y. Subject independent facial expression recognition with robust face detection using a convolutional neural network. Neural Netw. 2003, 16, 555–559. [Google Scholar] [CrossRef]
- Ciregan, D.; Meier, U.; Schmidhuber, J. Multi-column deep neural networks for image classification. In Proceedings of the 2012 IEEE Conference on Computer Vision and Pattern Recognition, Providence, RI, USA, 16–21 June 2012; pp. 3642–3649. [Google Scholar] [CrossRef]
- Zeiler, M.D.; Fergus, R. Visualizing and Understanding Convolutional Networks. In European Conference on Computer Vision; Springer: Cham, Switzerland, 2013. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. arXiv, 2014; arXiv:1409.1556. [Google Scholar]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.E.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going Deeper with Convolutions. arXiv, 2014; arXiv:1409.4842. [Google Scholar]
- Mamoshina, P.; Vieira, A.; Putin, E.; Zhavoronkov, A. Applications of Deep Learning in Biomedicine. Mol. Pharm. 2016, 13, 1445–1454. [Google Scholar] [CrossRef]
- Bagchi, A. Protein-protein Interactions: Basics, Characteristics, and Predictions. In Soft Computing for Biological Systems; Purohit, H.J., Kalia, V.C., More, R.P., Eds.; Springer: Singapore, 2018; pp. 111–120. [Google Scholar] [CrossRef]
- Nath, A.; Kumari, P.; Chaube, R. Prediction of Human Drug Targets and Their Interactions Using Machine Learning Methods: Current and Future Perspectives. In Computational Drug Discovery and Design; Gore, M., Jagtap, U.B., Eds.; Springer: New York, NY, USA, 2018; pp. 21–30. [Google Scholar] [CrossRef]
- Shehu, A.; Barbará, D.; Molloy, K. A Survey of Computational Methods for Protein Function Prediction. In Big Data Analytics in Genomics; Wong, K.C., Ed.; Springer International Publishing: Cham, Switerland, 2016; pp. 225–298. [Google Scholar] [CrossRef]
- Leung, M.K.K.; Delong, A.; Alipanahi, B.; Frey, B.J. Machine Learning in Genomic Medicine: A Review of Computational Problems and Data Sets. Proc. IEEE 2016, 104, 176–197. [Google Scholar] [CrossRef]
- Xiong, Y.; Zhu, X.; Dai, H.; Wei, D.Q. Survey of Computational Approaches for Prediction of DNA-Binding Residues on Protein Surfaces. In Computational Systems Biology: Methods and Protocols; Huang, T., Ed.; Springer: New York, NY, USA, 2018; pp. 223–234. [Google Scholar] [CrossRef]
- Libbrecht, M.W.; Noble, W.S. Machine learning in genetics and genomics. Nat. Rev. Genet. 2015, 16, 321–332. [Google Scholar] [CrossRef]
- Khosravi, P.; Kazemi, E.; Imielinski, M.; Elemento, O.; Hajirasouliha, I. Deep Convolutional Neural Networks Enable Discrimination of Heterogeneous Digital Pathology Images. EBioMedicine 2018, 27, 317–328. [Google Scholar] [CrossRef]
- Veta, M.; Pluim, J.P.W.; van Diest, P.J.; Viergever, M.A. Breast Cancer Histopathology Image Analysis: A Review. IEEE Trans. Biomed. Eng. 2014, 61, 1400–1411. [Google Scholar] [CrossRef]
- Weese, J.; Lorenz, C. Four challenges in medical image analysis from an industrial perspective. Med. Image Anal. 2016, 33, 44–49. [Google Scholar] [CrossRef]
- Greenspan, H.; van Ginneken, B.; Summers, R.M. Guest Editorial Deep Learning in Medical Imaging: Overview and Future Promise of an Exciting New Technique. IEEE Trans. Med. Imaging 2016, 35, 1153–1159. [Google Scholar] [CrossRef]
- Wells, W.M. Medical Image Analysis—Past, present, and future. Med. Image Anal. 2016, 33, 4–6. [Google Scholar] [CrossRef]
- Zhang, S.; Metaxas, D. Large-Scale medical image analytics: Recent methodologies, applications and Future directions. Med. Image Anal. 2016, 33, 98–101. [Google Scholar] [CrossRef]
- Criminisi, A. Machine learning for medical images analysis. Med. Image Anal. 2016, 33, 91–93. [Google Scholar] [CrossRef]
- De Bruijne, M. Machine learning approaches in medical image analysis: From detection to diagnosis. Med. Image Anal. 2016, 33, 94–97. [Google Scholar] [CrossRef]
- Kergosien, Y.L.; Racoceanu, D. Semantic knowledge for histopathological image analysis: From ontologies to processing portals and deep learning. In Proceedings of the 13th International Conference on Medical Information Processing and Analysis, San Andres Island, Colombia, 5–7 October 2017; p. 105721F. [Google Scholar] [CrossRef]
- Gandomkar, Z.; Brennan, P.; Mello-Thoms, C. Computer-based image analysis in breast pathology. J. Pathol. Inform. 2016, 7, 43. [Google Scholar] [CrossRef]
- Chen, J.M.; Li, Y.; Xu, J.; Gong, L.; Wang, L.W.; Liu, W.L.; Liu, J. Computer-aided prognosis on breast cancer with hematoxylin and eosin histopathology images: A review. Tumor Biol. 2017, 39, 1010428317694550. [Google Scholar] [CrossRef]
- Aswathy, M.; Jagannath, M. Detection of breast cancer on digital histopathology images: Present status and future possibilities. Inform. Med. Unlocked 2016, 8, 74–79. [Google Scholar] [CrossRef]
- Hamilton, P.W.; Bankhead, P.; Wang, Y.; Hutchinson, R.; Kieran, D.; McArt, D.G.; James, J.; Salto-Tellez, M. Digital pathology and image analysis in tissue biomarker research. Methods 2014, 70, 59–73. [Google Scholar] [CrossRef]
- Irshad, H.; Veillard, A.; Roux, L.; Racoceanu, D. Methods for Nuclei Detection, Segmentation, and Classification in Digital Histopathology: A Review-Current Status and Future Potential. IEEE Rev. Biomed. Eng. 2014, 7, 97–114. [Google Scholar] [CrossRef]
- Saha, M.; Chakraborty, C.; Racoceanu, D. Efficient deep learning model for mitosis detection using breast histopathology images. Comput. Med. Imaging Graph. 2018, 64, 29–40. [Google Scholar] [CrossRef]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; van der Laak, J.A.; van Ginneken, B.; Sánchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef]
- Pouliakis, A.; Karakitsou, E.; Margari, N.; Bountris, P.; Haritou, M.; Panayiotides, J.; Koutsouris, D.; Karakitsos, P. Artificial Neural Networks as Decision Support Tools in Cytopathology: Past, Present, and Future. Biomed. Eng. Comput. Biol. 2016, 7, 1–18. [Google Scholar] [CrossRef]
- Janowczyk, A.; Madabhushi, A. Deep learning for digital pathology image analysis: A comprehensive tutorial with selected use cases. J. Pathol. Inform. 2016, 7, 29. [Google Scholar] [CrossRef]
- Erickson, B.J.; Korfiatis, P.; Akkus, Z.; Kline, T.L. Machine Learning for Medical Imaging. RadioGraphics 2017, 37, 505–515. [Google Scholar] [CrossRef]
- Akkus, Z.; Galimzianova, A.; Hoogi, A.; Rubin, D.L.; Erickson, B.J. Deep Learning for Brain MRI Segmentation: State of the Art and Future Directions. J. Digit. Imaging 2017, 30, 449–459. [Google Scholar] [CrossRef]
- Mahmud, M.; Vassanelli, S. Processing and Analysis of Multichannel Extracellular Neuronal Signals: State-of-the-Art and Challenges. Front. Neurosci. 2016, 10, 248. [Google Scholar] [CrossRef]
- Major, T.C.; Conrad, J.M. A survey of brain computer interfaces and their applications. In Proceedings of the IEEE SOUTHEASTCON 2014, Lexington, KY, USA, 13–16 March 2014; pp. 1–8. [Google Scholar] [CrossRef]
- Kerous, B.; Liarokapis, F. Brain-Computer Interfaces—A Survey on Interactive Virtual Environments. In Proceedings of the 2016 8th International Conference on Games and Virtual Worlds for Serious Applications (VS-GAMES), Barcelona, Spain, 7–9 September 2016; pp. 1–4. [Google Scholar] [CrossRef]
- Abdulkader, S.N.; Atia, A.; Mostafa, M.S.M. Brain computer interfacing: Applications and challenges. Egypt. Inform. J. 2015, 16, 213–230. [Google Scholar] [CrossRef]
- Tan, J.H.; Hagiwara, Y.; Pang, W.; Lim, I.; Oh, S.L.; Adam, M.; Tan, R.S.; Chen, M.; Acharya, U.R. Application of stacked convolutional and long short-term memory network for accurate identification of CAD ECG signals. Comput. Biol. Med. 2018, 94, 19–26. [Google Scholar] [CrossRef]
- Acharya, U.R.; Fujita, H.; Oh, S.L.; Hagiwara, Y.; Tan, J.H.; Adam, M. Application of deep convolutional neural network for automated detection of myocardial infarction using ECG signals. Inform. Sci. 2017, 415, 190–198. [Google Scholar] [CrossRef]
- Turner, J.T.; Page, A.; Mohsenin, T.; Oates, T. Deep Belief Networks used on High Resolution Multichannel Electroencephalography Data for Seizure Detection; CoRR: Leawood, KS, USA, 2017. [Google Scholar]
- Zhao, Y.; He, L. Deep learning in the eeg diagnosis of alzheimer’s disease. In Asian Conference on Computer Vision; Springer: Cham, Switerland, 2015; pp. 340–353. [Google Scholar] [CrossRef]
- Atzori, M.; Cognolato, M.; Müller, H. Deep learning with convolutional neural networks applied to emg data: A resource for the classification of movements for prosthetic hands. Front. Neurorobot. 2016, 10, 9. [Google Scholar] [CrossRef]
- Geng, W.; Du, Y.; Jin, W.; Wei, W.; Hu, Y.; Li, J. Gesture recognition by instantaneous surface EMG images. Sci. Rep. 2016, 6, 36571. [Google Scholar] [CrossRef]
- Lotte, F.; Bougrain, L.; Cichocki, A.; Clerc, M.; Congedo, M.; Rakotomamonjy, A.; Yger, F. A review of classification algorithms for EEG-based brain-computer interfaces: A 10 year update. J. Neural Eng. 2018, 15, 031005. [Google Scholar] [CrossRef]
- Faust, O.; Hagiwara, Y.; Hong, T.J.; Lih, O.S.; Acharya, U.R. Deep learning for healthcare applications based on physiological signals: A review. Comput. Methods Programs Biomed. 2018, 161, 1–13. [Google Scholar] [CrossRef]
- Bhaskar, H.; Hoyle, D.C.; Singh, S. Machine learning in bioinformatics: A brief survey and recommendations for practitioners. Comput. Biol. Med. 2006, 36, 1104–1125. [Google Scholar] [CrossRef]
- Ong, B.T.; Sugiura, K.; Zettsu, K. Dynamically pre-trained deep recurrent neural networks using environmental monitoring data for predicting PM2.5. Neural Comput. Appl. 2016, 27, 1553–1566. [Google Scholar] [CrossRef]
- Liang, Z.; Zhang, G.; Huang, J.X.; Hu, Q.V. Deep learning for healthcare decision making with EMRs. In Proceedings of the 2014 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Belfast, UK, 2–5 November 2014; pp. 556–559. [Google Scholar] [CrossRef]
- Che, Z.; Purushotham, S.; Khemani, R.; Liu, Y. Distilling Knowledge from Deep Networks with Applications to Healthcare Domain. arXiv, 2015; arXiv:1512.03542. [Google Scholar]
- Mehrabi, S.; Sohn, S.; Li, D.; Pankratz, J.J.; Therneau, T.; Sauver, J.L.S.; Liu, H.; Palakal, M. Temporal Pattern and Association Discovery of Diagnosis Codes Using Deep Learning. In Proceedings of the 2015 International Conference on Healthcare Informatics, Dallas, TX, USA, 21–23 October2015; pp. 408–416. [Google Scholar] [CrossRef]
- Lipton, Z.C.; Kale, D.C.; Elkan, C.; Wetzel, R.C. Learning to Diagnose with LSTM Recurrent Neural Networks. arXiv, 2015; arXiv:1511.03677. [Google Scholar]
- Zou, B.; Lampos, V.; Gorton, R.; Cox, I.J. On Infectious Intestinal Disease Surveillance Using Social Media Content. In Proceedings of the 6th International Conference on Digital Health Conference, Montréal, QC, Canada, 11–13 April 2016; ACM: New York, NY, USA, 2016; pp. 157–161. [Google Scholar] [CrossRef]
- Phan, N.; Dou, D.; Piniewski, B.; Kil, D. Social restricted Boltzmann Machine: Human behavior prediction in health social networks. In Proceedings of the 2015 IEEE/ACM International Conference on Advances in Social Networks Analysis and Mining (ASONAM), Paris, France, 25–28 August 2015; pp. 424–431. [Google Scholar] [CrossRef]
- Garimella, V.R.K.; Alfayad, A.; Weber, I. Social Media Image Analysis for Public Health. In Proceedings of the 2016 CHI Conference on Human Factors in Computing Systems, San Jose, CA, USA, 7–12 May 2016. [Google Scholar]
- Zhao, L.; Chen, J.; Chen, F.; Wang, W.; Lu, C.; Ramakrishnan, N. SimNest: Social Media Nested Epidemic Simulation via Online Semi-Supervised Deep Learning. In Proceedings of the 2015 IEEE International Conference on Data Mining, Atlantic City, NJ, USA, 14–17 November 2015; pp. 639–648. [Google Scholar] [CrossRef]
- Ibrahim, R.; Yousri, N.A.; Ismail, M.A.; El-Makky, N.M. Multi-level gene/MiRNA feature selection using deep belief nets and active learning. In Proceedings of the 2014 36th Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Chicago, IL, USA, 26–30 August 2014; pp. 3957–3960. [Google Scholar] [CrossRef]
- Fakoor, R.; Ladhak, F.; Nazi, A.; Huber, M. Using Deep Learning to Enhance Cancer Diagnosis and Classification. In Proceedings of the International Conference on Machine Learning (ICML), WHEALTH Workshop, Atlanta, GA, USA, 16–21 June 2013. [Google Scholar]
- Danaee, P.; Ghaeini, R.; Hendrix, D.A. A Deep Learning Approach for Cancer Detection and Relevant Gene Identification. In Proceedings of the Pacific Symposium on Biocomputing, Kohala Coast, HI, USA, 3–7 January 2017; Volume 22, pp. 219–229. [Google Scholar]
- Yuan, Y.; Shi, Y.; Li, C.; Kim, J.; Cai, W.; Han, Z.; Feng, D.D. DeepGene: An advanced cancer type classifier based on deep learning and somatic point mutations. BMC Bioinform. 2016, 17, 476. [Google Scholar] [CrossRef]
