Featured Application
Developed fluorescent assay and expression system can be used for obtaining improved cellobiose dehydrogenase whole cell biocatalysts for lactobionic acid production and building of biosensors and biofuel cells.
Abstract
Cellobiose dehydrogenase (CDH) from Phanerochaete chrysosporium can be used in lactobionic acid production, biosensor for lactose, biofuel cells, lignocellulose degradation, and wound-healing applications. To make it a better biocatalyst, CDH with higher activity in an immobilized form is desirable. For this purpose, CDH was expressed for the first time on the surface of S. cerevisiae EBY100 cells in an active form as a triple mutant tmCDH (D20N, A64T, V592M) and evolved further for higher activity using resazurin-based fluorescent assay. In order to decrease blank reaction of resazurin with yeast cells and to have linear correlation between enzyme activity on the cell surface and fluorescence signal, the assay was optimized with respect to resazurin concentration (0.1 mM), substrate concentration (10 mM lactose and 0.08 mM cellobiose), and pH (6.0). Using optimized assay an error prone PCR gene library of tmCDH was screened. Two mutants with 5 (H5) and 7 mutations (H9) were found having two times higher activity than the parent tmCDH enzyme that already had improved activity compared to wild type CDH whose activity could not be detected on the surface of yeast cells.
1. Introduction
The cellobiose dehydrogenase (CDH; EC 1.1.99.18) is a monomeric, an extracellular hemoflavoprotein with two domains, flavin and heme domain, connected via flexible peptide linker [1]. CDH catalyzes the oxidation of the reducing end of cellobiose and other β-1,4-linked disaccharides with a β-glucose moiety, to their corresponding lactones [2]. Different electron acceptors can reoxidize the reduced enzyme, such as 2,6-dicholoro-indophenol (DCIP), or metal ions, for example Fe(III) or even oxygen, albeit the latter is a very poor electron acceptor [2]. Although diverse of white-rot and other fungi have been reported to secret CDH, and a lot have been purified and characterized, the in vivo function of CDH is not sufficiently understood [3]. It has been suggested that CDH plays a role in lignocellulosic biomass hydrolysis, such as relief of product inhibition of cellulases by oxidation of cellobiose, generation of peroxide for inhibition of opposing microbes, and regeneration of redox metal ions (such as Fe2+) to sustain the Fenton-type chemistry that produces hydroxyl radicals who can modify polysaccharides causing partial hydrolysis and swelling of insoluble fibers which helps breakdown of lignocellulose by glycoside hydrolase [1,2,3]. Due to its properties, CDH has been applied in biosensors for detection of lactose, in enzymatic biofuel cells as anode catalyst [4,5], for lactobionic acid production [6], dye removal [7], and bioremediation [1].
Improvement of activity and stability of CDH would benefit all of these applications and directed evolution based on random introduction of mutations resulting in a protein library, where mutants having desired properties can be screened is used very often [8]. In order to make screening process more efficient during directed evolution, various expression systems that keep connection between genotype and phenotype are developed, such as phage [9], bacterial [10], yeast [11], and mammalian cell display [12]. The advantage of yeast display is the existence of eukaryotic protein production machinery that is essential for post-translational modifications of expressed proteins [8] and compatibility with flow cytometric analysis, which allows quantitative measurements of equilibrium binding constants, dissociation kinetics, and specificity of the displayed proteins without requisite of soluble protein expression and purification [13]. Screening methods usually involve phenotypic selection based on colorimetric or fluorometric assays in microtiter plates (MTPs) [14] and solid-phase screening on agar plates or membranes [15]. Regardless, during colony screening on solid-phase, or individual clones in MTPs, usually 103–105 clones and seldom more than 106 can be screened with sophisticated automated systems [16]. One of emerging ultra-high-throughput screening systems to this day is fluorescence-activated cell sorting (FACS) which can routinely sort >107 cells/h, if there is an appropriate fluorescence assay [17,18].
Resazurin (7-hydroxy-3H-phenoxazin-3-one-10-oxide) is a tetrazolium-based, non-toxic, water-soluble dye that can be reduced by cellular metabolic activity to the highly fluorescent resorufin [19]. Resazurin also changes color from blue to pink upon reduction [20]. This reduction causes the loss of an oxygen atom loosely bound to the nitrogen of the phenaxazine nucleus. Atmospheric oxygen cannot reverse this change and it is mostly independent of both oxygen content and reduction potential [19]. Resazurin has been used to examine the viability of microorganisms (non-destructively) [21], for biofilm quantification [22], and anti-biofilm disinfectant testing of clinical devices [23]. Several intracellular oxidoreductases, for instance diaphorases and NADH dehydrogenase, were reported to be responsible for the reduction of resazurin to resorufin during cell viability assay [24].
In this study, fluorescent assay was developed for detection of CDH expressed on the surface of yeast cells, using resazurin as an electron acceptor. For the first time CDH was expressed in an active form on the surface of yeast cells as a triple mutant (tmCDH), while wild type CDH activity could not be detected. Fluorescent assay was optimized and used for directed evolution of tmCDH on the surface of yeast cells.
2. Materials and Methods
2.1. Materials and Strains
S. cerevisiae strain EBY100 and pCTCON2 vector were kindly provided by Prof. Dane Wittrup [25]. Protocols and media for EBY100 were described in Invitrogen (Carlsbad, CA, USA) manual. All chemicals used were of analytical reagent grade or higher and obtained from Sigma-Aldrich Chemie GmbH, Taufkirchen, Germany. Resazurin sodium salt was obtained from Invitrogen, Carlsbad, CA, USA. Enzymes and nucleotides were purchased from Fermentas, Waltham, MA, USA, if not stated otherwise. Soluble CDH was wild type CDH expressed and purified from Pichia pastoris KM71 using published procedure [26].