- Stormo, G.D. DNA binding sites: Representation and discovery. Bioinformatics 2000, 16, 16–23. [Google Scholar] [CrossRef]
- Hassanzadeh, H.R.; Wang, M.D. DeeperBind: Enhancing prediction of sequence specificities of DNA binding proteins. In Proceedings of the 2016 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Shenzhen, China, 15–18 December 2016; pp. 178–183. [Google Scholar] [CrossRef]
- Zeng, H.; Edwards, M.D.; Liu, G.; Gifford, D.K. Convolutional neural network architectures for predicting DNA–protein binding. Bioinformatics 2016, 32, i121–i127. [Google Scholar] [CrossRef]
- Zhang, S.; Zhou, J.; Hu, H.; Gong, H.; Chen, L.; Cheng, C.; Zeng, J. A deep learning framework for modeling structural features of RNA-binding protein targets. Nucleic Acids Res. 2016, 44, e32. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Shen, H.B. RNA-protein binding motifs mining with a new hybrid deep learning based cross-domain knowledge integration approach. BMC Bioinform. 2017, 18, 136. [Google Scholar] [CrossRef]
- Pan, X.; Fan, Y.X.; Yan, J.; Shen, H.B. IPMiner: Hidden ncRNA-protein interaction sequential pattern mining with stacked autoencoder for accurate computational prediction. BMC Genom. 2016, 17, 582. [Google Scholar] [CrossRef] [PubMed]
- Haberal, I.; Ogul, H. DeepMBS: Prediction of Protein Metal Binding-Site Using Deep Learning Networks. In Proceedings of the 2017 Fourth International Conference on Mathematics and Computers in Sciences and in Industry (MCSI), Corfu, Greece, 24–27 August 2017; pp. 21–25. [Google Scholar] [CrossRef]
- Li, Y.; Shi, W.; Wasserman, W.W. Genome-wide prediction of cis-regulatory regions using supervised deep learning methods. BMC Bioinform. 2018, 19, 202. [Google Scholar] [CrossRef] [PubMed]
- Min, X.; Chen, N.; Chen, T.; Jiang, R. DeepEnhancer: Predicting enhancers by convolutional neural networks. In Proceedings of the 2016 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Shenzhen, China, 15–18 December 2016; pp. 637–644. [Google Scholar] [CrossRef]
- Liu, F.; Li, H.; Ren, C.; Bo, X.; Shu, W. PEDLA: Predicting enhancers with a deep learning-based algorithmic framework. Sci. Rep. 2016, 6, 28517. [Google Scholar] [CrossRef] [PubMed]
- Umarov, R.K.; Solovyev, V.V. Recognition of prokaryotic and eukaryotic promoters using convolutional deep learning neural networks. PLoS ONE 2017, 12, e0171410. [Google Scholar] [CrossRef]
- Qin, Q.; Feng, J. Imputation for transcription factor binding predictions based on deep learning. PLoS Comput. Biol. 2017, 13, e1005403. [Google Scholar] [CrossRef]
- Leung, M.K.K.; Xiong, H.Y.; Lee, L.J.; Frey, B.J. Deep learning of the tissue-regulated splicing code. Bioinformatics 2014, 30, i121–i129. [Google Scholar] [CrossRef]
- Dutta, A.; Dubey, T.; Singh, K.K.; Anand, A. SpliceVec: Distributed feature representations for splice junction prediction. Comput. Biol. Chem. 2018, 74, 434–441. [Google Scholar] [CrossRef]
- Lee, T.; Yoon, S. Boosted Categorical Restricted Boltzmann Machine for Computational Prediction of Splice Junctions. In Proceedings of the 32nd International Conference on Machine Learning, Lille, France, 6–11 July 2015; Bach, F., Blei, D., Eds.; PMLR: Lille, France, 2015; Volume 37, pp. 2483–2492. [Google Scholar]
- Frasca, M.; Pavesi, G. A neural network based algorithm for gene expression prediction from chromatin structure. In Proceedings of the 2013 International Joint Conference on Neural Networks (IJCNN), Dallas, TX, USA, 4–9 August 2013; pp. 1–8. [Google Scholar] [CrossRef]
- Singh, R.; Lanchantin, J.; Robins, G.; Qi, Y. DeepChrome: Deep-learning for predicting gene expression from histone modifications. Bioinformatics 2016, 32, i639–i648. [Google Scholar] [CrossRef]
- Chen, Y.; Li, Y.; Narayan, R.; Subramanian, A.; Xie, X. Gene expression inference with deep learning. Bioinformatics 2016, 32, 1832–1839. [Google Scholar] [CrossRef]
- Xie, R.; Wen, J.; Quitadamo, A.; Cheng, J.; Shi, X. A deep auto-encoder model for gene expression prediction. BMC Genom. 2017, 18, 845. [Google Scholar] [CrossRef]
- Kelley, D.R.; Snoek, J.; Rinn, J.L. Basset: Learning the regulatory code of the accessible genome with deep convolutional neural networks. Genome Res. 2016, 26, 990–999. [Google Scholar] [CrossRef]
- Quang, D.; Xie, X. DanQ: A hybrid convolutional and recurrent deep neural network for quantifying the function of DNA sequences. Nucleic Acids Res. 2016, 44, e107. [Google Scholar] [CrossRef]
- Zhou, J.; Troyanskaya, O.G. Predicting effects of noncoding variants with deep learning–based sequence model. Nat. Methods 2015, 12, 931. [Google Scholar] [CrossRef]
- Liu, F.; Ren, C.; Li, H.; Zhou, P.; Bo, X.; Shu, W. De novo identification of replication-timing domains in the human genome by deep learning. Bioinformatics 2016, 32, 641–649. [Google Scholar] [CrossRef]
- Zeng, H.; Gifford, D.K. Predicting the impact of non-coding variants on DNA methylation. Nucleic Acids Res. 2017, 45, e99. [Google Scholar] [CrossRef]
- Angermueller, C.; Lee, H.J.; Reik, W.; Stegle, O. Deepcpg: Accurate prediction of single-cell dna methylation states using deep learning. Genome Biol. 2017, 18, 67. [Google Scholar] [CrossRef]
- Li, H.; Hou, J.; Adhikari, B.; Lyu, Q.; Cheng, J. Deep learning methods for protein torsion angle prediction. BMC Bioinform. 2017, 18, 417. [Google Scholar] [CrossRef]
- Hattori, L.T.; Benitez, C.M.V.; Lopes, H.S. A deep bidirectional long short-term memory approach applied to the protein secondary structure prediction problem. In Proceedings of the 2017 IEEE Latin American Conference on Computational Intelligence (LA-CCI), Arequipa, Peru, 8–10 November 2017; pp. 1–6. [Google Scholar] [CrossRef]
- James, L.; Abdollah, D.; Rhys, H.; Alok, S.; Kuldip, P.; Abdul, S.; Yaoqi, Z.; Yuedong, Y. Predicting backbone C angles and dihedrals from protein sequences by stacked sparse auto-encoder deep neural network. J. Comput. Chem. 2014, 35, 2040–2046. [Google Scholar] [CrossRef]
- Fang, C.; Shang, Y.; Xu, D. Prediction of Protein Backbone Torsion Angles Using Deep Residual Inception Neural Networks. IEEE/ACM Trans. Comput. Biol. Bioinform. 2018, 1. [Google Scholar] [CrossRef]
- Gao, Y.; Wang, S.; Deng, M.; Xu, J. RaptorX-Angle: Real-value prediction of protein backbone dihedral angles through a hybrid method of clustering and deep learning. BMC Bioinform. 2018, 19, 100. [Google Scholar] [CrossRef]
- Heffernan, R.; Paliwal, K.; Lyons, J.; Dehzangi, A.; Sharma, A.; Wang, J.; Sattar, A.; Yang, Y.; Zhou, Y. Improving prediction of secondary structure, local backbone angles, and solvent accessible surface area of proteins by iterative deep learning. Sci. Rep. 2015, 5, 11476. [Google Scholar] [CrossRef]
- Fang, C.; Shang, Y.; Xu, D. A New Deep Neighbor Residual Network for Protein Secondary Structure Prediction. In Proceedings of the 2017 IEEE 29th International Conference on Tools with Artificial Intelligence (ICTAI), Boston, MA, USA, 6–8 November 2017; pp. 66–71. [Google Scholar] [CrossRef]
- Hu, Y.; Nie, T.; Shen, D.; Yu, G. Sequence Translating Model Using Deep Neural Block Cascade Network: Taking Protein Secondary Structure Prediction as an Example. In Proceedings of the 2018 IEEE International Conference on Big Data and Smart Computing (BigComp), Shanghai, China, 15–17 January 2018; pp. 58–65. [Google Scholar] [CrossRef]
- Liu, Y.; Cheng, J.; Ma, Y.; Chen, Y. Protein secondary structure prediction based on two dimensional deep convolutional neural networks. In Proceedings of the 2017 3rd IEEE International Conference on Computer and Communications (ICCC), Chengdu, China, 13–16 December 2017; pp. 1995–1999. [Google Scholar] [CrossRef]
- Aydin, Z.; Uzut, O.G. Combining classifiers for protein secondary structure prediction. In Proceedings of the 2017 9th International Conference on Computational Intelligence and Communication Networks (CICN), Girne, Cyprus, 16–17 September 2017; pp. 29–33. [Google Scholar] [CrossRef]
- Wang, S.; Peng, J.; Ma, J.; Xu, J. Protein Secondary Structure Prediction Using Deep Convolutional Neural Fields. Sci. Rep. 2016, 6, 18962. [Google Scholar] [CrossRef]
- Li, Y.; Shibuya, T. Malphite: A convolutional neural network and ensemble learning based protein secondary structure predictor. In Proceedings of the 2015 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Washington, DC, USA, 9–12 November 2015; pp. 1260–1266. [Google Scholar] [CrossRef]
- Zhou, J.; Troyanskaya, O. Deep Supervised and Convolutional Generative Stochastic Network for Protein Secondary Structure Prediction. arXiv, 2014; arXiv:1403.1347. [Google Scholar]
- Chen, Y. Long sequence feature extraction based on deep learning neural network for protein secondary structure prediction. In Proceedings of the 2017 IEEE 3rd Information Technology and Mechatronics Engineering Conference (ITOEC), Chongqing, China, 3–5 October 2017; pp. 843–847. [Google Scholar] [CrossRef]
- Bai, L.; Yang, L. A Unified Deep Learning Model for Protein Structure Prediction. In Proceedings of the 2017 3rd IEEE International Conference on Cybernetics (CYBCONF), Exeter, UK, 21–23 June 2017; pp. 1–6. [Google Scholar] [CrossRef]
- Ibrahim, W.; Abadeh, M.S. Protein fold recognition using Deep Kernelized Extreme Learning Machine and linear discriminant analysis. Neural Comput. Appl. 2018, 1–14. [Google Scholar] [CrossRef]
- Deng, L.; Fan, C.; Zeng, Z. A sparse autoencoder-based deep neural network for protein solvent accessibility and contact number prediction. BMC Bioinform. 2017, 18, 569. [Google Scholar] [CrossRef]
- Li, H.; Lyu, Q.; Cheng, J. A Template-Based Protein Structure Reconstruction Method Using Deep Autoencoder Learning. J. Proteom. Bioinform. 2016, 9, 306–313. [Google Scholar] [CrossRef]
- Di Lena, P.; Nagata, K.; Baldi, P. Deep architectures for protein contact map prediction. Bioinformatics 2012, 28, 2449–2457. [Google Scholar] [CrossRef]
- Wu, H.; Wang, K.; Lu, L.; Xue, Y.; Lyu, Q.; Jiang, M. Deep Conditional Random Field Approach to Transmembrane Topology Prediction and Application to GPCR Three-Dimensional Structure Modeling. IEEE/ACM Trans. Comput. Biol. Bioinform. 2017, 14, 1106–1114. [Google Scholar] [CrossRef]
- Nguyen, S.P.; Shang, Y.; Xu, D. DL-PRO: A novel deep learning method for protein model quality assessment. In Proceedings of the 2014 International Joint Conference on Neural Networks (IJCNN), Beijing, China, 6–11 July 2014; pp. 2071–2078. [Google Scholar] [CrossRef]
- Nguyen, S.; Li, Z.; Shang, Y. Deep Networks and Continuous Distributed Representation of Protein Sequences for Protein Quality Assessment. In Proceedings of the 2017 IEEE 29th International Conference on Tools with Artificial Intelligence (ICTAI), Boston, MA, USA, 6–8 November 2017; pp. 527–534. [Google Scholar] [CrossRef]
- Wang, J.; Li, Z.; Shang, Y. New Deep Neural Networks for Protein Model Evaluation. In Proceedings of the 2017 IEEE 29th International Conference on Tools with Artificial Intelligence (ICTAI), Boston, MA, USA, 6–8 November 2017; pp. 309–313. [Google Scholar] [CrossRef]
- Li, Z.; Nguyen, S.P.; Xu, D.; Shang, Y. Protein Loop Modeling Using Deep Generative Adversarial Network. In Proceedings of the 2017 IEEE 29th International Conference on Tools with Artificial Intelligence (ICTAI), Boston, MA, USA, 6–8 November 2017; pp. 1085–1091. [Google Scholar] [CrossRef]
- Nguyen, S.P.; Li, Z.; Xu, D.; Shang, Y. New Deep Learning Methods for Protein Loop Modeling. IEEE/ACM Trans. Comput. Biol. Bioinform. 2017, 1. [Google Scholar] [CrossRef]
- Spencer, M.; Eickholt, J.; Cheng, J. A Deep Learning Network Approach to ab initio Protein Secondary Structure Prediction. IEEE/ACM Trans. Comput. Biol. Bioinform. 2015, 12, 103–112. [Google Scholar] [CrossRef]
- Eickholt, J.; Cheng, J. DNdisorder: Predicting protein disorder using boosting and deep networks. BMC Bioinform. 2013, 14, 88. [Google Scholar] [CrossRef]
- Sun, T.; Zhou, B.; Lai, L.; Pei, J. Sequence-based prediction of protein protein interaction using a deep-learning algorithm. BMC Bioinform. 2017, 18, 277. [Google Scholar] [CrossRef]
- Lei, H.; Wen, Y.; Elazab, A.; Tan, E.L.; Zhao, Y.; Lei, B. Protein-protein Interactions Prediction via Multimodal Deep Polynomial Network and Regularized Extreme Learning Machine. IEEE J. Biomed. Health Inform. 2018. [Google Scholar] [CrossRef]
- Chen, H.; Shen, J.; Wang, L.; Song, J. Leveraging Stacked Denoising Autoencoder in Prediction of Pathogen-Host Protein-Protein Interactions. In Proceedings of the 2017 IEEE International Congress on Big Data (BigData Congress), Honolulu, HI, USA, 25–30 June 2017; pp. 368–375. [Google Scholar] [CrossRef]
- Huang, L.; Liao, L.; Wu, C.H. Completing sparse and disconnected protein-protein network by deep learning. BMC Bioinform. 2018, 19, 103. [Google Scholar] [CrossRef]