2.2. Enzymatic Assays
Fluorescence measurements were obtained using FluoroMax-4 (Horiba Scientific, Kyoto, Japan) and Tecan Infinity 200 pro fluorimeter (Tecan Group Ltd, Mannedorf, Switzerland). Excitation wavelength was set on 540 nm with 590 nm as emission wavelength. Hellma fluorescence cuvettes (Sigma-Aldrich Chemie GmbH, Taufkirchen, Germany) were used for development of the assay. Greiner cellstar 96-well plate (Greiner Bio One GmbH, Frickenhausen, Germany) was used for library screening.
Fluorescence-based assay was optimized in 1.4 mL Hellma fluorescence cuvette, using FluoroMax-4 in final reaction volume of 1 mL. Fluorescence-based assay for cells was performed in 96-well FLUOTRAC 600 clear, flat bottom, black plate using Tecan Infinity 200 pro fluorimeter in a total reaction volume of 200 µL for 10 min. A mix of resazurin (0.1 mM; 7-hydroxy-3H-phenoxazin-3-one-10-oxide sodium salt; λex540 nm; λem590 nm) and lactose (10 mM) or cellobiose (8 mM) in sodium phosphate buffer (0.1 M; pH 5.5) was used for cuvette and MTP measurements, if not stated otherwise. All measurements were done in triplicate.
Spectrophotometric assay for measuring CDH activity was done using 0.3 mM DCIP (dichlorophenol–indophenol), 30 mM lactose, and 0.1 M sodium acetate pH 4.5, at 25 °C in total reaction volume of 1 mL. Reaction was started by adding purified CDH and reduction of DCIP was followed by measuring absorbance decrease at 520 nm (ε520 = 6.80 mM−1·cm−1) [27]. One unit of enzyme activity is defined as the amount of enzyme that reduces 1 µmol of DCIP per minute under the abovementioned conditions.
2.3. Cloning of cdh Gene in pCTCON2
A thermal cycler (Mastercycler personal; Eppendorf, Hamburg, Germany) and thin-wall PCR tubes (Multiply-Pro tubes; 0.2 mL; Sarstedt, Germany) were used for all PCRs. Volume for PCR was 50 µL. The concentration of DNA was quantified using a NanoDrop photometer (Thermo Fischer Scientific, Waltham, MA, USA).
The Pharenochaete chrysosporium cdh gene (U46081.1) for mature enzyme was synthesized and cloned in pUC57 plasmid by GenScript, Piscataway, NJ, USA. The PCR reaction (94 °C for 4 min, 1 cycle; 94 °C 1 min/50 °C 1 min/72 °C for 2.45 min, 30 cycles; 72 °C for 10 min, 1 cycle) contained Pfu DNA polymerase (0.04 u/µL), dNTP mix (0.2 mM), forward primer (0.5 µM)(5′-ATGCTAGCCAGAGTGCCTCACAGTTTACC-3′), reverse primer (0.5 µM) (5′- ATGGATCCTCAAGGACCTCCCGCAAG-3′), template CDH-pUC57 (30 ng). The purified fragments were restriction digested with NheI and BamHI enzyme and ligated in the pCTCON2 vector. The ligated gene was used to transform Echerichia coli DH5α. Gene length was checked with DNA electrophoresis and EBY100 yeast cells were transformed with CDH-pCTCON2 construct.
2.4. Library Construction
Plate Thermo-Shaker thermostat PST-60HL (Biosan, Riga, Latvia) was used for library growth in microtiter plates, and for flask fermentation a thermos-shaker Incubator Unimax 1010 (Heidolph GmbH, Schwabach, Germany) was used.
The PCR reaction (94 °C for 4 min, 1 cycle; 94 °C 1 min/50 °C 1 min/72°C for 2.45 min, 30 cycles; 72 °C for 10 min, 1 cycle) contained Taq DNA polymerase (0.04 u/µL), dNTP mix (0.2 mM), 0.05 mM Mn2+, forward primer (0.5 µM)( 5′-ATGCTAGCCAGAGTGCCTCACAGTTTACC-3′), reverse primer (0.5 µM) (5′- ATGGATCCTCAAGGACCTCCCGCAAG-3′), template CDH-pCTCON2 (30 ng). The purified fragments were restriction digested with NheI and BamHI enzyme and ligated in the pCTCON2 vector. The ligation mixture was transformed to DH5α E. coli cells. Size of the library in E. coli was determined by plating one tenth of the transformed E. coli DH5α cells and counting number of colonies on agar plates. The rest of the transformed E. coli cells was grown in liquid culture and mini prep plasmid DNA of gene library mixture was prepared. The total number of the E. coli transformants in the library was determined to be around 20,000. Afterwards, the gene library mixture was transformed into S. cerevisiae EBY100 competent cells, using the Gietz protocol [28]. Transformed yeast cells were grown in YNB-CAA glucose liquid culture and one tenth of the yeast library was plated on YNB-CAA glucose agar plates for library size determination. The size of yeast library was around 10,000 and CDH positive cells in the library were detected using the YNB-CAA galactose agar plates with 1 mM dichlorophenolindophenol (DCIP). Around 8000 colonies were prescreened using DCIP agar plate assay and around 2000 colonies that were positive for CDH activity were transferred to MTP for enzyme expression and screening.
Transferred cells were first grown in sterile MTPs (24 h, 30 °C, 200 rpm) in liquid YNB-CAA glucose media to saturation, centrifuged and resuspended in YNB-CAA galactose media (0.8 O.D. 600 nm) and incubated for 24 h (25 °C, 200 rpm) for induction of enzyme expression. Aliquots of cells suspension from MTPs were tested for CDH enzyme activity using resazurin assay. Mutants having higher activity than the tmCDH were kinetically characterized, for Km value.
2.5. Flow Cytometry Assay
Flow cytometry analysis was performed using a BD FACSDiVa flow cytometer (Becton Dickinson and Company, Franklin Lakes, NJ, USA). For emulsification, a MICCRA D-1 dispenser (ART Prozess- & Labortechnik Gmbh & Co. KG, Müllheim, Germany) was used.
In vitro compartmentalization of S. cerevisiae EBY100 cells was done by modified protocol of Aharoni et al. [17]. After 24 h of expression in Gal media, yeast cells were washed and together with the components required for the reaction (25 µL), were added to a 250 µL ice cold oil phase of 1.5% (v/v) ABIL EM 90 (Evonik Tego Chemie GmbH, Essen, Germany) in Light Mineral Oil. The two phases were homogenized on ice in a 2-mL round-bottom cryotube (3 min, 8000 rpm using a MICCRA D1 homogenizer. The second water phase (250 µL) comprising 1.5% (w/v) carboxy-methylcellulose sodium salt (CMC), 1% (v/v) Triton X100 in phosphate-buffered saline (PBS) was added to the primary emulsion and homogenized on ice (3 min, 7000 rpm). Emulsions were diluted in PBS and analyzed using a BD FACSDiVa flow cytometer. The trigger parameter was set to forward scattering. The analysis rate was around 8000 events/s. We used 568 nm laser for excitation and 630/22 nm bandpass filter for emission detection.
3. Results
3.1. Development of Fluorescent Assay
In order to develop screening system based on fluorescence for detection of CDH activity on the surface of yeast cells we tested resazurin as a substrate for soluble CDH enzyme. Resazurin was used previously as a probe for assessing cell viability during which it was reduced to fluorescent resorufin [24] and in this article it was tried as an electron acceptor in CDH-catalyzed oxidation of lactose, Scheme 1.
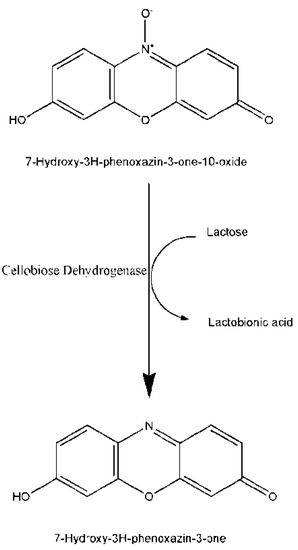
Scheme 1.
Mechanism of the resazurin-based fluorescent assay for cellobiose dehydrogenase (CDH).
In order to have the most sensitive detection and linear correlation between enzyme activity and fluorescence signal, we optimized assay conditions with respect to the pH, resazurin and substrate concentrations. The assay was optimized for detection of CDH expressed on the surface of yeast cells.
3.1.1. pH Optimization
Since resorufin fluorescence is higher at alkaline pH values [29] due to an acidic pH optimum of CDH [1] we tested influence of pH on fluorescence signal of resorufin produced by soluble CDH enzyme, Figure 1.
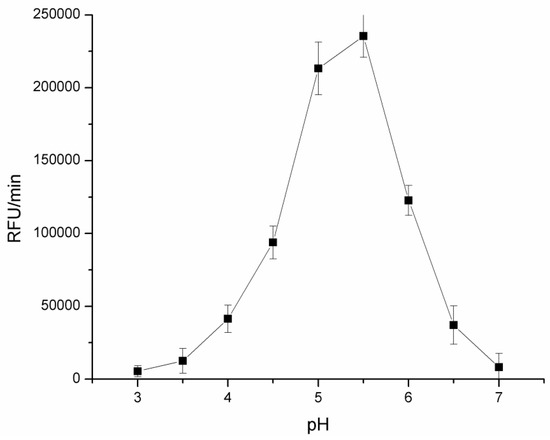
Figure 1.
Dependence of fluorescence signal (RFU/min) on pH in 0.1 M sodium phosphate buffer with 10 mM lactose, 0.1 mM resazurin and 0.29 IU/mL of CDH enzyme. Fluorescence measured at ex/em 540/590 nm.
Maximal fluorescence signal was detected around pH 5.0–6.0 which is higher than reported pH optimum of the enzyme at 4.5. This was result of higher fluorescence emission of resorufin at more alkaline pH values. Therefore, in further optimization experiments we used pH 5.5 for CDH activity measurements.
3.1.2. Optimization of Resazurin Concentration
Since the optimal pH for the assay was obtained, we tested influence of different resazurin concentrations (0.01–1 mM) on fluorescence signal (Figure 2).
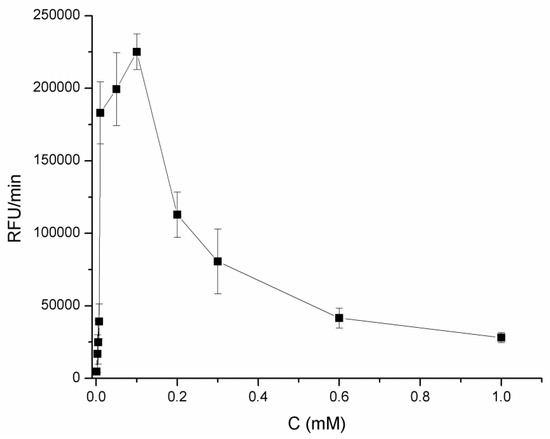
Figure 2.
Dependence of fluorescence signal (RFU/min) on resazurin concentration in 0.1 M sodium phosphate buffer pH 5.5 with 10 mM lactose and 0.29 IU/mL of CDH enzyme. Fluorescence measured at ex/em 540/590 nm.
The highest enzymatic activity and fluorescence signal was obtained for 0.1 mM resazurin concentration and that concentration was used for further experiments.
3.1.3. Optimization of Substrate Concentration
In this assay we used lactose and cellobiose as substrates. Concentrations of lactose varied from 0.1 mM till 30 mM, and cellobiose from 0.001 mM till 0.1 mM (Figure 3).
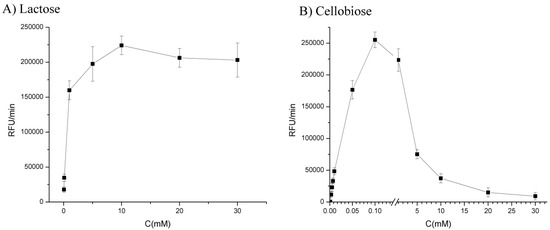
Figure 3.
Dependence of fluorescence signal (RFU/min) on (A) lactose (B) cellobiose concentration in 0.1 M sodium phosphate buffer pH 5.5 with 0.1 mM resazurin and 0.29 IU/mL of CDH enzyme. Fluorescence measured at ex/em 540/590 nm.