- Zhao, Z.; Gong, X. Protein-protein interaction interface residue pair prediction based on deep learning architecture. IEEE/ACM Trans. Comput. Biol. Bioinform. 2018, 1. [Google Scholar] [CrossRef]
- Farhoodi, R.; Akbal-Delibas, B.; Haspel, N. Accurate prediction of docked protein structure similarity using neural networks and restricted Boltzmann machines. In Proceedings of the 2015 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Washington, DC, USA, 9–12 November 2015; pp. 1296–1303. [Google Scholar] [CrossRef]
- Han, Y.; Kim, D. Deep convolutional neural networks for pan-specific peptide-MHC class I binding prediction. BMC Bioinform. 2017, 18, 585. [Google Scholar] [CrossRef]
- Kuksa, P.P.; Min, M.R.; Dugar, R.; Gerstein, M. High-order neural networks and kernel methods for peptide-MHC binding prediction. Bioinformatics 2015, 31, 3600–3607. [Google Scholar] [CrossRef]
- Wang, L.; You, Z.H.; Chen, X.; Xia, S.X.; Liu, F.; Yan, X.; Zhou, Y. Computational Methods for the Prediction of Drug-Target Interactions from Drug Fingerprints and Protein Sequences by Stacked Auto-Encoder Deep Neural Network. In Bioinformatics Research and Applications; Cai, Z., Daescu, O., Li, M., Eds.; Springer International Publishing: Cham, Switerland, 2017; pp. 46–58. [Google Scholar]
- Bahi, M.; Batouche, M. Drug-Target Interaction Prediction in Drug Repositioning Based on Deep Semi-Supervised Learning. In Computational Intelligence and Its Applications; Amine, A., Mouhoub, M., Ait Mohamed, O., Djebbar, B., Eds.; Springer International Publishing: Cham, Switerland, 2018; pp. 302–313. [Google Scholar]
- Peng-Wei; Chan, K.C.C.; You, Z.H. Large-scale prediction of drug-target interactions from deep representations. In Proceedings of the 2016 International Joint Conference on Neural Networks (IJCNN), Vancouver, BC, Canada, 24–29 July 2016; pp. 1236–1243. [Google Scholar] [CrossRef]
- Masatoshi, H.; Kei, T.; Hiroaki, I.; Jun, Y.; Jianguo, P.; Jinlong, H.; Yasushi, O. CGBVS-DNN: Prediction of Compound-protein Interactions Based on Deep Learning. Mol. Inform. 2016, 36, 1600045. [Google Scholar] [CrossRef]
- Tian, K.; Shao, M.; Zhou, S.; Guan, J. Boosting compound-protein interaction prediction by deep learning. In Proceedings of the 2015 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Washington, DC, USA, 9–12 November 2015; pp. 29–34. [Google Scholar] [CrossRef]
- Tian, K.; Shao, M.; Wang, Y.; Guan, J.; Zhou, S. Boosting compound-protein interaction prediction by deep learning. Methods 2016, 110, 64–72. [Google Scholar] [CrossRef]
- Ramadan, R.A.; Vasilakos, A.V. Brain computer interface: Control signals review. Neurocomputing 2017, 223, 26–44. [Google Scholar] [CrossRef]
- Al-Nafjan, A.; Hosny, M.; Al-Ohali, Y.; Al-Wabil, A. Review and Classification of Emotion Recognition Based on EEG Brain-Computer Interface System Research: A Systematic Review. Appl. Sci. 2017, 7, 1239. [Google Scholar] [CrossRef]
- Ahn, M.; Jun, S.C. Performance variation in motor imagery brain–computer interface: A brief review. J. Neurosci. Methods 2015, 243, 103–110. [Google Scholar] [CrossRef]
- Alonso-Valerdi, L.M.; Salido-Ruiz, R.A.; Ramirez-Mendoza, R.A. Motor imagery based brain–computer interfaces: An emerging technology to rehabilitate motor deficits. Neuropsychologia 2015, 79, 354–363. [Google Scholar] [CrossRef]
- Pattnaik, P.K.; Sarraf, J. Brain Computer Interface issues on hand movement. J. King Saud Univ. Comput. Inf. Sci. 2018, 30, 18–24. [Google Scholar] [CrossRef]
- McFarland, D.; Wolpaw, J. EEG-based brain–computer interfaces. Curr. Opin. Biomed. Eng. 2017, 4, 194–200. [Google Scholar] [CrossRef]
- Chan, H.L.; Kuo, P.C.; Cheng, C.Y.; Chen, Y.S. Challenges and Future Perspectives on Electroencephalogram-Based Biometrics in Person Recognition. Front. Neuroinform. 2018, 12, 66. [Google Scholar] [CrossRef]
- Langkvist, M.; Karlsson, L.; Loutfi, A. Sleep Stage Classification Using Unsupervised Feature Learning. Adv. Artif. Neural Syst. 2012, 2012, 9. [Google Scholar] [CrossRef]
- Li, K.; Li, X.; Zhang, Y.; Zhang, A. Affective state recognition from EEG with deep belief networks. In Proceedings of the 2013 IEEE International Conference on Bioinformatics and Biomedicine, Shanghai, China, 18–21 December 2013; pp. 305–310. [Google Scholar] [CrossRef]
- Jia, X.; Li, K.; Li, X.; Zhang, A. A Novel Semi-Supervised Deep Learning Framework for Affective State Recognition on EEG Signals. In Proceedings of the 2014 IEEE International Conference on Bioinformatics and Bioengineering, Boca Raton, FL, USA, 10–12 November 2014; pp. 30–37. [Google Scholar] [CrossRef]
- Xu, H.; Plataniotis, K.N. EEG-based affect states classification using Deep Belief Networks. In Proceedings of the 2016 Digital Media Industry Academic Forum (DMIAF), Santorini, Greece, 4–6 July 2016; pp. 148–153. [Google Scholar] [CrossRef]
- Zheng, W.L.; Guo, H.T.; Lu, B.L. Revealing critical channels and frequency bands for emotion recognition from EEG with deep belief network. In Proceedings of the 2015 7th International IEEE/EMBS Conference on Neural Engineering (NER), Montpellier, France, 22–24 April 2015; pp. 154–157. [Google Scholar] [CrossRef]
- Zheng, W.L.; Zhu, J.Y.; Peng, Y.; Lu, B.L. EEG-based emotion classification using deep belief networks. In Proceedings of the 2014 IEEE International Conference on Multimedia and Expo (ICME), Chengdu, China, 14–18 July 2014; pp. 1–6. [Google Scholar] [CrossRef]
- Zheng, W.L.; Lu, B.L. Investigating Critical Frequency Bands and Channels for EEG-Based Emotion Recognition with Deep Neural Networks. IEEE Trans. Auton. Ment. Dev. 2015, 7, 162–175. [Google Scholar] [CrossRef]
- Gao, Y.; Lee, H.J.; Mehmood, R.M. Deep learninig of EEG signals for emotion recognition. In Proceedings of the 2015 IEEE International Conference on Multimedia Expo Workshops (ICMEW), Turin, Italy, 29 June–3 July 2015; pp. 1–5. [Google Scholar] [CrossRef]
- Jirayucharoensak, S.; Pan-Ngum, S.; Israsena, P. Eeg-based emotion recognition using deep learning network with principal component based covariate shift adaptation. Sci. World J. 2014, 2014, 627892. [Google Scholar] [CrossRef]
- KB, S.K.; Krishna, G.; Bhalaji, N.; Chithra, S. BCI cinematics—A pre-release analyser for movies using H2O deep learning platform. Comput. Electr. Eng. 2018, 74, 547–556. [Google Scholar] [CrossRef]
- Tripathi, S.; Acharya, S.; Sharma, R.; Mittal, S.; Bhattacharya, S. Using deep and convolutional neural networks for accurate emotion classification on deap dataset. In Proceedings of the Twenty-Ninth IAAI Conference, San Francisco, CA, USA, 6–9 February 2017; pp. 4746–4752. [Google Scholar]
- Li, J.; Zhang, Z.; He, H. Hierarchical Convolutional Neural Networks for EEG-Based Emotion Recognition. Cogn. Comput. 2018, 10, 368–380. [Google Scholar] [CrossRef]
- Stober, S.; Cameron, D.J.; Grahn, J.A. Classifying EEG Recordings of Rhythm Perception. In Proceedings of the 15th International Society for Music Information Retrieval Conference (ISMIR 2014), Taipei, Taiwan, 27–31 Ocober 2014. [Google Scholar]
- Stober, S.; Cameron, D.J.; Grahn, J.A. Using Convolutional Neural Networks to Recognize Rhythm Stimuli from Electroencephalography Recordings. Adv. Neural Inf. Process. Syst. 2014, 27, 1449–1457. [Google Scholar]
- Stober, S.; Sternin, A.; Owen, A.M.; Grahn, J.A. Deep Feature Learning for EEG Recordings. arXiv, 2015; arXiv:1511.04306. [Google Scholar]
- Sun, X.; Qian, C.; Chen, Z.; Wu, Z.; Luo, B.; Pan, G. Remembered or Forgotten?—An EEG-Based Computational Prediction Approach. PLoS ONE 2016, 11, 1–20. [Google Scholar] [CrossRef]
- Wand, M.; Schultz, T. Pattern learning with deep neural networks in EMG-based speech recognition. In Proceedings of the 2014 36th Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Chicago, IL, USA, 26–30 August 2014; pp. 4200–4203. [Google Scholar] [CrossRef]
- Cecotti, H.; Graeser, A. Convolutional Neural Network with embedded Fourier Transform for EEG classification. In Proceedings of the 2008 19th International Conference on Pattern Recognition, Tampa, FL, USA, 8–11 December 2008; pp. 1–4. [Google Scholar] [CrossRef]
- Bashivan, P.; Rish, I.; Yeasin, M.; Codella, N. Learning Representations from EEG with Deep Recurrent-Convolutional Neural Networks. arXiv, 2015; arXiv:1511.06448. [Google Scholar]
- Spampinato, C.; Palazzo, S.; Kavasidis, I.; Giordano, D.; Souly, N.; Shah, M. Deep Learning Human Mind for Automated Visual Classification. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 4503–4511. [Google Scholar] [CrossRef]
- Kwak, N.S.; Müller, K.R.; Lee, S.W. A convolutional neural network for steady state visual evoked potential classification under ambulatory environment. PLoS ONE 2017, 12, 1–20. [Google Scholar] [CrossRef]
- Thomas, J.; Maszczyk, T.; Sinha, N.; Kluge, T.; Dauwels, J. Deep learning-based classification for brain-computer interfaces. In Proceedings of the 2017 IEEE International Conference on Systems, Man, and Cybernetics (SMC), Banff, AB, Canada, 5–8 October 2017; pp. 234–239. [Google Scholar] [CrossRef]
- Ahn, M.H.; Min, B.K. Applying deep-learning to a top-down SSVEP BMI. In Proceedings of the 2018 6th International Conference on Brain-Computer Interface (BCI), GangWon, Korea, 15–17 January 2018; pp. 1–3. [Google Scholar] [CrossRef]
- Ma, T.; Li, H.; Yang, H.; Lv, X.; Li, P.; Liu, T.; Yao, D.; Xu, P. The extraction of motion-onset VEP BCI features based on deep learning and compressed sensing. J. Neurosci. Methods 2017, 275, 80–92. [Google Scholar] [CrossRef]
- Zhao, S.; Rudzicz, F. Classifying phonological categories in imagined and articulated speech. In Proceedings of the 2015 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Brisbane, Australia, 19–24 April 2015; pp. 992–996. [Google Scholar] [CrossRef]
- Ahmed, S.; Merino, L.M.; Mao, Z.; Meng, J.; Robbins, K.; Huang, Y. A Deep Learning method for classification of images RSVP events with EEG data. In Proceedings of the 2013 IEEE Global Conference on Signal and Information Processing, Austin, TX, USA, 3–5 December 2013; pp. 33–36. [Google Scholar] [CrossRef]
- Jiao, Z.; Gao, X.; Wang, Y.; Li, J.; Xu, H. Deep Convolutional Neural Networks for mental load classification based on EEG data. Pattern Recognit. 2018, 76, 582–595. [Google Scholar] [CrossRef]
- Cecotti, H.; Graser, A. Convolutional Neural Networks for P300 Detection with Application to Brain-Computer Interfaces. IEEE Trans. Pattern Anal. Mach. Intell. 2011, 33, 433–445. [Google Scholar] [CrossRef]
- Kshirsagar, G.B.; Londhe, N.D. Deep convolutional neural network based character detection in devanagari script input based P300 speller. In Proceedings of the 2017 International Conference on Electrical, Electronics, Communication, Computer, and Optimization Techniques (ICEECCOT), Mysuru, India, 5–16 December 2017; pp. 507–511. [Google Scholar] [CrossRef]
- Liu, M.; Wu, W.; Gu, Z.; Yu, Z.; Qi, F.; Li, Y. Deep learning based on Batch Normalization for P300 signal detection. Neurocomputing 2018, 275, 288–297. [Google Scholar] [CrossRef]
- Lawhern, V.J.; Solon, A.J.; Waytowich, N.R.; Gordon, S.M.; Hung, C.P.; Lance, B.J. EEGNet: A Compact Convolutional Network for EEG-based Brain-Computer Interfaces. J. Neural Eng. 2018, 15, 056013. [Google Scholar] [CrossRef]
- Nashed, N.N.; Eldawlatly, S.; Aly, G.M. A deep learning approach to single-trial classification for P300 spellers. In Proceedings of the 2018 IEEE 4th Middle East Conference on Biomedical Engineering (MECBME), Tunis, Tunisi, 28–30 March 2018; pp. 11–16. [Google Scholar] [CrossRef]
- Manor, R.; Geva, A.B. Convolutional Neural Network for Multi-Category Rapid Serial Visual Presentation BCI. Front. Comput. Neurosci. 2015, 9, 146. [Google Scholar] [CrossRef]
- Hajinoroozi, M.; Mao, Z.; Huang, Y. Prediction of driver’s drowsy and alert states from EEG signals with deep learning. In Proceedings of the 2015 IEEE 6th International Workshop on Computational Advances in Multi-Sensor Adaptive Processing (CAMSAP), Cancun, Mexico, 13–16 December 2015; pp. 493–496. [Google Scholar] [CrossRef]
- Mao, Z.; Yao, W.X.; Huang, Y. EEG-based biometric identification with deep learning. In Proceedings of the 2017 8th International IEEE/EMBS Conference on Neural Engineering (NER), Shanghai, China, 25–28 May 2017; pp. 609–612. [Google Scholar] [CrossRef]
- Hajinoroozi, M.; Mao, Z.; Jung, T.P.; Lin, C.T.; Huang, Y. EEG-based prediction of driver’s cognitive performance by deep convolutional neural network. Signal Process. Image Commun. 2016, 47, 549–555. [Google Scholar] [CrossRef]
- Chai, R.; Ling, S.H.; San, P.P.; Naik, G.R.; Nguyen, T.N.; Tran, Y.; Craig, A.; Nguyen, H.T. Improving EEG-Based Driver Fatigue Classification Using Sparse-Deep Belief Networks. Front. Neurosci. 2017, 11, 103. [Google Scholar] [CrossRef]
- Deep Feature Learning Using Target Priors with Applications in ECoG Signal Decoding for BCI; AAAI Press: Menlo Park, CA, USA, 2013.