It could be seen that the highest fluorescence signal was obtained for 10 mM lactose concentration, while in the case of cellobiose there was maximum of fluorescence signal between 0.05 and 0.1 mM more precisely at 0.08 mM cellobiose concentration and therefore for all further experiments optimal concentrations of both substrates were used.
3.1.4. Influence of Enzyme Amount
Different amounts of CDH were added to reaction mixture and the fluorescence signal was measured. The assay showed a linear response to CDH enzyme concentrations in the range of 0.1–0.45 IU/mL (Figure 4).
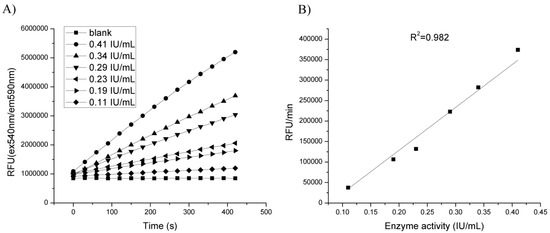
Figure 4.
Dependence of fluorescence (RFU) change in time for different amounts of enzyme in 0.1 M sodium-phosphate at pH 5.5 and 10 mM lactose and 0.1 mM resazurin. (A) Reaction kinetics in time; (B) Calibration curve. Fluorescence measured at ex/em 540/590 nm.
There was no increase of fluorescence in control reaction, and by increasing enzyme amount there was also increase in the slope of fluorescence change. This was confirmation that resazurin assay could be used for detection of CDH mutants with different ranges of activities.
3.2. Optimization of Resazurin Fluorescence Assay with CDH Expressed on the Surface of Yeast Cells
3.2.1. Expression of CDH on the Surface of Yeast Cells
Cellobiose dehydrogenase gene was cloned in pCTCON2 vector for expression as a chimera with Aga2 protein on the surface of yeast cells. This was verified by plasmid restriction mapping (see Supplementary material; Figure S1), colony PCR and DNA sequencing. We screened colonies on several agar plates for CDH activity and none of them having wild type (wt) CDH gene showed any activity. After that one round of epPCR was performed using wtCDH gene and after screening several agar plates we found a clone that showed measurable activity on the surface of yeast cells. Sequencing results showed that the clone had three mutations within cdh gene (tmCDH): D20N, A64T, and V592M. In order to confirm agar plate screening results fermentations in liquid cultures were done for cells having wtCDH-pCTCON2, tmCDH-pCTCON2, and empty pCTCON2 vector. Only tmCDH showed measurable activity on the cell surface during fermentation. Fermentation conditions for maximum production of CDH activity were at 25 °C after 24–36 h galactose induction (see Supplementary material; Figure S2).
3.2.2. Influence of Substrate Concentration
Since enzymes immobilized on the surface of yeast cells can have higher Km values than their soluble counterparts, we have tested influence of substrate concentration on fluorescence signal (see Supplementary material; Figures S3 and S4). It could be seen that maximal enzymatic activity was also detected around 10–20 mM lactose, while with cellobiose maximal activity was detected at slightly higher substrate concentration of 0.1 mM compared to 0.08 mM for the soluble enzyme.
3.2.3. Influence of Yeast Cells Density
Having established optimized reaction conditions, we tested assay in a cuvette using S. cerevisiae EBY100 cells expressing tmCDH at high and low concentrations per reaction volume. We used EBY100 cells transformed with the empty vector as a negative control. After 24 h of induction, different amounts of cells were added to the reaction mixtures so that final OD600nm was 0.1, 0.2, and 0.4 (1.28×106, 2.55x106, and 5.3x106 cells, respectively).
It could be seen that assay showed a linear response to the amount of added cells confirming that it can be used for detection of the CDH mutants expressed on the surface of yeast cells (see Supplementary material; Figure S5).
3.2.4. Screening of tmCDH Gene Library in MTP by Resazurin Assay
Gene library of tmCDH parent gene made by error prone PCR was expressed in S. cerevisiae EBY100 cells. Clones were transferred from agar plates into wells of MTP in liquid YNB-CAA Glc medium and grown for 48 h at 30 °C till saturation, diluted 4 times with YNB-CAA Gal medium to induce expression of CDH, grown for 24 h and activity of cell suspension was measured in MTP (Figure 5).
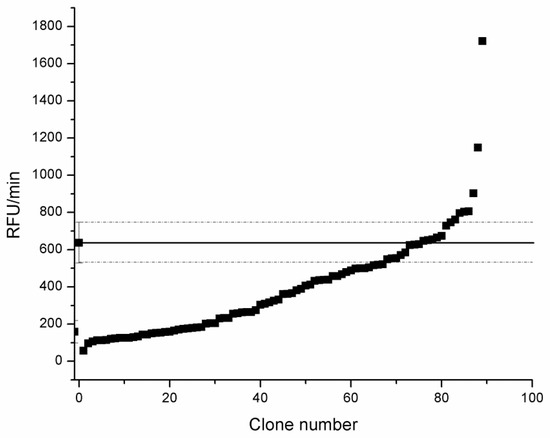
Figure 5.
Screening of error prone tmCDH-pCTCON2 gene library in MTPs. Activity was measured in 0.1 M sodium phosphate buffer pH 5.5 with 0.1 mM resazurin and 10 mM lactose for 30 minutes. Relative fluorescence units (RFU)/min are at excitation/emission (ex/em) wavelengths of 540/590 nm. -▪- Parental gene activity (tmCDH).
From this library, two mutants were found higher activity than the parent tmCDH mutant, H5 and H9.
3.3. Kinetic Characterization of CDH Mutants on the Surface of Yeast Cells
Mutants showing higher activity than parent gene were more detailed kinetically characterized in the immobilized form on the surface of yeast cells and their genes were sequenced (Table 1).

Table 1.
Kinetics properties of CDH mutants. Enzymatic assay was done with resazurin (0.1 mM), lactose/cellobiose (different concentrations), sodium-phosphate buffer (0.1 M; pH 5.5), and fluorescence measured at ex/em 540/590 nm.
It could be seen from kinetic data that H5 and H9 mutants had 1.6 and 2 times increased Vmax compared to the parent tmCDH that already was improved compared to wtCDH since it was showing activity on the cell surface while wtCDH did not have any measurable activity. Both mutants had increased Km values, but since screening was performed under high substrate concentrations, they were selected from the library for increased Vmax value.
3.4. Detection of CDH by Flow Cytometry
In order to test if there is potential to use developed resazurin assay for detecting CDH activity by flow cytometry in water-in-oil-in-water double emulsions series of enzymatic reactions was set up and analyzed by flow cytometry (Figure 6).
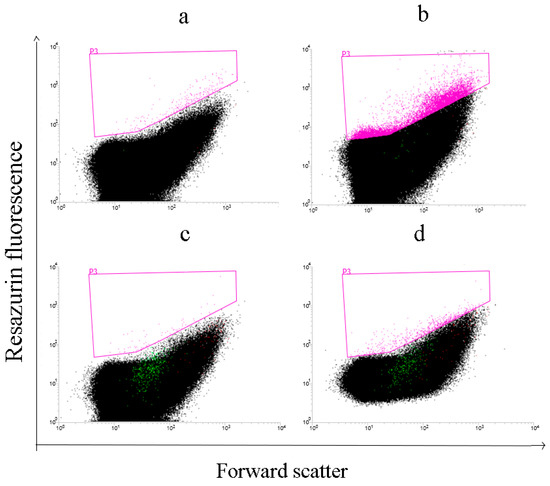
Figure 6.
Flow cytometry analysis of the resazurin assay for cellobiose dehydrogenase activity in double emulsions. (a) Emulsions with resazurin; (b) resazurin plus soluble enzyme; (c) resazurin plus cells not expressing CDH; (d) resazurin plus cells expressing tmCDH. The plots show bivariate histograms of forward scattering and resorufin fluorescence.
It could be seen that population of droplet with increased red fluorescence was visible in enzymatic reaction mixtures with soluble enzyme compared to the blank reaction. Also, smaller population of droplets with increased fluorescence was observed in the case of cells encapsulated within double emulsion expressing active tmCDH compared with cells without expressed CDH. Cross-boundary reactions during analyzing cells were not significant due to the small number of encapsulated cells compared to the number of double emulsion compartments.
4. Discussion
In order to develop CDH expression system suitable for whole-cell biocatalysis and directed evolution by high-throughput screening systems, cdh gene from Phanerochaete chrysosporium was cloned in pCTCON2 vector for expression as a chimera with Aga2 protein on the surface of Saccharomyces cerevisiae EBY100 yeast cells. After expression on the cell surface activity of the wild type CDH could not be detected. Significant decrease in specific activity of enzymes expressed on the surface of yeast cells compared with their soluble counterparts was described previously for other enzymes like glucose oxidase [30]. Since CDH has much lower specific activity compared to glucose oxidase [31] further decrease in specific activity of CDH due to the expression in the immobilized form on the surface of yeast cells could be the reason why we did not detect wild type CDH activity. During agar plate screening of yeast colonies expressing epPCR gene library of wtCDH, a triple mutant (D20N, A64T, and V592M) of CDH that was showing activity was found. All three substitutions were located on the surface of CDH protein, with two of them in heme domain (D20N and A64T) near the active site containing heme, Figure 7.
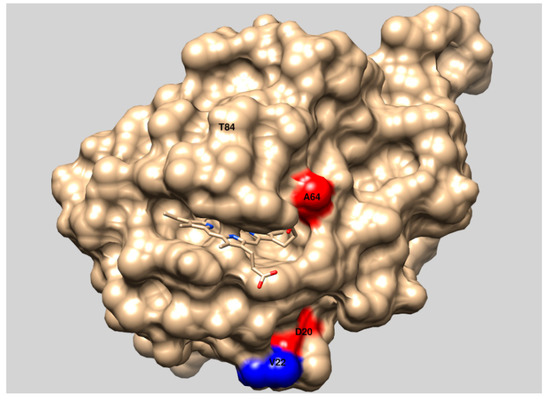
Figure 7.
Presentation of the structure of the heme domain of CDH from P. chrysosporium (PDB accession code 1D7C) with labelled substitutions. The picture was made with UCSF Chimera 1.13.1. (University of California, San Francisco, CA, US).
Third substitution V592M was within FAD domain and also very close to the entrance of the active site containing FAD in it, Figure 8.
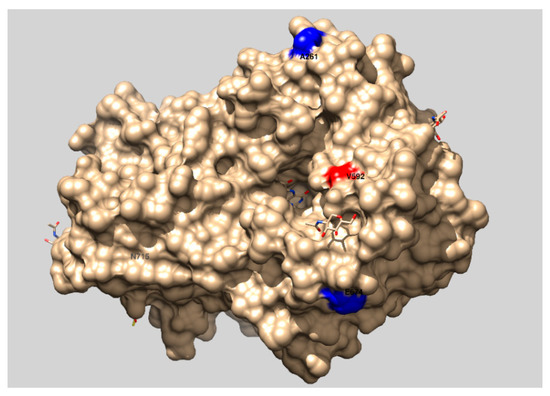
Figure 8.
Presentation of the structure of the FAD domain of CDH from P.chrysosporium (PDB accession code 1NAA) with labelled substitutions. The picture was made with UCSF Chimera 1.13.1. (University of California, San Francisco, CA, US).
Both A64T and V592M substitutions are in the region where there two domains interact with each other and these interactions are important for the activity of CDH [3]. Any change in this region can significantly influence activity of CDH and these two substitutions could be the reason tmCDH showed measurable activity on the surface of yeast cells, while at the same time wtCDH activity could not be detected.