- Yang, H.; Sakhavi, S.; Ang, K.K.; Guan, C. On the use of convolutional neural networks and augmented csp features for multi-class motor imagery of eeg signals classification. In Proceedings of the 2015 37th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Milan, Italy, 25–29 August 2015; pp. 2620–2623. [Google Scholar]
- Hartmann, K.G.; Schirrmeister, R.T.; Ball, T. Hierarchical internal representation of spectral features in deep convolutional networks trained for EEG decoding. In Proceedings of the 2018 6th International Conference on Brain-Computer Interface (BCI), GangWon, Korea, 15–17 January 2018; pp. 1–6. [Google Scholar] [CrossRef]
- Sakhavi, S.; Guan, C.; Yan, S. Learning Temporal Information for Brain-Computer Interface Using Convolutional Neural Networks. IEEE Trans. Neural Netw. Learn. Syst. 2018, 29, 1–11. [Google Scholar] [CrossRef]
- Carvalho, S.R.; Filho, I.C.; Resende, D.O.D.; Siravenha, A.C.; Souza, C.D.; Debarba, H.G.; Gomes, B.; Boulic, R. A Deep Learning Approach for Classification of Reaching Targets from EEG Images. In Proceedings of the 2017 30th SIBGRAPI Conference on Graphics, Patterns and Images (SIBGRAPI), Niteroi, Brazil, 17–20 October 2017; pp. 178–184. [Google Scholar] [CrossRef]
- Tabar, Y.R.; Halici, U. A novel deep learning approach for classification of EEG motor imagery signals. J. Neural Eng. 2017, 14, 016003. [Google Scholar] [CrossRef]
- Ren, Y.; Wu, Y. Convolutional deep belief networks for feature extraction of EEG signal. In Proceedings of the 2014 International Joint Conference on Neural Networks (IJCNN), Beijing, China, 6–11 July 2014; pp. 2850–2853. [Google Scholar] [CrossRef]
- Jingwei, L.; Yin, C.; Weidong, Z. Deep learning EEG response representation for brain computer interface. In Proceedings of the 2015 34th Chinese Control Conference (CCC), Hangzhou, China, 28–30 July 2015; pp. 3518–3523. [Google Scholar] [CrossRef]
- Sakhavi, S.; Guan, C.; Yan, S. Parallel convolutional-linear neural network for motor imagery classification. In Proceedings of the 2015 23rd European Signal Processing Conference (EUSIPCO), Nice, France, 31 August–4 September 2015; pp. 2736–2740. [Google Scholar] [CrossRef]
- Tang, Z.; Li, C.; Sun, S. Single-trial EEG classification of motor imagery using deep convolutional neural networks. Opt. Int. J. Light Electron. Opt. 2017, 130, 11–18. [Google Scholar] [CrossRef]
- Lu, N.; Li, T.; Ren, X.; Miao, H. A Deep Learning Scheme for Motor Imagery Classification based on Restricted Boltzmann Machines. IEEE Trans. Neural Syst. Rehabil. Eng. 2017, 25, 566–576. [Google Scholar] [CrossRef]
- An, X.; Kuang, D.; Guo, X.; Zhao, Y.; He, L. A Deep Learning Method for Classification of EEG Data Based on Motor Imagery. In Intelligent Computing in Bioinformatics; Huang, D.S., Han, K., Gromiha, M., Eds.; Springer International Publishing: Cham, Switerland, 2014; pp. 203–210. [Google Scholar] [CrossRef]
- Li, J.; Cichocki, A. Deep Learning of Multifractal Attributes from Motor Imagery Induced EEG. In Neural Information Processing; Loo, C.K., Yap, K.S., Wong, K.W., Teoh, A., Huang, K., Eds.; Springer International Publishing: Cham, Switerland, 2014; pp. 503–510. [Google Scholar] [CrossRef]
- Kumar, S.; Sharma, A.; Mamun, K.; Tsunoda, T. A Deep Learning Approach for Motor Imagery EEG Signal Classification. In Proceedings of the 2016 3rd Asia-Pacific World Congress on Computer Science and Engineering (APWC on CSE), Nadi, Fiji, 5–6 December 2016; pp. 34–39. [Google Scholar] [CrossRef]
- Sturm, I.; Lapuschkin, S.; Samek, W.; Müller, K.R. Interpretable deep neural networks for single-trial EEG classification. J. Neurosci. Methods 2016, 274, 141–145. [Google Scholar] [CrossRef]
- Hennrich, J.; Herff, C.; Heger, D.; Schultz, T. Investigating deep learning for fNIRS based BCI. In Proceedings of the 2015 37th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Milan, Italy, 25–29 August 2015; pp. 2844–2847. [Google Scholar] [CrossRef]
- Völker, M.; Schirrmeister, R.T.; Fiederer, L.D.J.; Burgard, W.; Ball, T. Deep transfer learning for error decoding from non-invasive EEG. In Proceedings of the 2018 6th International Conference on Brain-Computer Interface (BCI), GangWon, Korea, 15–17 January 2018; pp. 1–6. [Google Scholar] [CrossRef]
- Huve, G.; Takahashi, K.; Hashimoto, M. Brain activity recognition with a wearable fNIRS using neural networks. In Proceedings of the 2017 IEEE International Conference on Mechatronics and Automation (ICMA), Takamatsu, Japan, 6–9 August 2017; pp. 1573–1578. [Google Scholar] [CrossRef]
- Yin, Z.; Zhao, M.; Wang, Y.; Yang, J.; Zhang, J. Recognition of emotions using multimodal physiological signals and an ensemble deep learning model. Comput. Methods Programs Biomed. 2017, 140, 93–110. [Google Scholar] [CrossRef]
- Du, L.H.; Liu, W.; Zheng, W.L.; Lu, B.L. Detecting driving fatigue with multimodal deep learning. In Proceedings of the 2017 8th International IEEE/EMBS Conference on Neural Engineering (NER), Shanghai, China, 25–28 May 2017; pp. 74–77. [Google Scholar] [CrossRef]
- Acharya, U.R.; Oh, S.L.; Hagiwara, Y.; Tan, J.H.; Adeli, H.; Subha, D.P. Automated EEG-based screening of depression using deep convolutional neural network. Comput. Methods Programs Biomed. 2018, 161, 103–113. [Google Scholar] [CrossRef]
- Fraiwan, L.; Lweesy, K. Neonatal sleep state identification using deep learning autoencoders. In Proceedings of the 2017 IEEE 13th International Colloquium on Signal Processing its Applications (CSPA), Batu Ferringhi, Malaysia, 10–12 March 2017; pp. 228–231. [Google Scholar] [CrossRef]
- Wu, Z.; Ding, X.; Zhang, G. A Novel Method for Classification of ECG Arrhythmias Using Deep Belief Networks. Int. J. Comput. Intell. Appl. 2016, 15, 1650021. [Google Scholar] [CrossRef]
- Rahhal, M.A.; Bazi, Y.; AlHichri, H.; Alajlan, N.; Melgani, F.; Yager, R. Deep learning approach for active classification of electrocardiogram signals. Inf. Sci. 2016, 345, 340–354. [Google Scholar] [CrossRef]
- Sannino, G.; Pietro, G.D. A deep learning approach for ECG-based heartbeat classification for arrhythmia detection. Future Gener. Comput. Syst. 2018, 86, 446–455. [Google Scholar] [CrossRef]
- Acharya, U.R.; Oh, S.L.; Hagiwara, Y.; Tan, J.H.; Adam, M.; Gertych, A.; Tan, R.S. A deep convolutional neural network model to classify heartbeats. Comput. Biol. Med. 2017, 89, 389–396. [Google Scholar] [CrossRef]
- Kiranyaz, S.; Ince, T.; Gabbouj, M. Real-Time Patient-Specific ECG Classification by 1-D Convolutional Neural Networks. IEEE Trans. Biomed. Eng. 2016, 63, 664–675. [Google Scholar] [CrossRef]
- Acharya, U.R.; Fujita, H.; Lih, O.S.; Hagiwara, Y.; Tan, J.H.; Adam, M. Automated detection of arrhythmias using different intervals of tachycardia ECG segments with convolutional neural network. Inf. Sci. 2017, 405, 81–90. [Google Scholar] [CrossRef]
- Acharya, U.R.; Fujita, H.; Oh, S.L.; Raghavendra, U.; Tan, J.H.; Adam, M.; Gertych, A.; Hagiwara, Y. Automated identification of shockable and non-shockable life-threatening ventricular arrhythmias using convolutional neural network. Future Gener. Comput. Syst. 2018, 79, 952–959. [Google Scholar] [CrossRef]
- Majumdar, A.; Ward, R. Robust greedy deep dictionary learning for ECG arrhythmia classification. In Proceedings of the 2017 International Joint Conference on Neural Networks (IJCNN), Anchorage, AK, USA, 14–19 May 2017; pp. 4400–4407. [Google Scholar] [CrossRef]
- Zheng, G.; Ji, S.; Dai, M.; Sun, Y. ECG Based Identification by Deep Learning. In Biometric Recognition; Zhou, J., Wang, Y., Sun, Z., Xu, Y., Shen, L., Feng, J., Shan, S., Qiao, Y., Guo, Z., Yu, S., Eds.; Springer International Publishing: Cham, Switerland, 2017; pp. 503–510. [Google Scholar] [CrossRef]
- Luo, K.; Li, J.; Wang, Z.; Cuschieri, A. Patient-Specific Deep Architectural Model for ECG Classification. J. Healthc. Eng. 2017, 2017, 13. [Google Scholar] [CrossRef]
- Huanhuan, M.; Yue, Z. Classification of Electrocardiogram Signals with Deep Belief Networks. In Proceedings of the 2014 IEEE 17th International Conference on Computational Science and Engineering, Chengdu, China, 19–21 December 2014; pp. 7–12. [Google Scholar] [CrossRef]
- Yan, Y.; Qin, X.; Wu, Y.; Zhang, N.; Fan, J.; Wang, L. A restricted Boltzmann machine based two-lead electrocardiography classification. In Proceedings of the 2015 IEEE 12th International Conference on Wearable and Implantable Body Sensor Networks (BSN), Cambridge, MA, USA, 9–12 June 2015; pp. 1–9. [Google Scholar] [CrossRef]
- Taji, B.; Chan, A.D.C.; Shirmohammadi, S. Classifying measured electrocardiogram signal quality using deep belief networks. In Proceedings of the 2017 IEEE International Instrumentation and Measurement Technology Conference (I2MTC), Turin, Italy, 22–25 May 2017; pp. 1–6. [Google Scholar] [CrossRef]
- Mirowski, P.; Madhavan, D.; LeCun, Y.; Kuzniecky, R. Classification of patterns of EEG synchronization for seizure prediction. Clin. Neurophysiol. 2009, 120, 1927–1940. [Google Scholar] [CrossRef]
- Mirowski, P.W.; LeCun, Y.; Madhavan, D.; Kuzniecky, R. Comparing SVM and convolutional networks for epileptic seizure prediction from intracranial EEG. In Proceedings of the 2008 IEEE Workshop on Machine Learning for Signal Processing, Cancun, Mexico, 16–19 October 2008; pp. 244–249. [Google Scholar] [CrossRef]
- Thodoroff, P.; Pineau, J.; Lim, A. Learning Robust Features using Deep Learning for Automatic Seizure Detection. In Machine Learning for Healthcare Conference; Doshi-Velez, F., Fackler, J., Kale, D., Wallace, B., Wiens, J., Eds.; PMLR: Los Angeles, CA, USA, 2016; Volume 56, pp. 178–190. [Google Scholar]
- Acharya, U.R.; Oh, S.L.; Hagiwara, Y.; Tan, J.H.; Adeli, H. Deep convolutional neural network for the automated detection and diagnosis of seizure using EEG signals. Comput. Biol. Med. 2017, 100, 270–278. [Google Scholar] [CrossRef]
- Antoniades, A.; Spyrou, L.; Took, C.C.; Sanei, S. Deep learning for epileptic intracranial EEG data. In Proceedings of the 2016 IEEE 26th International Workshop on Machine Learning for Signal Processing (MLSP), Vietri sul Mare, Italy, 13–16 September 2016; pp. 1–6. [Google Scholar] [CrossRef]
- Liang, J.; Lu, R.; Zhang, C.; Wang, F. Predicting Seizures from Electroencephalography Recordings: A Knowledge Transfer Strategy. In Proceedings of the 2016 IEEE International Conference on Healthcare Informatics (ICHI), Chicago, IL, USA, 4–7 October 2016; pp. 184–191. [Google Scholar] [CrossRef]
- Page, A.; Shea, C.; Mohsenin, T. Wearable seizure detection using convolutional neural networks with transfer learning. In Proceedings of the 2016 IEEE International Symposium on Circuits and Systems (ISCAS), Montreal, QC, Canada, 22–25 May 2016; pp. 1086–1089. [Google Scholar] [CrossRef]
- Hosseini, M.P.; Tran, T.X.; Pompili, D.; Elisevich, K.; Soltanian-Zadeh, H. Deep Learning with Edge Computing for Localization of Epileptogenicity Using Multimodal rs-fMRI and EEG Big Data. In Proceedings of the 2017 IEEE International Conference on Autonomic Computing (ICAC), Columbus, OH, USA, 17–21 July 2017; pp. 83–92. [Google Scholar] [CrossRef]
- Wulsin, D.F.; Gupta, J.R.; Mani, R.; Blanco, J.A.; Litt, B. Modeling electroencephalography waveforms with semi-supervised deep belief nets: Fast classification and anomaly measurement. J. Neural Eng. 2011, 8, 036015. [Google Scholar] [CrossRef]
- Wulsin, D.; Blanco, J.; Mani, R.; Litt, B. Semi-Supervised Anomaly Detection for EEG Waveforms Using Deep Belief Nets. In Proceedings of the 2010 Ninth International Conference on Machine Learning and Applications, Washington, DC, USA, 2–14 December 2010; pp. 436–441. [Google Scholar] [CrossRef]
- San, P.P.; Ling, S.H.; Nguyen, H.T. Deep learning framework for detection of hypoglycemic episodes in children with type 1 diabetes. In Proceedings of the 2016 38th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Orlando, FL, USA, 16–20 August 2016; pp. 3503–3506. [Google Scholar] [CrossRef]
- Van Putten, M.J.A.M.; Hofmeijer, J.; Ruijter, B.J.; Tjepkema-Cloostermans, M.C. Deep Learning for outcome prediction of postanoxic coma. In EMBEC & NBC 2017; Eskola, H., Väisänen, O., Viik, J., Hyttinen, J., Eds.; Springer: Singapore, 2018; pp. 506–509. [Google Scholar] [CrossRef]
- Pourbabaee, B.; Roshtkhari, M.J.; Khorasani, K. Deep Convolutional Neural Networks and Learning ECG Features for Screening Paroxysmal Atrial Fibrillation Patients. IEEE Trans. Syst. Man Cybern. Syst. 2017, 48, 1–10. [Google Scholar] [CrossRef]
- Cheng, M.; Sori, W.J.; Jiang, F.; Khan, A.; Liu, S. Recurrent Neural Network Based Classification of ECG Signal Features for Obstruction of Sleep Apnea Detection. In Proceedings of the 2017 IEEE International Conference on Computational Science and Engineering (CSE) and IEEE International Conference on Embedded and Ubiquitous Computing (EUC), Guangzhou, China, 21–24 July 2017; Volume 2, pp. 199–202. [Google Scholar] [CrossRef]
- Shashikumar, S.P.; Shah, A.J.; Li, Q.; Clifford, G.D.; Nemati, S. A deep learning approach to monitoring and detecting atrial fibrillation using wearable technology. In Proceedings of the 2017 IEEE EMBS International Conference on Biomedical Health Informatics (BHI), Orlando, FL, USA, 16–19 February 2017; pp. 141–144. [Google Scholar] [CrossRef]
- Muduli, P.R.; Gunukula, R.R.; Mukherjee, A. A deep learning approach to fetal-ECG signal reconstruction. In Proceedings of the 2016 Twenty Second National Conference on Communication (NCC), Guwahati, India, 4–6 March 2016; pp. 1–6. [Google Scholar] [CrossRef]
- Zhu, X.; Zheng, W.L.; Lu, B.L.; Chen, X.; Chen, S.; Wang, C. EOG-based drowsiness detection using convolutional neural networks. In Proceedings of the 2014 International Joint Conference on Neural Networks (IJCNN), Beijing, China, 6–11 July 2014; pp. 128–134. [Google Scholar] [CrossRef]
- Schirrmeister, R.T.; Springenberg, J.T.; Fiederer, L.D.J.; Glasstetter, M.; Eggensperger, K.; Tangermann, M.; Hutter, F.; Burgard, W.; Ball, T. Deep learning with convolutional neural networks for EEG decoding and visualization. Hum. Brain Mapp. 2017, 38, 5391–5420. [Google Scholar] [CrossRef]
- Christodoulidis, S.; Anthimopoulos, M.; Ebner, L.; Christe, A.; Mougiakakou, S. Multisource Transfer Learning with Convolutional Neural Networks for Lung Pattern Analysis. IEEE J. Biomed. Health Inform. 2017, 21, 76–84. [Google Scholar] [CrossRef]
- Chang, H.; Han, J.; Zhong, C.; Snijders, A.M.; Mao, J.H. Unsupervised Transfer Learning via Multi-Scale Convolutional Sparse Coding for Biomedical Applications. IEEE Trans. Pattern Anal. Mach. Intell. 2018, 40, 1182–1194. [Google Scholar] [CrossRef]