Expression of tmCDH in liquid culture was further optimized and the highest activity was obtained at 25 °C after 24 h of galactose induction (Supplementary material; Figure S2). Optimal temperature for enzyme expression was lower than usually reported 30 °C, while expression time of 24 h was longer than optimal 16 h previously determined for glucose oxidase expression in the same host [32]. This could be result of higher molecular weight of CDH protein compared to GOx that very often requires lower expression rate of protein in order to properly fold. Some other proteins like eGFP when expressed in yeast also require lower expression temperature of 20 °C [33].
Due to the decrease in specific activity of surface displayed enzymes, we wanted to have more sensitive enzymatic assay than usually used spectrophotometric assays based on 2,6-dichlorophenol-indophenol and methylene green [34] and that could also be used with flow cytometry. Therefore, we tested fluorogenic resazurin dye as a substrate for detection of CDH activity on the surface of yeast cells. Resazurin reduction to resorufin that can be measured by red fluorescence was used previously for measuring viability of living cells [24] and blood glucose concentration [35]. The CDH assay using resazurin was optimized with respect to resazurin, cellobiose/lactose concentration, and pH of the reaction medium.
Since resorufin has the highest fluorescence at pH above 7.5 when it is in anionic form [36] and CDH has an acidic pH optimum around pH 4–5 with only 20% of residual activity at pH 7.5 [37], optimal pH for resazurin assay was determined to be at pH 5.5 (Figure 1). Above this pH resorufin has higher fluorescence but enzyme is less active, while below this pH the enzyme is more active but due to protonation of resorufin fluorescence of that product is decreased. Similar results were obtained also during development of resazurin based assay for dihydroorotate dehydrogenase that also has an acidic pH optimum [38].
Optimal concentration of resazurin dye was determined to be 0.1 mM since at higher concentrations it behaves as an inhibitor of CDH activity (Figure 2). Drop in fluorescence signal produced by enzymatic action at higher concentrations of resazurin (above 0.025 mM) was also previously reported for diaphorase coupled detection of glucose [35]. Drop in the enzyme activity at higher resazurin concentrations were results of substrate inhibition phenomenon that is often caused by nonproductive binding of the substrate for the enzyme active site.
Lactose and cellobiose that were used as CDH substrates in the assay also showed inhibition of fluorescence signal at higher concentrations. Substrate inhibition of CDH at higher concentrations due to non-productive binding of two disaccharide units is probably the result of two binding sites for hexoses that are present in the active site of CDH [39]. Substrate inhibition was observed only with cellobiose and not for lactose, probably because of much higher affinity of binding subsites for glucose than for galactose, leading to higher probability of non-productive binding of two molecules of cellobiose for one molecule of the enzyme. This difference in affinity was confirmed by much lower CDH Km value for cellobiose than for lactose. Therefore, optimal substrate concentration was determined to be 10 mM for lactose and 0.08 mM for cellobiose. The resazurin assay showed linear correlation between amount of added enzyme and fluorescence intensity, showing that it could be used for detection of enzymatic variants with different activities usually created in directed evolution experiments.
The assay was also tested and optimized for measuring CDH activity on the surface of yeast cells that is particularly important for developing high-throughput screening systems based on flow cytometry. Similar for soluble enzyme, substrate inhibition was detected, but due to the diffusional limitations that immobilized enzymes usually show, optimal substrate concentrations were higher, and for lactose it was 20 mM, while for cellobiose was 0.1 mM. Also, with CDH expressed on the cell surface the assay showed a linear response to the number of added cells.
In order to test the resazurin assay as a high-throughput screening system in MTP, a tmCDH gene library was generated by epPCR using 0.05 mM MnCl2 and around 2000 colonies were screened in MTPs by resazurin assay for higher activity. Two mutants with activity higher than the parent gene were found and characterized by sequencing and measuring kinetic parameters in immobilized form on the surface of yeast cells. The expression rate of Aga1 protein that acts as an anchor for CDH did not change in the CDH gene library and was reported to be 105 copies per single cell [13]. Therefore, the number of CDH mutants expressed on the surface of yeast cells should remain constant (105 per cell) and all kinetic data normalized to cell density of washed cells (OD600nm) should not be influenced by mutant expression rate. Mutant H5 showed additional mutation V22A on the surface of CDH heme domain (Figure 7) that was very close to the already mutated D20N position in tmCDH. This could mean that mutations in this region can increase activity of CDH. Mutant H9 showed additional T84A substitution in heme domain that is located inside the protein molecule and A261P, E674G, N715S substitutions on the surface of FAD domain of CDH compared to the parent tmCDH (D20N, A64T, and V592M). Since substitutions in internal part of protein molecule can have profound influence on activity and stability, change of T84 residue to alanine, as one of the most acceptable substitutions after serine based on BLOSUM62 matrix table, can have positive influence on activity of CDH.
The H5 and H9 mutants had higher activity at high lactose concentrations compared to the parent tmCDH and therefore showed potential to be used as a whole-cell biocatalyst for production of lactobionic acid [6].
Resazurin assay in double emulsions could detect activity of CDH by flow cytometry both for the soluble enzyme and for the enzyme expressed on the surface of yeast cells.
5. Conclusions
The cdh gene from Phanerochaete chrysosporium was cloned in pCTCON2 plasmid and triple mutant of CDH (D20N, A64T, and V592M) was expressed in an active form on the surface of yeast cells for the first time, while wtCDH did not show any measurable activity. A resazurin-based fluorescent assay for the detection of CDH activity on the surface of yeast cells was developed and used in MTP for finding mutants with improved activity. The assay also showed potential for detection of CDH activity by flow cytometry. Therefore, developed resazurin assay and expression system could be used for finding better CDH whole-cell biocatalyst in directed evolution experiments. The H5 and H9 mutants that we found showed increased activity for lactose on the surface of yeast cells and could be used as whole cell biocatalysts in biotechnology.
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-3417/9/7/1413/s1, Figure S1. DNA electrophoresis on 0.6% agarose gel.; Figure S2. Fermentation graph after galactose induction at 25 °C.; Figure S3. Dependence of fluorescence slope in time with lactose concentration.; Figure S4. Dependence of fluorescence slope in time with cellobiose concentration.; Figure S5. Dependence of fluorescence signal (RFU/min) on OD at 600 nm of yeast cells expressing cellobiose dehydrogenase.; Table S1. CDH synthetic gene sequence (U46081.1).