- Chen, H.; Ni, D.; Qin, J.; Li, S.; Yang, X.; Wang, T.; Heng, P.A. Standard Plane Localization in Fetal Ultrasound via Domain Transferred Deep Neural Networks. IEEE J. Biomed. Health Inform. 2015, 19, 1627–1636. [Google Scholar] [CrossRef]
- Van Opbroek, A.; Ikram, M.A.; Vernooij, M.W.; de Bruijne, M. Transfer Learning Improves Supervised Image Segmentation Across Imaging Protocols. IEEE Trans. Med. Imaging 2015, 34, 1018–1030. [Google Scholar] [CrossRef]
- Tan, C.; Sun, F.; Zhang, W. Deep Transfer Learning for EEG-Based Brain Computer Interface. In Proceedings of the 2018 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Calgary, AB, Canada, 15–20 April 2018; pp. 916–920. [Google Scholar] [CrossRef]
- Zemouri, A.; Omri, N.; Fnaiech, F.; Zerhouni, N.; Fnaiech, N. A New Growing Pruning Deep Learning Neural Network Algorithm (GP-DLNN). Neural Comput. Appl. 2019, in press. [Google Scholar]
- Selvaraju, R.R.; Das, A.; Vedantam, R.; Cogswell, M.; Parikh, D.; Batra, D. Grad-CAM: Why did you say that? Visual Explanations from Deep Networks via Gradient-based Localization. In Proceedings of the NIPS 2016 Workshop on Interpretable Machine Learning for Complex Systems, Barcelona, Spain, 9 December 2016. [Google Scholar]
- Zeiler, M.D.; Fergus, R. Visualizing and Understanding Convolutional Networks. In Computer Vision—ECCV 2014; Fleet, D., Pajdla, T., Schiele, B., Tuytelaars, T., Eds.; Springer International Publishing: Cham, Switerland, 2014; pp. 818–833. [Google Scholar]
- Montavon, G.; Lapuschkin, S.; Binder, A.; Samek, W.; Müller, K.R. Explaining nonlinear classification decisions with deep Taylor decomposition. Pattern Recognit. 2017, 65, 211–222. [Google Scholar] [CrossRef]
- Van der Maaten, L.; Hinton, G. Visualizing data using t-SNE. J. Mach. Learn. Res. 2008, 9, 2579–2605. [Google Scholar]
- Creswell, A.; White, T.; Dumoulin, V.; Arulkumaran, K.; Sengupta, B.; Bharath, A.A. Generative Adversarial Networks: An Overview. IEEE Signal Process. Mag. 2018, 35, 53–65. [Google Scholar] [CrossRef]
- Goodfellow, I.; Pouget-Abadie, J.; Mirza, M.; Xu, B.; Warde-Farley, D.; Ozair, S.; Courville, A.; Bengio, Y. Generative Adversarial Nets. In Advances in Neural Information Processing Systems 27; Ghahramani, Z., Welling, M., Cortes, C., Lawrence, N.D., Weinberger, K.Q., Eds.; Curran Associates, Inc.: Lake Tahoe, NV, USA, 2014; pp. 2672–2680. [Google Scholar]
- Liu, W.; Luo, Z.; Li, S. Improving deep ensemble vehicle classification by using selected adversarial samples. Knowl.-Based Syst. 2018, 160, 167–175. [Google Scholar] [CrossRef]
- Li, Y.; Xiao, N.; Ouyang, W. Improved Generative Adversarial Networks with Reconstruction Loss. Neurocomputing 2018, 323, 363–372. [Google Scholar] [CrossRef]
- Ji, Y.; Zhang, H.; Wu, Q.J. Saliency detection via conditional adversarial image-to-image network. Neurocomputing 2018, 316, 357–368. [Google Scholar] [CrossRef]
- Kiasari, M.A.; Moirangthem, D.S.; Lee, M. Coupled generative adversarial stacked Auto-encoder: CoGASA. Neural Netw. 2018, 100, 1–9. [Google Scholar] [CrossRef]
- Dhamala, J.; Ghimire, S.; Sapp, J.L.; Horáček, B.M.; Wang, L. High-Dimensional Bayesian Optimization of Personalized Cardiac Model Parameters via an Embedded Generative Model. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2018; Frangi, A.F., Schnabel, J.A., Davatzikos, C., Alberola-López, C., Fichtinger, G., Eds.; Springer International Publishing: Cham, Switerland, 2018; pp. 499–507. [Google Scholar]
- Ghimire, S.; Dhamala, J.; Gyawali, P.K.; Sapp, J.L.; Horacek, M.; Wang, L. Generative Modeling and Inverse Imaging of Cardiac Transmembrane Potential. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2018; Frangi, A.F., Schnabel, J.A., Davatzikos, C., Alberola-López, C., Fichtinger, G., Eds.; Springer International Publishing: Cham, Switerland, 2018; pp. 508–516. [Google Scholar]
- Ren, J.; Hacihaliloglu, I.; Singer, E.A.; Foran, D.J.; Qi, X. Adversarial Domain Adaptation for Classification of Prostate Histopathology Whole-Slide Images. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2018; Frangi, A.F., Schnabel, J.A., Davatzikos, C., Alberola-López, C., Fichtinger, G., Eds.; Springer International Publishing: Cham, Switerland, 2018; pp. 201–209. [Google Scholar]
- Han, L.; Yin, Z. A Cascaded Refinement GAN for Phase Contrast Microscopy Image Super Resolution. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2018; Frangi, A.F., Schnabel, J.A., Davatzikos, C., Alberola-López, C., Fichtinger, G., Eds.; Springer International Publishing: Cham, Switerland, 2018; pp. 347–355. [Google Scholar]
- Wang, J.; Zhao, Y.; Noble, J.H.; Dawant, B.M. Conditional Generative Adversarial Networks for Metal Artifact Reduction in CT Images of the Ear. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2018; Frangi, A.F., Schnabel, J.A., Davatzikos, C., Alberola-López, C., Fichtinger, G., Eds.; Springer International Publishing: Cham, Switerland, 2018; pp. 3–11. [Google Scholar]
- Wang, Y.; Yu, B.; Wang, L.; Zu, C.; Lalush, D.S.; Lin, W.; Wu, X.; Zhou, J.; Shen, D.; Zhou, L. 3D conditional generative adversarial networks for high-quality PET image estimation at low dose. NeuroImage 2018, 174, 550–562. [Google Scholar] [CrossRef]
- Liao, H.; Huo, Z.; Sehnert, W.J.; Zhou, S.K.; Luo, J. Adversarial Sparse-View CBCT Artifact Reduction. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2018; Frangi, A.F., Schnabel, J.A., Davatzikos, C., Alberola-López, C., Fichtinger, G., Eds.; Springer International Publishing: Cham, Switerland, 2018; pp. 154–162. [Google Scholar]
- Chen, Y.; Shi, F.; Christodoulou, A.G.; Xie, Y.; Zhou, Z.; Li, D. Efficient and Accurate MRI Super-Resolution Using a Generative Adversarial Network and 3D Multi-level Densely Connected Network. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2018; Frangi, A.F., Schnabel, J.A., Davatzikos, C., Alberola-López, C., Fichtinger, G., Eds.; Springer International Publishing: Cham, Switerland, 2018; pp. 91–99. [Google Scholar]
- Mishra, D.; Chaudhury, S.; Sarkar, M.; Soin, A.S. Ultrasound Image Enhancement Using Structure Oriented Adversarial Network. IEEE Signal Process. Lett. 2018, 25, 1349–1353. [Google Scholar] [CrossRef]
- Wolterink, J.M.; Leiner, T.; Viergever, M.A.; Išgum, I. Generative Adversarial Networks for Noise Reduction in Low-Dose CT. IEEE Trans. Med. Imaging 2017, 36, 2536–2545. [Google Scholar] [CrossRef]
- Cerrolaza, J.J.; Li, Y.; Biffi, C.; Gomez, A.; Sinclair, M.; Matthew, J.; Knight, C.; Kainz, B.; Rueckert, D. 3D Fetal Skull Reconstruction from 2DUS via Deep Conditional Generative Networks. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2018; Frangi, A.F., Schnabel, J.A., Davatzikos, C., Alberola-López, C., Fichtinger, G., Eds.; Springer International Publishing: Cham, Switerland, 2018; pp. 383–391. [Google Scholar]
- Seitzer, M.; Yang, G.; Schlemper, J.; Oktay, O.; Würfl, T.; Christlein, V.; Wong, T.; Mohiaddin, R.; Firmin, D.; Keegan, J.; et al. Adversarial and Perceptual Refinement for Compressed Sensing MRI Reconstruction. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2018; Frangi, A.F., Schnabel, J.A., Davatzikos, C., Alberola-López, C., Fichtinger, G., Eds.; Springer International Publishing: Cham, Switerland, 2018; pp. 232–240. [Google Scholar]
- Zhang, P.; Wang, F.; Xu, W.; Li, Y. Multi-channel Generative Adversarial Network for Parallel Magnetic Resonance Image Reconstruction in K-space. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2018; Frangi, A.F., Schnabel, J.A., Davatzikos, C., Alberola-López, C., Fichtinger, G., Eds.; Springer International Publishing: Cham, Switerland, 2018; pp. 180–188. [Google Scholar]
- Quan, T.M.; Nguyen-Duc, T.; Jeong, W. Compressed Sensing MRI Reconstruction Using a Generative Adversarial Network With a Cyclic Loss. IEEE Trans. Med. Imaging 2018, 37, 1488–1497. [Google Scholar] [CrossRef]
- Zhao, H.; Li, H.; Maurer-Stroh, S.; Cheng, L. Synthesizing retinal and neuronal images with generative adversarial nets. Med. Image Anal. 2018, 49, 14–26. [Google Scholar] [CrossRef]
- Frid-Adar, M.; Diamant, I.; Klang, E.; Amitai, M.; Goldberger, J.; Greenspan, H. GAN-based synthetic medical image augmentation for increased CNN performance in liver lesion classification. Neurocomputing 2018, 321, 321–331. [Google Scholar] [CrossRef]
- Paschali, M.; Conjeti, S.; Navarro, F.; Navab, N. Generalizability vs. Robustness: Investigating Medical Imaging Networks Using Adversarial Examples. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2018; Frangi, A.F., Schnabel, J.A., Davatzikos, C., Alberola-López, C., Fichtinger, G., Eds.; Springer International Publishing: Cham, Switerland, 2018; pp. 493–501. [Google Scholar]
- Jin, D.; Xu, Z.; Tang, Y.; Harrison, A.P.; Mollura, D.J. CT-Realistic Lung Nodule Simulation from 3D Conditional Generative Adversarial Networks for Robust Lung Segmentation. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2018; Frangi, A.F., Schnabel, J.A., Davatzikos, C., Alberola-López, C., Fichtinger, G., Eds.; Springer International Publishing: Cham, Switerland, 2018; pp. 732–740. [Google Scholar]
- Chen, J.; Xie, Y.; Wang, K.; Wang, Z.H.; Lahoti, G.; Zhang, C.; Vannan, M.A.; Wang, B.; Qian, Z. Generative Invertible Networks (GIN): Pathophysiology-Interpretable Feature Mapping and Virtual Patient Generation. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2018; Frangi, A.F., Schnabel, J.A., Davatzikos, C., Alberola-López, C., Fichtinger, G., Eds.; Springer International Publishing: Cham, Switerland, 2018; pp. 537–545. [Google Scholar]
- Shams, S.; Platania, R.; Zhang, J.; Kim, J.; Lee, K.; Park, S.J. Deep Generative Breast Cancer Screening and Diagnosis. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2018; Frangi, A.F., Schnabel, J.A., Davatzikos, C., Alberola-López, C., Fichtinger, G., Eds.; Springer International Publishing: Cham, Switerland, 2018; pp. 859–867. [Google Scholar]
- Mahapatra, D.; Bozorgtabar, B.; Thiran, J.P.; Reyes, M. Efficient Active Learning for Image Classification and Segmentation Using a Sample Selection and Conditional Generative Adversarial Network. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2018; Frangi, A.F., Schnabel, J.A., Davatzikos, C., Alberola-López, C., Fichtinger, G., Eds.; Springer International Publishing: Cham, Switerland, 2018; pp. 580–588. [Google Scholar]
- Pan, Y.; Liu, M.; Lian, C.; Zhou, T.; Xia, Y.; Shen, D. Synthesizing Missing PET from MRI with Cycle-consistent Generative Adversarial Networks for Alzheimer’s Disease Diagnosis. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2018; Frangi, A.F., Schnabel, J.A., Davatzikos, C., Alberola-López, C., Fichtinger, G., Eds.; Springer International Publishing: Cham, Switerland, 2018; pp. 455–463. [Google Scholar]
- Fan, J.; Cao, X.; Xue, Z.; Yap, P.T.; Shen, D. Adversarial Similarity Network for Evaluating Image Alignment in Deep Learning Based Registration. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2018; Frangi, A.F., Schnabel, J.A., Davatzikos, C., Alberola-López, C., Fichtinger, G., Eds.; Springer International Publishing: Cham, Switerland, 2018; pp. 739–746. [Google Scholar]
- Hu, Y.; Gibson, E.; Ghavami, N.; Bonmati, E.; Moore, C.M.; Emberton, M.; Vercauteren, T.; Noble, J.A.; Barratt, D.C. Adversarial Deformation Regularization for Training Image Registration Neural Networks. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2018; Frangi, A.F., Schnabel, J.A., Davatzikos, C., Alberola-López, C., Fichtinger, G., Eds.; Springer International Publishing: Cham, Switerland, 2018; pp. 774–782. [Google Scholar]
- Dong, S.; Luo, G.; Wang, K.; Cao, S.; Mercado, A.; Shmuilovich, O.; Zhang, H.; Li, S. VoxelAtlasGAN: 3D Left Ventricle Segmentation on Echocardiography with Atlas Guided Generation and Voxel-to-Voxel Discrimination. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2018; Frangi, A.F., Schnabel, J.A., Davatzikos, C., Alberola-López, C., Fichtinger, G., Eds.; Springer International Publishing: Cham, Switerland, 2018; pp. 622–629. [Google Scholar]
- Zhao, M.; Wang, L.; Chen, J.; Nie, D.; Cong, Y.; Ahmad, S.; Ho, A.; Yuan, P.; Fung, S.H.; Deng, H.H.; et al. Craniomaxillofacial Bony Structures Segmentation from MRI with Deep-Supervision Adversarial Learning. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2018; Frangi, A.F., Schnabel, J.A., Davatzikos, C., Alberola-López, C., Fichtinger, G., Eds.; Springer International Publishing: Cham, Switerland, 2018; pp. 720–727. [Google Scholar]
- Li, Y.; Shen, L. cC-GAN: A Robust Transfer-Learning Framework for HEp-2 Specimen Image Segmentation. IEEE Access 2018, 6, 14048–14058. [Google Scholar] [CrossRef]
- Han, Z.; Wei, B.; Mercado, A.; Leung, S.; Li, S. Spine-GAN: Semantic segmentation of multiple spinal structures. Med. Image Anal. 2018, 50, 23–35. [Google Scholar] [CrossRef]
- Jiang, J.; Hu, Y.C.; Tyagi, N.; Zhang, P.; Rimner, A.; Mageras, G.S.; Deasy, J.O.; Veeraraghavan, H. Tumor-Aware, Adversarial Domain Adaptation from CT to MRI for Lung Cancer Segmentation. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2018; Frangi, A.F., Schnabel, J.A., Davatzikos, C., Alberola-López, C., Fichtinger, G., Eds.; Springer International Publishing: Cham, Switerland, 2018; pp. 777–785. [Google Scholar]
- Singh, V.K.; Romani, S.; Rashwan, H.A.; Akram, F.; Pandey, N.; Sarker, M.M.K.; Abdulwahab, S.; Torrents-Barrena, J.; Saleh, A.; Arquez, M.; et al. Conditional Generative Adversarial and Convolutional Networks for X-ray Breast Mass Segmentation and Shape Classification. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2018; Frangi, A.F., Schnabel, J.A., Davatzikos, C., Alberola-López, C., Fichtinger, G., Eds.; Springer International Publishing: Cham, Switerland, 2018; pp. 833–840. [Google Scholar]
- Biffi, C.; Oktay, O.; Tarroni, G.; Bai, W.; De Marvao, A.; Doumou, G.; Rajchl, M.; Bedair, R.; Prasad, S.; Cook, S.; et al. Learning Interpretable Anatomical Features Through Deep Generative Models: Application to Cardiac Remodeling. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2018; Frangi, A.F., Schnabel, J.A., Davatzikos, C., Alberola-López, C., Fichtinger, G., Eds.; Springer International Publishing: Cham, Switerland, 2018; pp. 464–471. [Google Scholar]
- Xu, C.; Xu, L.; Brahm, G.; Zhang, H.; Li, S. MuTGAN: Simultaneous Segmentation and Quantification of Myocardial Infarction Without Contrast Agents via Joint Adversarial Learning. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2018; Frangi, A.F., Schnabel, J.A., Davatzikos, C., Alberola-López, C., Fichtinger, G., Eds.; Springer International Publishing: Cham, Switerland, 2018; pp. 525–534. [Google Scholar]