Author Contributions
Conceptualization, R.F. and R.P.; methodology, R.O. and O.P.; validation, R.O. and O.P.; formal analysis, M.B. and R.P.; investigation, M.B., A.M.B., and N.P.; writing—original draft preparation, M.B. and R.P.; writing—review and editing, R.O., R.F., and R.P.; visualization, O.P. and N.P.
Funding
This research was funded by Ministry of Education, Science and Technological Development of Republic of Serbia, grant number ON172049 and ON173017. M.B. acknowledges support provided by “FEBS Short-Term Fellowship”.
Acknowledgments
We thank Professor Dane Wittrup for providing us with S. cerevisiae EBY100 strain and pCTCON2 plasmid.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Cameron, M.D.; Aust, S.D. Cellobiose dehydrogenase-an extracellular fungal flavocytochrome. Enzyme Microb. Technol. 2001, 28, 129–138. [Google Scholar] [CrossRef]
- Henriksson, G.; Johansson, G.; Pettersson, G. A critical review of cellobiose dehydrogenases. J. Biotechnol. 2000, 78, 93–113. [Google Scholar] [CrossRef]
- Zamocky, M.; Ludwig, R.; Peterbauer, C.; Hallberg, B.M.; Divne, C.; Nicholls, P.; Haltrich, D. Cellobiose dehydrogenase—A flavocytochrome from wood-degrading, phytopathogenic and saprotropic fungi. Curr. Protein Pept. Sci. 2006, 7, 255–280. [Google Scholar] [CrossRef] [PubMed]
- Ludwig, R.; Ortiz, R.; Schulz, C.; Harreither, W.; Sygmund, C.; Gorton, L. Cellobiose dehydrogenase modified electrodes: Advances by materials science and biochemical engineering. Anal. Bioanal. Chem. 2013, 405, 3637–3658. [Google Scholar] [CrossRef]
- Ludwig, R.; Harreither, W.; Tasca, F.; Gorton, L. Cellobiose dehydrogenase: a versatile catalyst for electrochemical applications. Chemphyschem 2010, 11, 2674–2697. [Google Scholar] [CrossRef]
- Ludwig, R.; Ozga, M.; Zámocky, M.; Peterbauer, C.; Kulbe, K.D.; Haltrich, D. Continuous Enzymatic Regeneration of Electron Acceptors Used by Flavoenzymes: Cellobiose Dehydrogenase-Catalyzed Production of Lactobionic Acid as an Example. Biocatal. Biotransfor. 2004, 22, 97–104. [Google Scholar] [CrossRef]
- Ciullini, I.; Tilli, S.; Scozzafava, A.; Briganti, F. Fungal laccase, cellobiose dehydrogenase, and chemical mediators: combined actions for the decolorization of different classes of textile dyes. Bioresour. Technol. 2008, 99, 7003–7010. [Google Scholar] [CrossRef]
- Traxlmayr, M.W.; Obinger, C. Directed evolution of proteins for increased stability and expression using yeast display. Arch. Biochem. Biophys. 2012, 526, 174–180. [Google Scholar] [CrossRef] [PubMed]
- McCafferty, J.; Griffiths, A.D.; Winter, G.; Chiswell, D.J. Phage antibodies: filamentous phage displaying antibody variable domains. Nature 1990, 348, 552–554. [Google Scholar] [CrossRef]
- Francisco, J.A.; Campbell, R.; Iverson, B.L.; Georgiou, G. Production and fluorescence-activated cell sorting of Escherichia coli expressing a functional antibody fragment on the external surface. Proc. Natl. Acad. Sci. USA 1993, 90, 10444–10448. [Google Scholar] [CrossRef] [PubMed]
- Antipov, E.; Cho, A.E.; Wittrup, K.D.; Klibanov, A.M. Highly L and D enantioselective variants of horseradish peroxidase discovered by an ultrahigh-throughput selection method. Proc. Natl. Acad. Sci. USA 2008, 105, 17694–17699. [Google Scholar] [CrossRef] [PubMed]
- Beerli, R.R.; Bauer, M.; Buser, R.B.; Gwerder, M.; Muntwiler, S.; Maurer, P.; Saudan, P.; Bachmann, M.F. Isolation of human monoclonal antibodies by mammalian cell display. Proc. Natl. Acad. Sci. USA 2008, 105, 14336–14341. [Google Scholar] [CrossRef]
- Cherf, G.M.; Cochran, J.R. Applications of Yeast Surface Display for Protein Engineering. Methods Mol. Biol. 2015, 1319, 155–175. [Google Scholar] [CrossRef] [PubMed]
- Cohen, N.; Abramov, S.; Dror, Y.; Freeman, A. In vitro enzyme evolution: the screening challenge of isolating the one in a million. Trends Biotechnol. 2001, 19, 507–510. [Google Scholar] [CrossRef]
- Lin, H.; Cornish, V.W. Screening and selection methods for large-scale analysis of protein function. Angew. Chem. Int. Ed. Engl. 2002, 41, 4402–4425. [Google Scholar] [CrossRef]
- Mastrobattista, E.; Taly, V.; Chanudet, E.; Treacy, P.; Kelly, B.T.; Griffiths, A.D. High-throughput screening of enzyme libraries: in vitro evolution of a beta-galactosidase by fluorescence-activated sorting of double emulsions. Chem. Biol. 2005, 12, 1291–1300. [Google Scholar] [CrossRef] [PubMed]
- Aharoni, A.; Amitai, G.; Bernath, K.; Magdassi, S.; Tawfik, D.S. High-throughput screening of enzyme libraries: thiolactonases evolved by fluorescence-activated sorting of single cells in emulsion compartments. Chem. Biol. 2005, 12, 1281–1289. [Google Scholar] [CrossRef] [PubMed]
- Georgiou, G. Analysis of large libraries of protein mutants using flow cytometry. Adv. Protein Chem. 2000, 55, 293–315. [Google Scholar]