- Zhang, Y.; Miao, S.; Mansi, T.; Liao, R. Task Driven Generative Modeling for Unsupervised Domain Adaptation: Application to X-ray Image Segmentation. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2018; Frangi, A.F., Schnabel, J.A., Davatzikos, C., Alberola-López, C., Fichtinger, G., Eds.; Springer International Publishing: Cham, Switerland, 2018; pp. 599–607. [Google Scholar]
- Giger, A.; Sandkühler, R.; Jud, C.; Bauman, G.; Bieri, O.; Salomir, R.; Cattin, P.C. Respiratory Motion Modelling Using cGANs. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2018; Frangi, A.F., Schnabel, J.A., Davatzikos, C., Alberola-López, C., Fichtinger, G., Eds.; Springer International Publishing: Cham, Switerland, 2018; pp. 81–88. [Google Scholar]
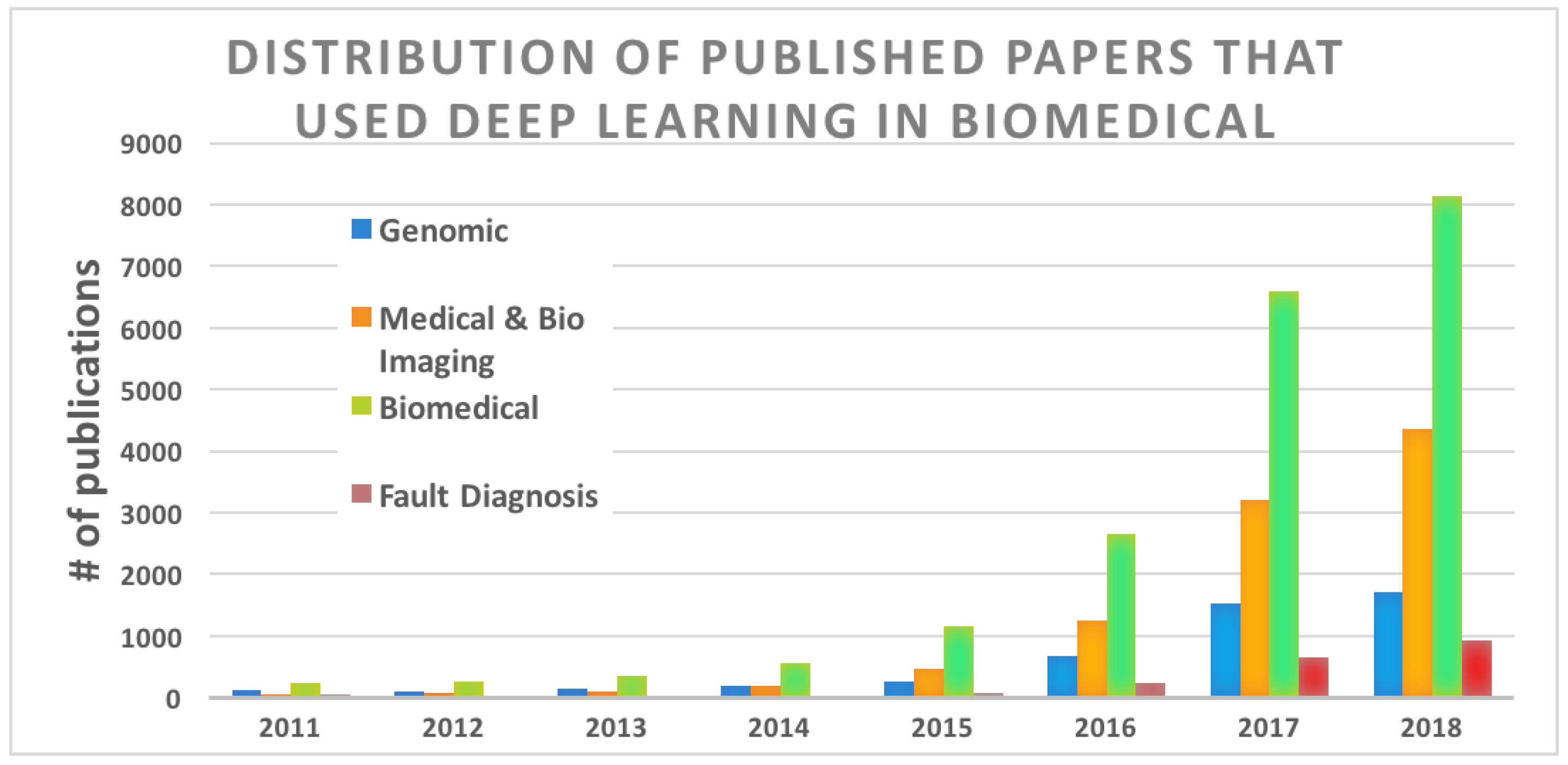
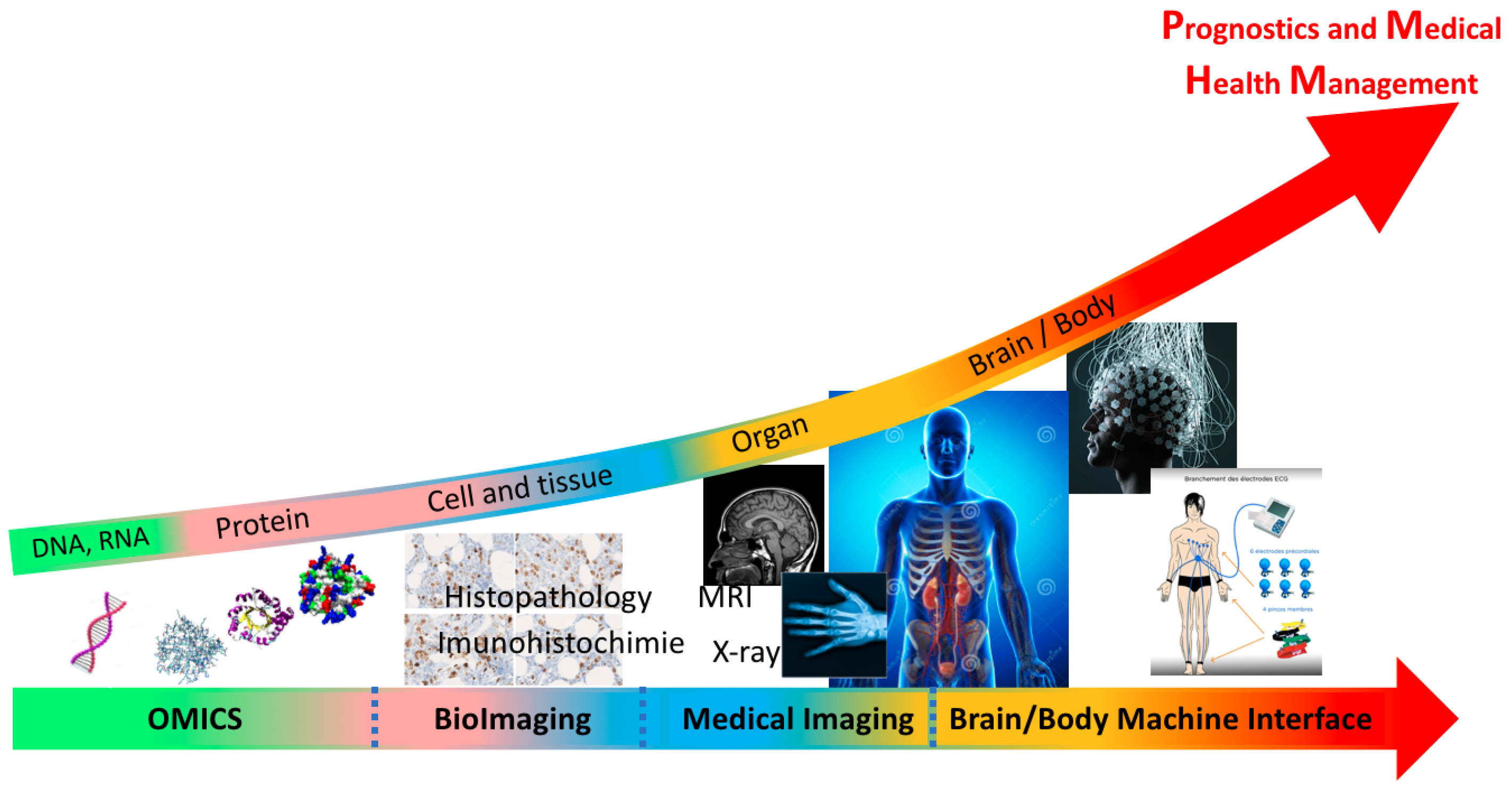
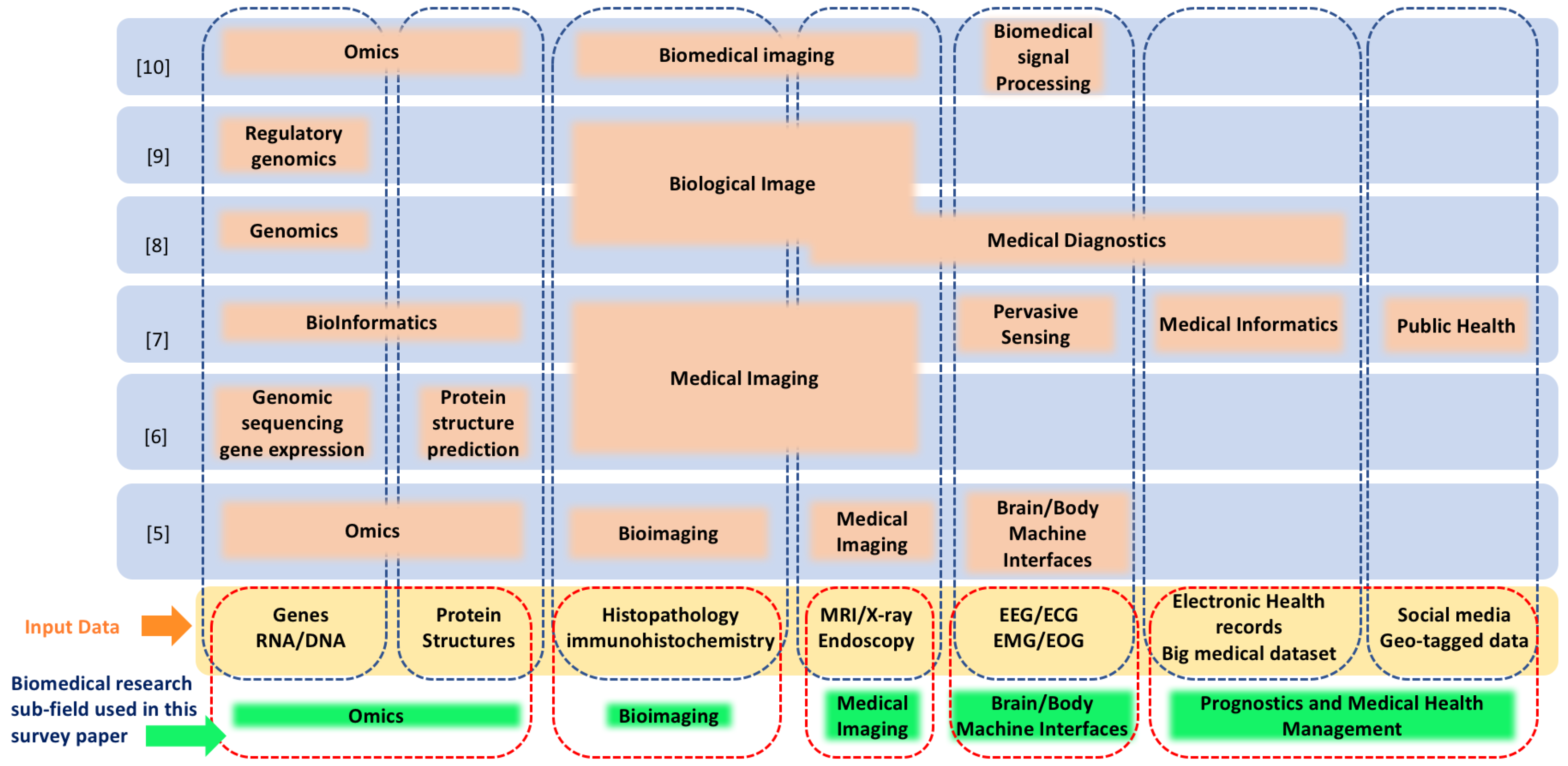
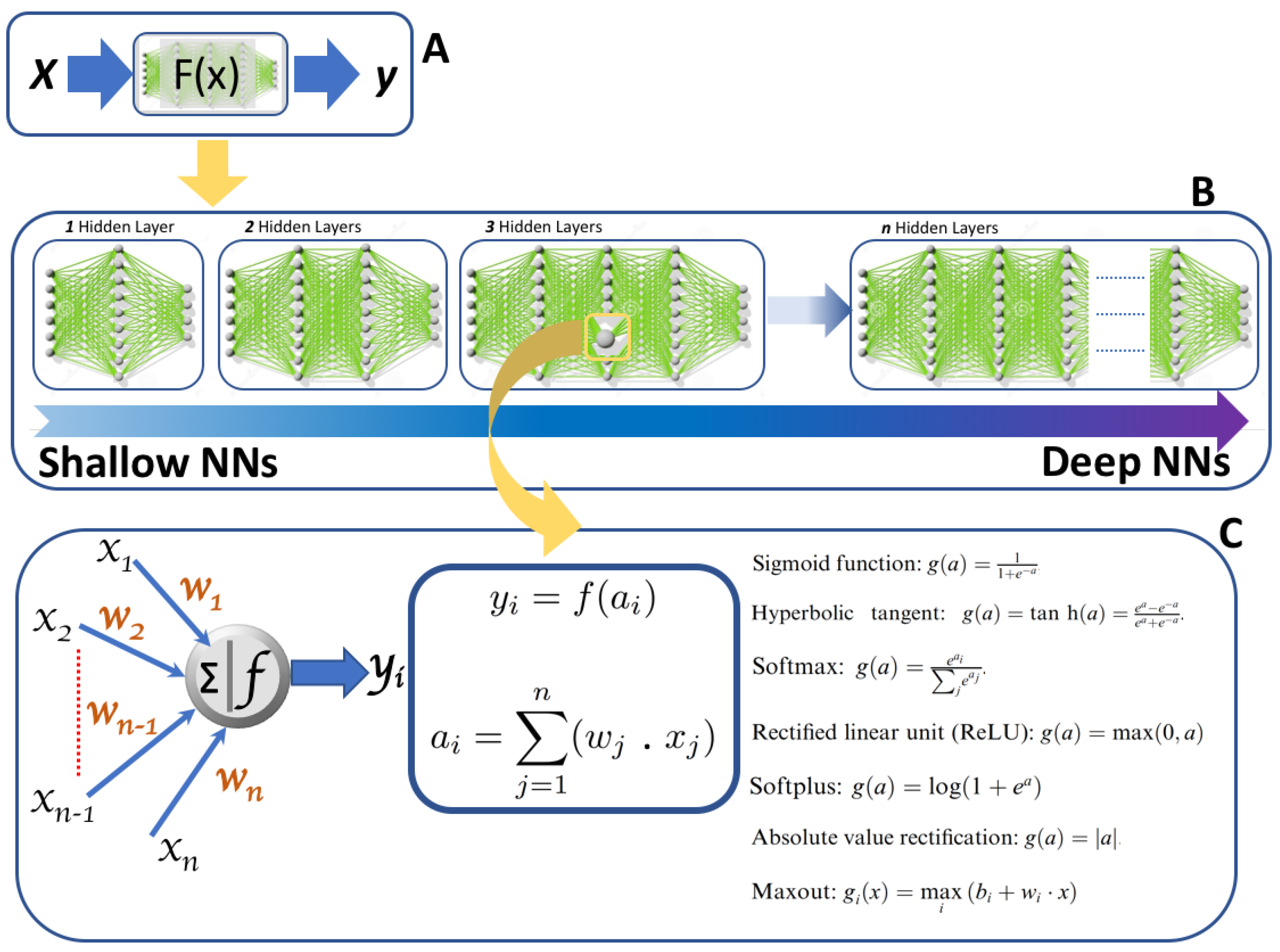

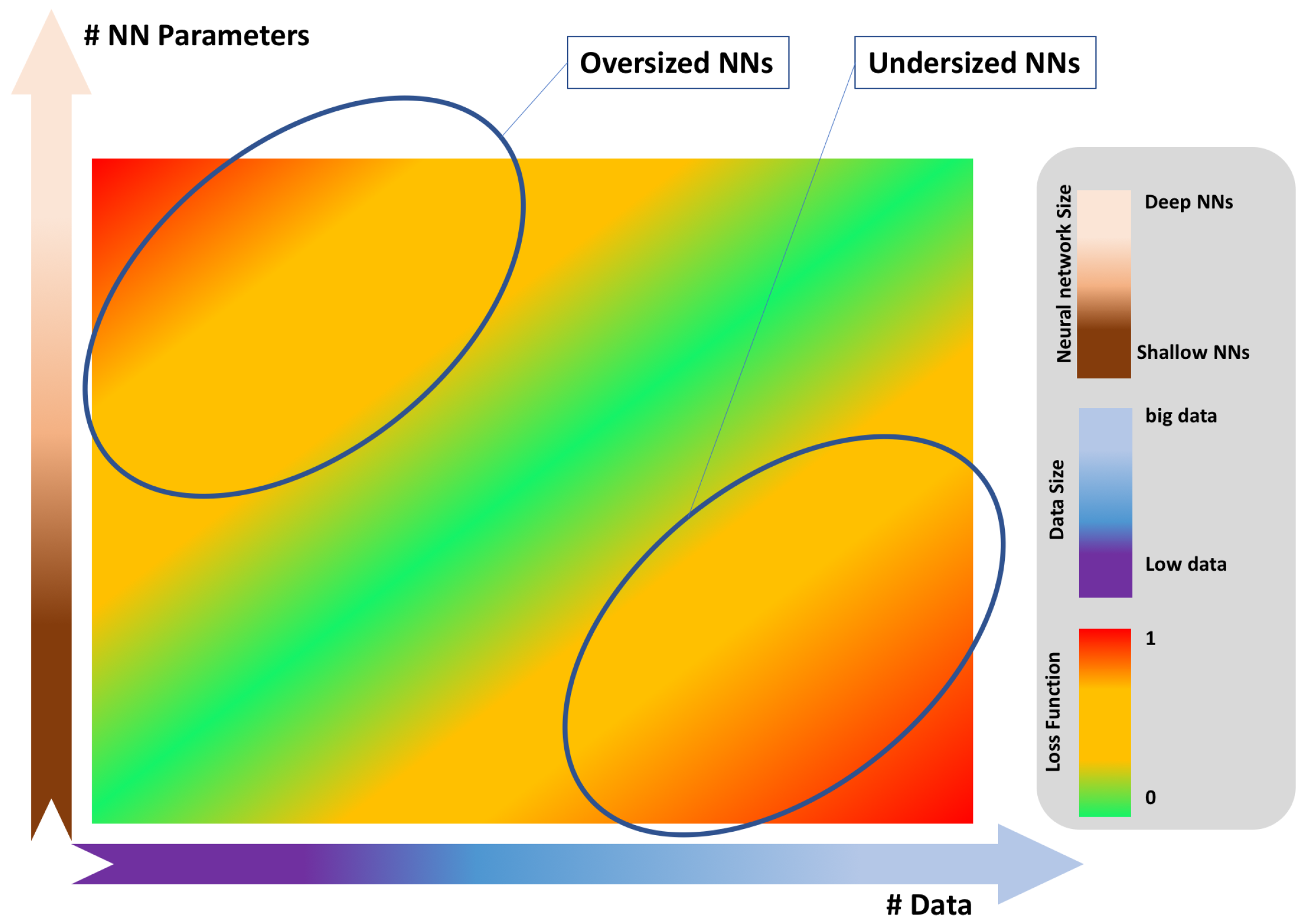
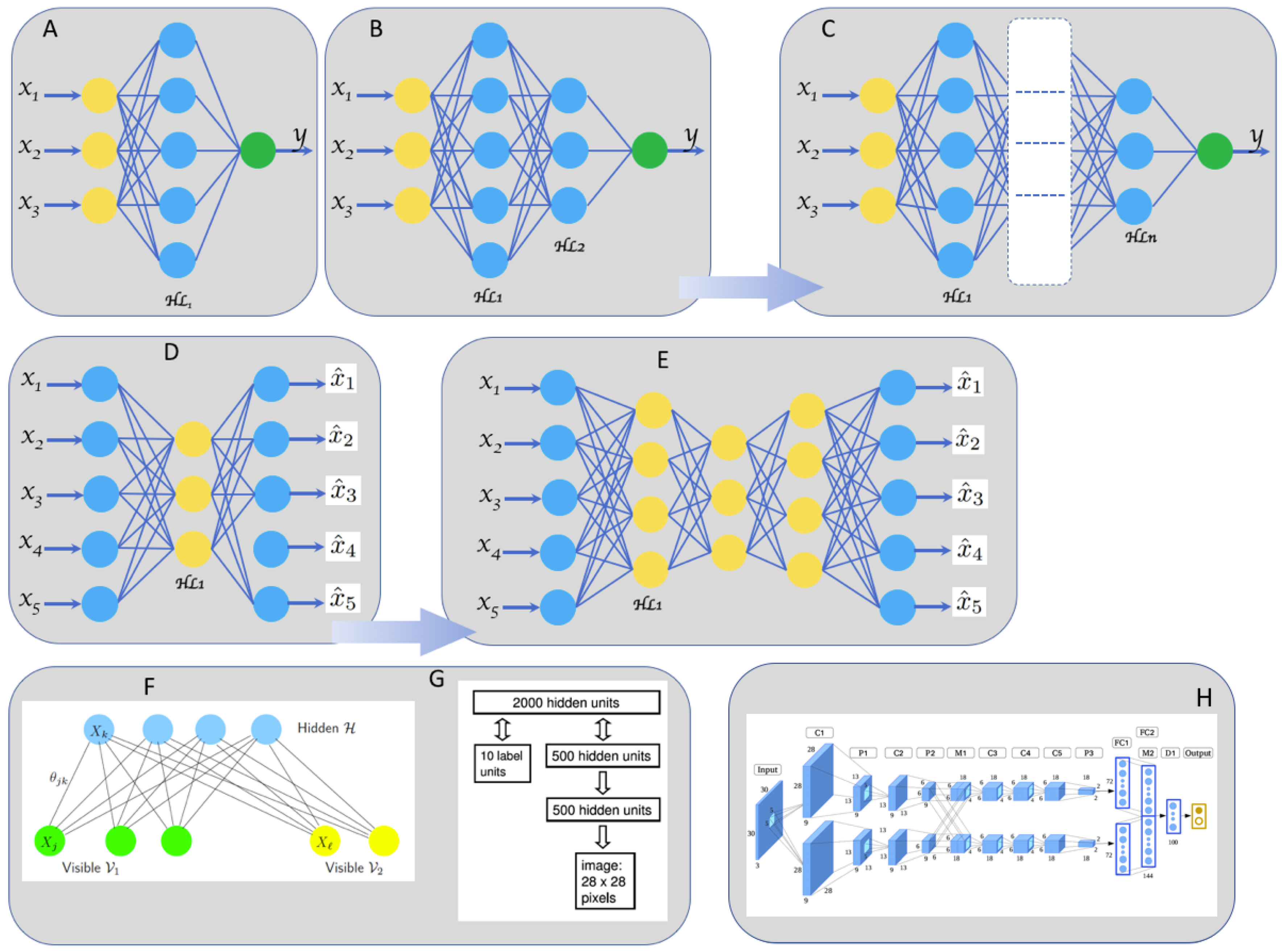

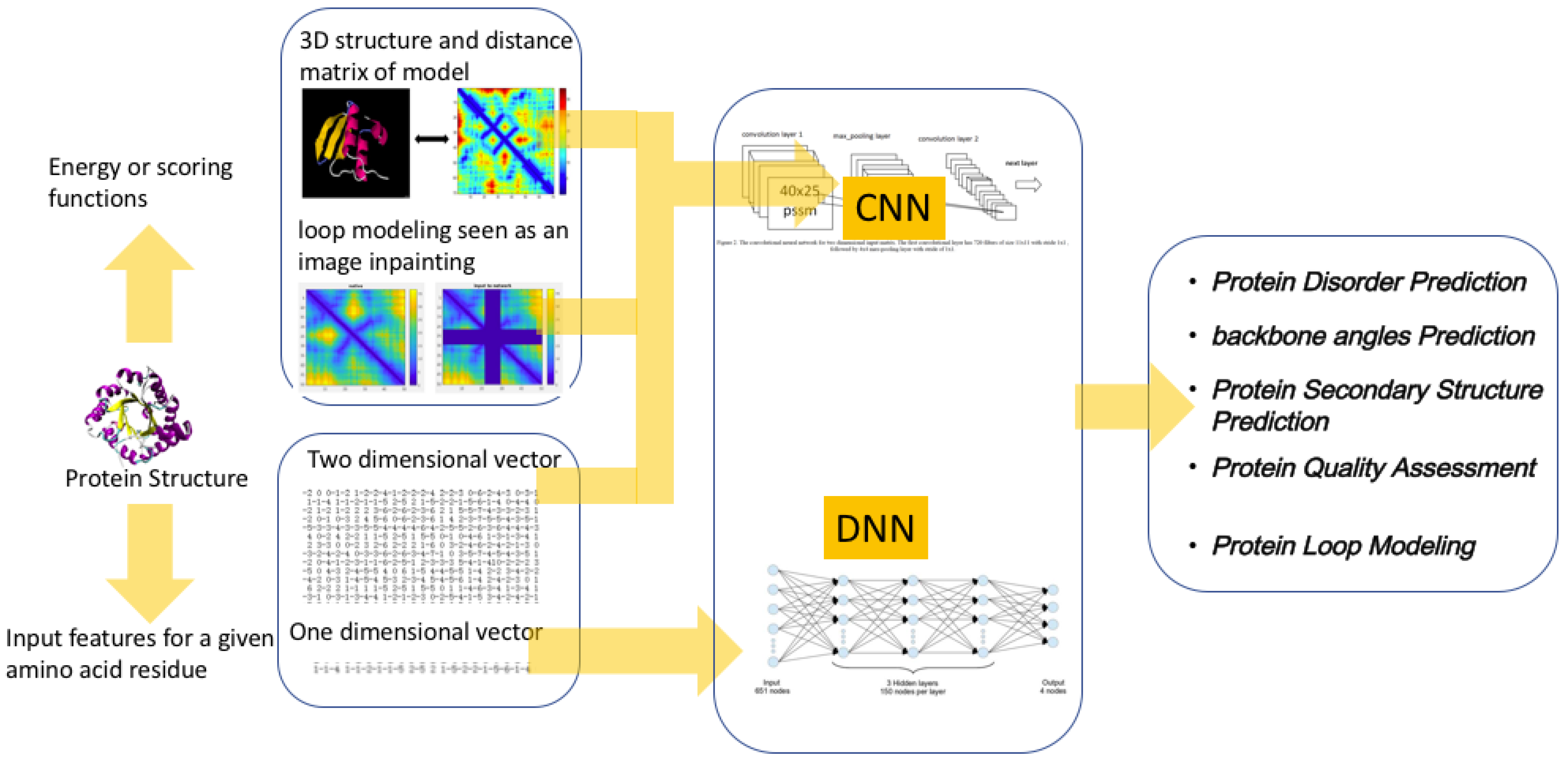
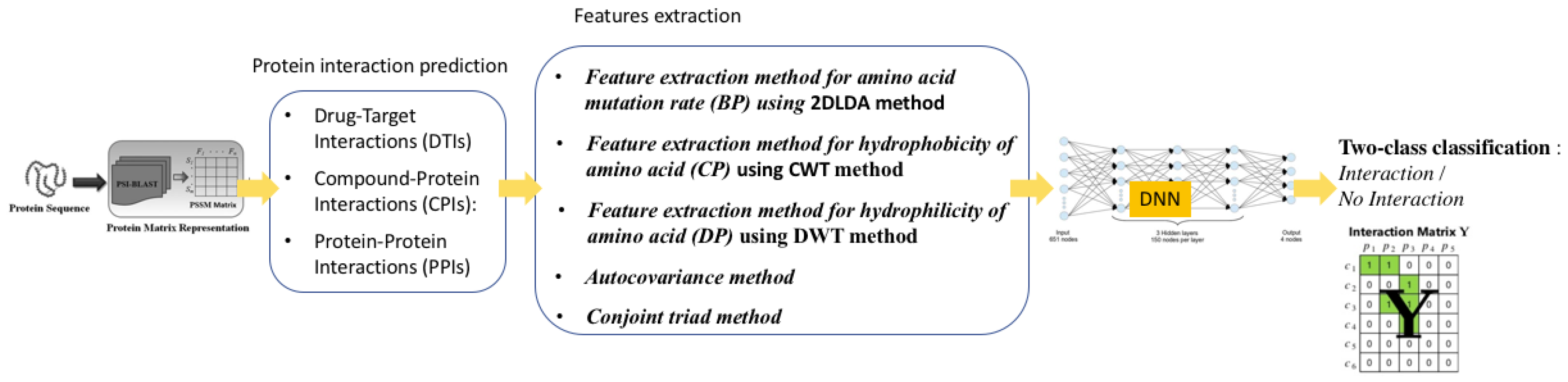
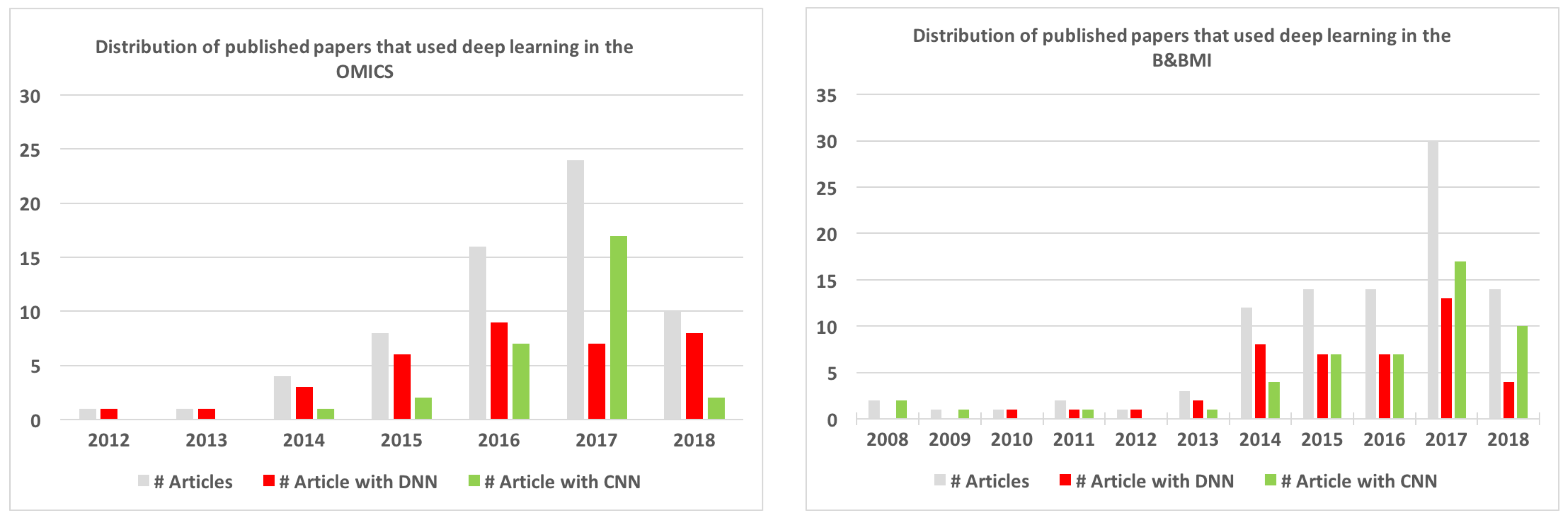
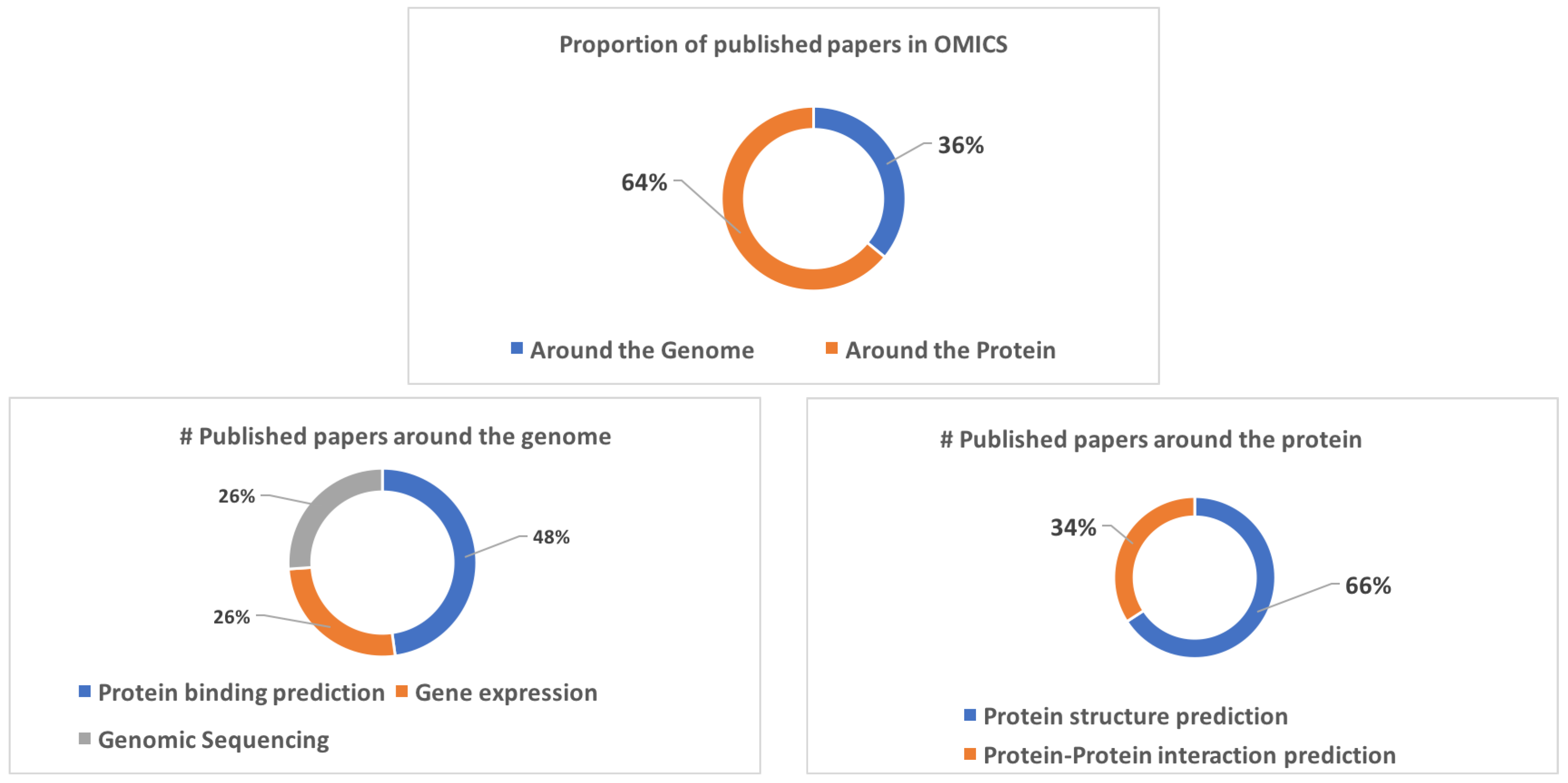
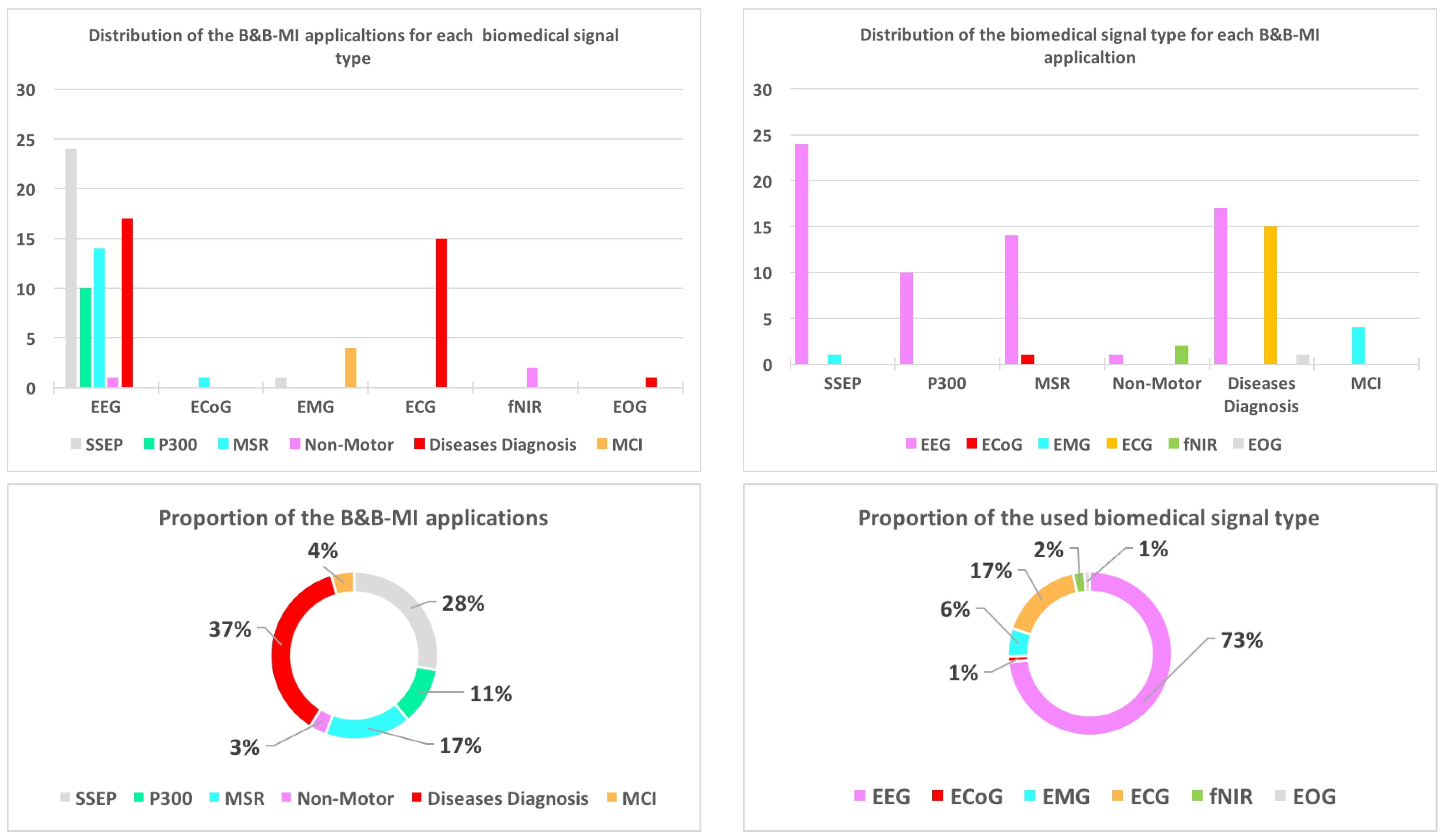
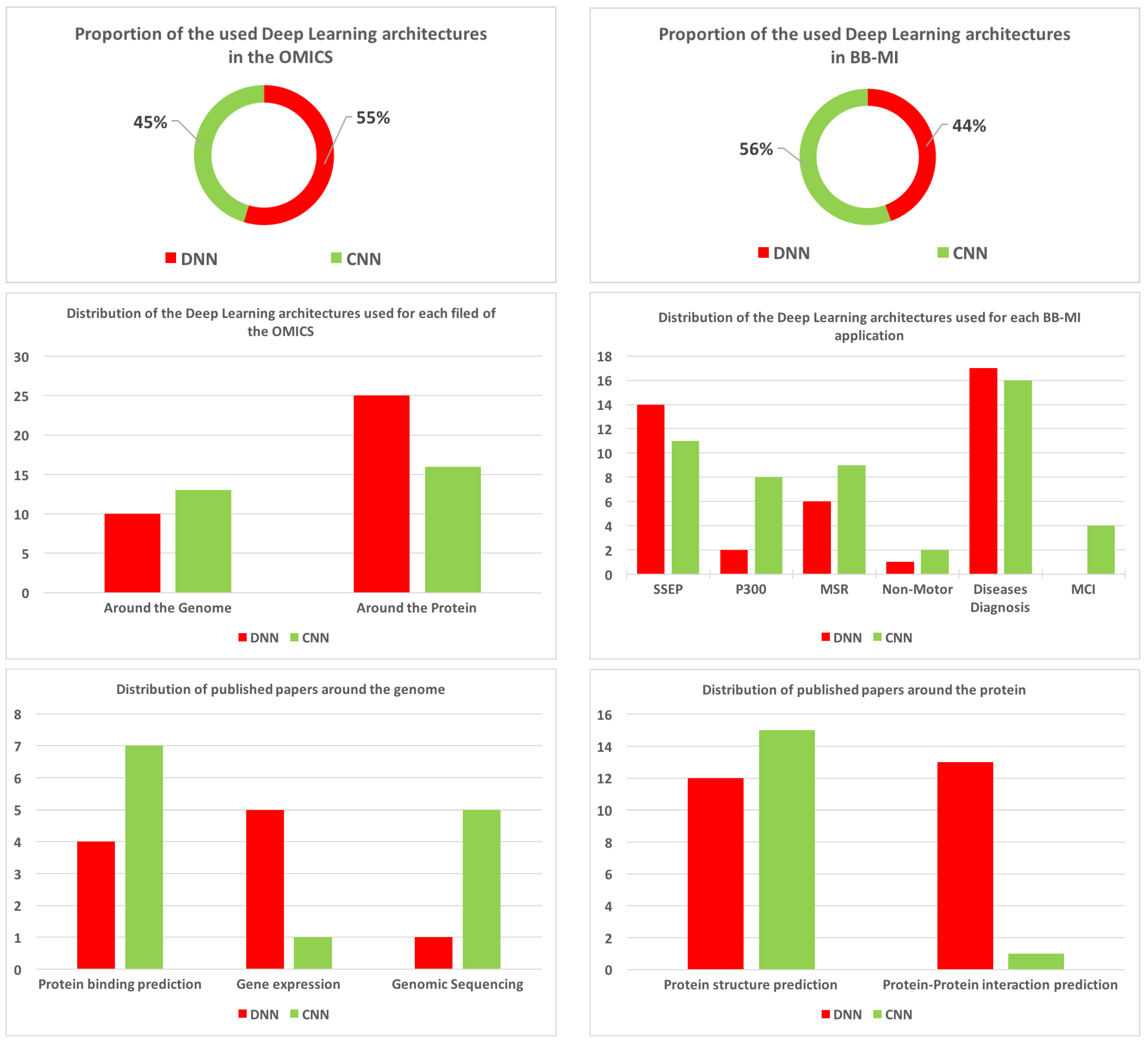
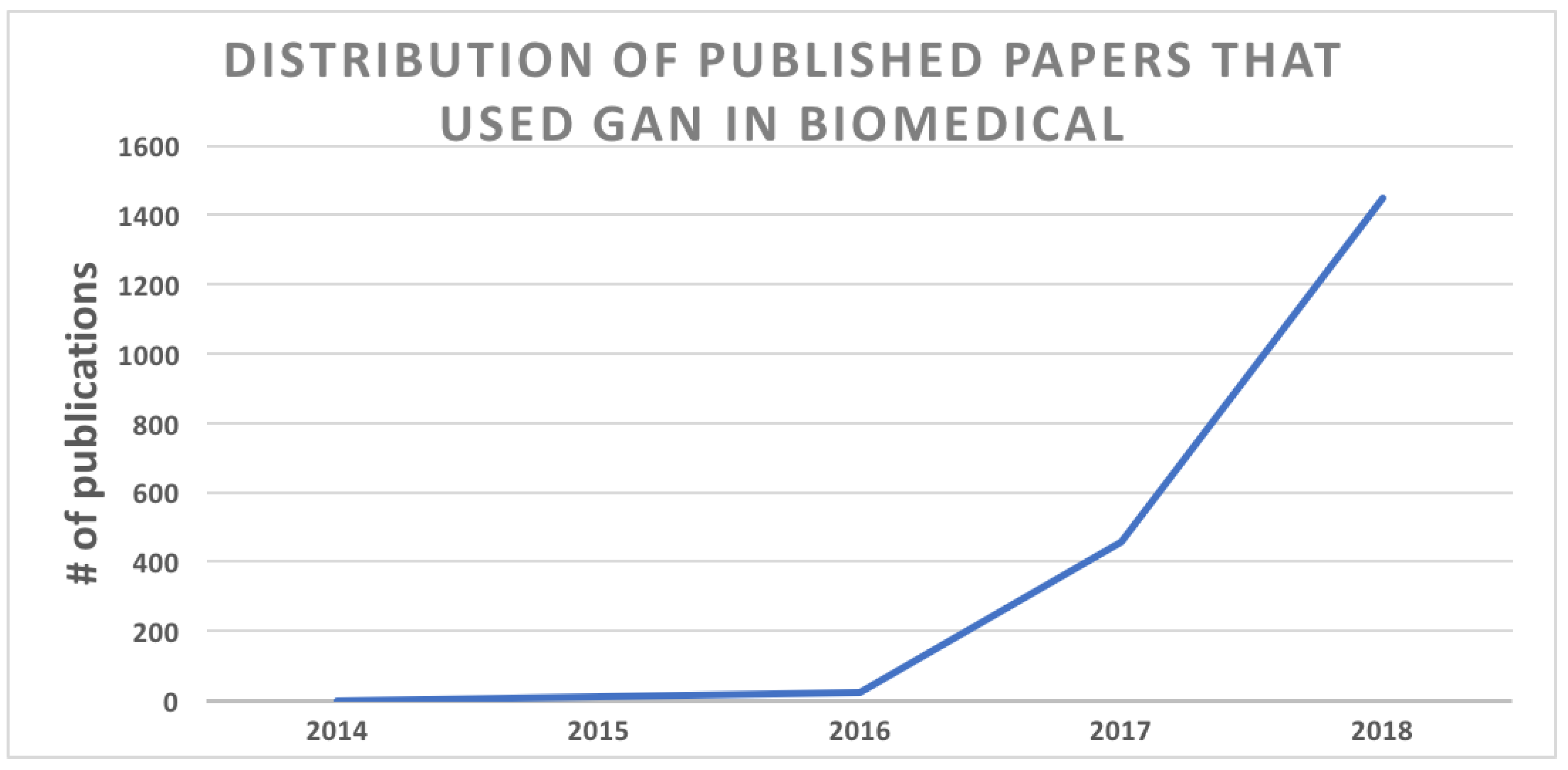
| Ref. | Year of Publi. | # of Citations | Research Topics |
|---|---|---|---|
| [60] | 2018 | 1 | Brief survey on machine learning used for the prediction of DNA-binding residues on protein surfaces |
| [55] | 2016 | 122 | Biomarkers, genOmics, transcriptOmics, proteOmics |
| Structural biology and chemistry | |||
| Drug discovery and repurposing | |||
| Multiplatform data (MultiOmics) | |||
| Challenges and limitations of deep learning systems | |||
| [59] | 2016 | 58 | A complete survey paper on how machine learning can be used to solve key problems in genomic |
| [61] | 2015 | 304 | Provides an overview of machine learning as it is applied to practical problems and major challenges in genOmics |
| Ref. | Year of Publi. | # of Citations | Research Topics |
|---|---|---|---|
| [77] | 2017 | 563 | A complete survey on deep learning in bio and medical imaging: |
| - Image/exam classification | |||
| - Object or lesion classification | |||
| - Organ, region and landmark localization | |||
| - Organ and substructure segmentation | |||
| - Lesion segmentation | |||
| - Anatomical application areas (brain, eye, chest, digital pathology and microscopy, breast, cardiac, abdomen, musculoskeletal | |||
| - Anatomical/cell structures detection, | |||
| - Tissue segmentation, | |||
| - Computer aided disease diagnosis/prognosis | |||
| [78] | 2016 | 18 | ANNs as decision support Tools in cytopathology: past, present, and future: |
| - Gynecological cytopathology, the PAPNET System | |||
| - Cytopathology of gastrointestinal system | |||
| - Cytopathology of thyroid gland | |||
| - Cytopathology of the breast | |||
| - Cytopathology of the urinary system | |||
| - Cytopathology of effusions | |||
| [79] | 2016 | 130 | DL for digital pathology image analysis |
| - Nuclei segmentation use case | |||
| - Epithelium segmentation use case | |||
| - Tubule segmentation use case | |||
| - Invasive ductal carcinoma segmentation use case | |||
| - Lymphocyte detection use case | |||
| - Mitosis detection use case | |||
| - Lymphoma subtype classification use case | |||
| [80] | 2017 | 55 | Machine learning for medical imaging |
| [81] | 2017 | 58 | DL for brain magnetic resonance imaging (MRI) Segmentation: state of the art and future directions |
| Ref. | Year of Publi. | # of Citations | Research Topics |
|---|---|---|---|
| [92] | 2018 | 19 | A review of classification algorithms for EEG based brain–computer interfaces: a 10-year update |
| [93] | 2018 | 21 | A complete survey on DL for healthcare applications based on physiological signals |
| [94] | 2006 | 85 | Machine learning in bioinformatics: a brief survey and recommendations for practitioners |
| Purpose | Signal Type | NN Type | Ref. |
|---|---|---|---|
| Steady state evoked potentials (SSEP) applications | |||
| Human’s affective and emotion states classification | EEG | DBN | [182,183,184,185,186,187,188] |
| DMLP | [189,190] | ||
| CNN | [191,192] | ||
| Auditory evoked potential | EEG | CNN | [193,194,195,196] |
| EMG | DMLP | [197] | |
| Visual evoked potential | EEG | CNN | [198,199,200,201,202] |
| DMLP | [203] | ||
| DBN | [204,205,206] | ||
| Classification of mental load | EEG | CNN | [207] |
| P300 applications | |||
| The P300 speller | EEG | CNN | [208,209,210,211] |
| DNN | [212] | ||
| Rapid serial visual presentation | EEG | CNN | [213] |
| Driver fatigue classification | EEG | CNN | [214,215,216] |
| DBN | [217] | ||
| Motor and sensorimotor rhythms (MSR) applications | |||
| Motor imagery tasks | ECoG | CNN | [218] |
| EEG | CNN | [219,220,221,222,223,224,225,226,227] | |
| DBN | [228,229] | ||
| DMLP | [230,231,232] | ||
| Non-motor cognitive tasks | |||
| Mental arithmetics (MA), word generation (WG) and mental rotation (MR) | fNIR | DMLP | [233] |
| Mental task | EEG | CNN | [234] |
| Mental subtractions | fNIR | DMLP | [235] |
| Hybrid signals | |||
| Sleep stage classification | EEG, EOG, EMG | DBN | [181] |
| Emotion analysis | EEG, EOG, EMG, skin temperature, GSR, blood volume pressure, and respiration signals | DMLP | [236] |
| Detecting driving fatigue | EEG, EOG | DMLP | [237] |
| Purpose | Signal Type | NN Type | Ref. |
|---|---|---|---|
| Automated detection of arrhythmia and cardiac abnormalities | EEG | DBN | [240] |
| DNN | [241,242] | ||
| ECG | CNN | [243,244,245,246] | |
| DNN | [247,248,249] | ||
| DBN | [250,251,252] | ||
| Automated detection and diagnosis of seizure | EEG | DBN | [88] |
| CNN | [253,254,255,256,257,258,259] | ||
| Localization of epileptogenicity | EEG | CNN | [260] |
| Epileptiform discharge anomaly detection | EEG | DBN | [261] |
| Alzheimer’s disease diagnosis | EEG | DBN | [89] |
| Neurophysiological clinical monitoring | EEG | DBN | [262] |
| Detection of hypoglycemic episodes in Children | ECG | DBN | [263] |
| Automated detection of myocardial infarction | EEG | CNN | [87] |
| Screening of depression | EEG | CNN | [238] |
| Prediction of post anoxic coma | EEG | CNN | [264] |
| Coronary artery disease (CAD) Diagnosis | EEG | CNN | [86] |
| Screening paroxysmal atrial fibrillation | ECG | CNN | [265] |
| Detection sleep apnea | ECG | DNN | [266] |
| Detecting atrial fibrillation | ECG | CNN | [267] |
| Fetal electrocardiogram (FECG) monitoring | ECG | DNN | [268] |
| Drowsiness detection | EOG | CNN | [269] |
| Neonatal Sleep State Identification | EEG | DNN | [239] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zemouri, R.; Zerhouni, N.; Racoceanu, D. Deep Learning in the Biomedical Applications: Recent and Future Status. Appl. Sci. 2019, 9, 1526. https://doi.org/10.3390/app9081526
Zemouri R, Zerhouni N, Racoceanu D. Deep Learning in the Biomedical Applications: Recent and Future Status. Applied Sciences. 2019; 9(8):1526. https://doi.org/10.3390/app9081526
Chicago/Turabian StyleZemouri, Ryad, Noureddine Zerhouni, and Daniel Racoceanu. 2019. "Deep Learning in the Biomedical Applications: Recent and Future Status" Applied Sciences 9, no. 8: 1526. https://doi.org/10.3390/app9081526
APA StyleZemouri, R., Zerhouni, N., & Racoceanu, D. (2019). Deep Learning in the Biomedical Applications: Recent and Future Status. Applied Sciences, 9(8), 1526. https://doi.org/10.3390/app9081526