- Guerin, T.F.; Mondido, M.; McClenn, B.; Peasley, B. Application of resazurin for estimating abundance of contaminant-degrading micro-organisms. Lett. Appl. Microbiol. 2001, 32, 340–345. [Google Scholar] [CrossRef] [PubMed]
- Twigg, R.S. Oxidation-Reduction Aspects of Resazurin. Nature 1945, 155, 401. [Google Scholar] [CrossRef]
- Pfaller, M.A.; Barry, A.L. Evaluation of a novel colorimetric broth microdilution method for antifungal susceptibility testing of yeast isolates. J. Clin. Microbiol. 1994, 32, 1992–1996. [Google Scholar]
- Peeters, E.; Nelis, H.J.; Coenye, T. Comparison of multiple methods for quantification of microbial biofilms grown in microtiter plates. J. Microbiol. Methods 2008, 72, 157–165. [Google Scholar] [CrossRef]
- Mariscal, A.; Lopez-Gigosos, R.M.; Carnero-Varo, M.; Fernandez-Crehuet, J. Fluorescent assay based on resazurin for detection of activity of disinfectants against bacterial biofilm. Appl. Microbiol. Biotechnol. 2009, 82, 773–783. [Google Scholar] [CrossRef]
- O’Brien, J.; Wilson, I.; Orton, T.; Pognan, F. Investigation of the Alamar Blue (resazurin) fluorescent dye for the assessment of mammalian cell cytotoxicity. Eur. J. Biochem. 2000, 267, 5421–5426. [Google Scholar] [CrossRef]
- Chao, G.; Lau, W.L.; Hackel, B.J.; Sazinsky, S.L.; Lippow, S.M.; Wittrup, K.D. Isolating and engineering human antibodies using yeast surface display. Nat. Protoc. 2006, 1, 755–768. [Google Scholar] [CrossRef]
- Yoshida, M.; Ohira, T.; Igarashi, K.; Nagasawa, H.; Aida, K.; Hallberg, B.M.; Divne, C.; Nishino, T.; Samejima, M. Production and characterization of recombinant Phanerochaete chrysosporium cellobiose dehydrogenase in the methylotrophic yeast Pichia pastoris. Biosci. Biotechnol. Biochem. 2001, 65, 2050–2057. [Google Scholar] [CrossRef]
- Baminger, U.; Nidetzky, B.; Kulbe, K.D.; Haltrich, D. A simple assay for measuring cellobiose dehydrogenase activity in the presence of laccase. J. Microbiol. Methods 1999, 35, 253–259. [Google Scholar] [CrossRef]
- Gietz, R.D.; Schiestl, R.H. High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2007, 2, 31–34. [Google Scholar] [CrossRef]
- Rampersad, S.N. Multiple Applications of Alamar Blue as an Indicator of Metabolic Function and Cellular Health in Cell Viability Bioassays. Sensors 2012, 12, 12347–12360. [Google Scholar] [CrossRef] [PubMed]
- Blazic, M.; Kovacevic, G.; Prodanovic, O.; Ostafe, R.; Gavrovic-Jankulovic, M.; Fischer, R.; Prodanovic, R. Yeast surface display for the expression, purification and characterization of wild-type and B11 mutant glucose oxidases. Protein Expr. Purif. 2013, 89, 175–180. [Google Scholar] [CrossRef]
- Gibson, Q.H.; Swoboda, B.E.; Massey, V. Kinetics and Mechanism of Action of Glucose Oxidase. J. Biol. Chem. 1964, 239, 3927–3934. [Google Scholar]
- Ostafe, R.; Prodanovic, R.; Nazor, J.; Fischer, R. Ultra-High-Throughput Screening Method for the Directed Evolution of Glucose Oxidase. Chem. Biol. 2014, 21, 414–421. [Google Scholar] [CrossRef]
- Huang, D.; Shusta, E.V. Secretion and surface display of green fluorescent protein using the yeast Saccharomyces cerevisiae. Biotechnol. Progr. 2005, 21, 349–357. [Google Scholar] [CrossRef]
- Brugger, D.; Krondorfer, I.; Zahma, K.; Stoisser, T.; Bolivar, J.M.; Nidetzky, B.; Peterbauer, C.K.; Haltrich, D. Convenient microtiter plate-based, oxygen-independent activity assays for flavin-dependent oxidoreductases based on different redox dyes. Biotechnol. J. 2014, 9, 474–482. [Google Scholar] [CrossRef]
- Matsu-ura, S.; Yamauchi, Y.; Ohmori, H.; Maeda, H. Blood glucose determination with the reduction of resazurin as a fluorometric indicator reaction. Bunseki Kagaku 2002, 51, 111–115. [Google Scholar] [CrossRef]
- Bueno, C.; Villegas, M.L.; Bertolotti, S.G.; Previtali, C.M.; Neumann, M.G.; Encinas, M.V. The excited-state interaction of resazurin and resorufin with amines in aqueous solutions. Photophysics and photochemical reaction. Photochem. Photobiol. 2002, 76, 385–390. [Google Scholar] [CrossRef]
- Harreither, W.; Sygmund, C.; Augustin, M.; Narciso, M.; Rabinovich, M.L.; Gorton, L.; Haltrich, D.; Ludwig, R. Catalytic Properties and Classification of Cellobiose Dehydrogenases from Ascomycetes. Appl. Environ. Microbiol. 2011, 77, 1804–1815. [Google Scholar] [CrossRef]
- Caballero, I.; Lafuente, M.J.; Gamo, F.J.; Cid, C. A high-throughput fluorescence-based assay for Plasmodium dihydroorotate dehydrogenase inhibitor screening. Anal. Biochem. 2016, 506, 13–21. [Google Scholar] [CrossRef]
- Henriksson, G.; Sild, V.; Szabo, I.J.; Pettersson, G.; Johansson, G. Substrate specificity of cellobiose dehydrogenase from Phanerochaete chrysosporium. Biochim. Biophys. Acta Protein Struct. Molec. Enzym. 1998, 1383, 48–54. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).