Biodegradation of Picolinic Acid by Rhodococcus sp. PA18
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals and Media
2.2. Isolation and Identification of PA-Degrading Bacteria
2.3. Biodegradation of PA by a Rhodococcus Strain
2.4. Effects of Different Factors on PA Biodegradation
2.5. Identification of Metabolites during PA Degradation
2.6. Cell-Free Extract Activity Assays
2.7. Analytical Methods
3. Results
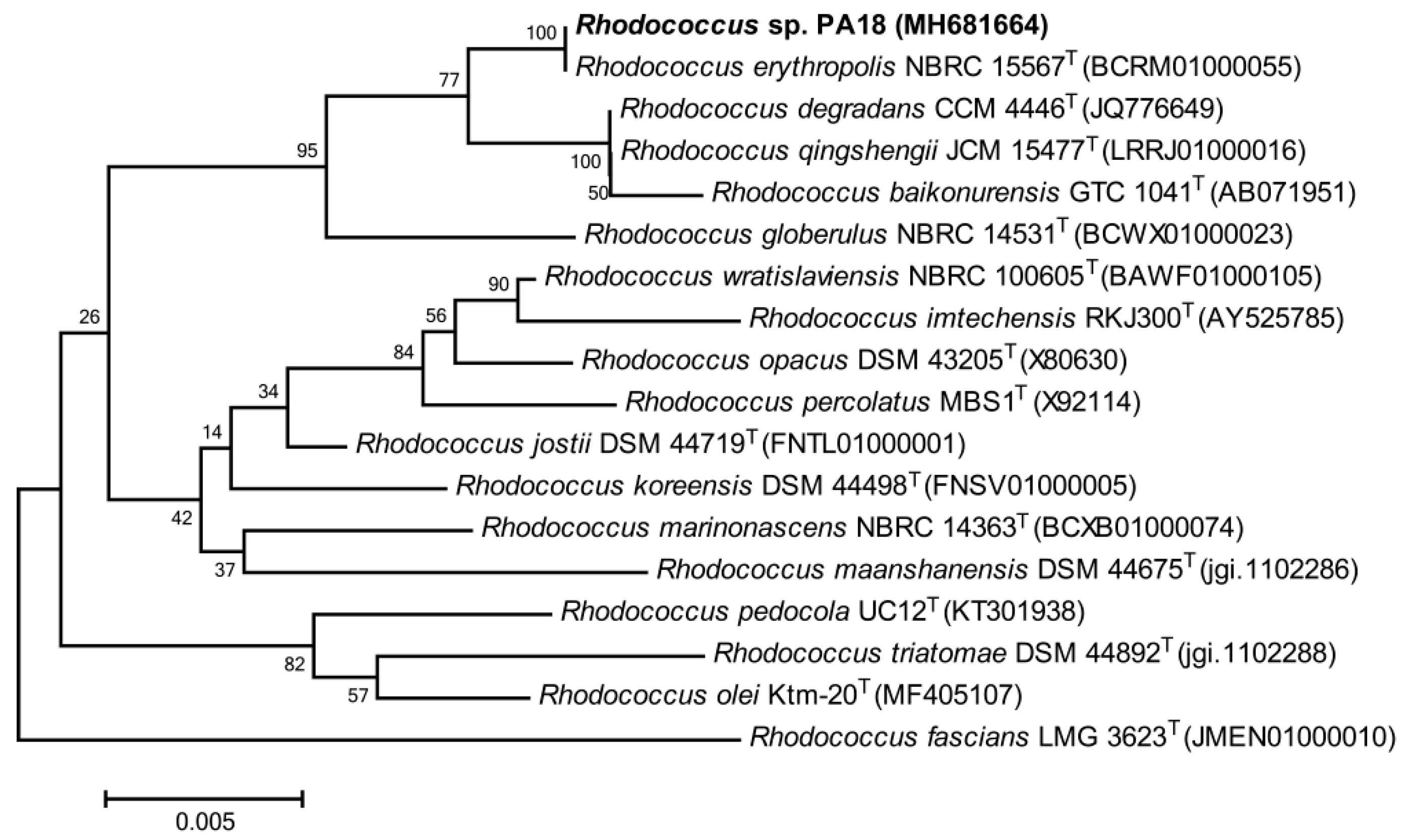
3.1. Isolation and Identification of the PA-Degrading Strains
3.2. Growth of Strain PA18 and PA Biodegradation
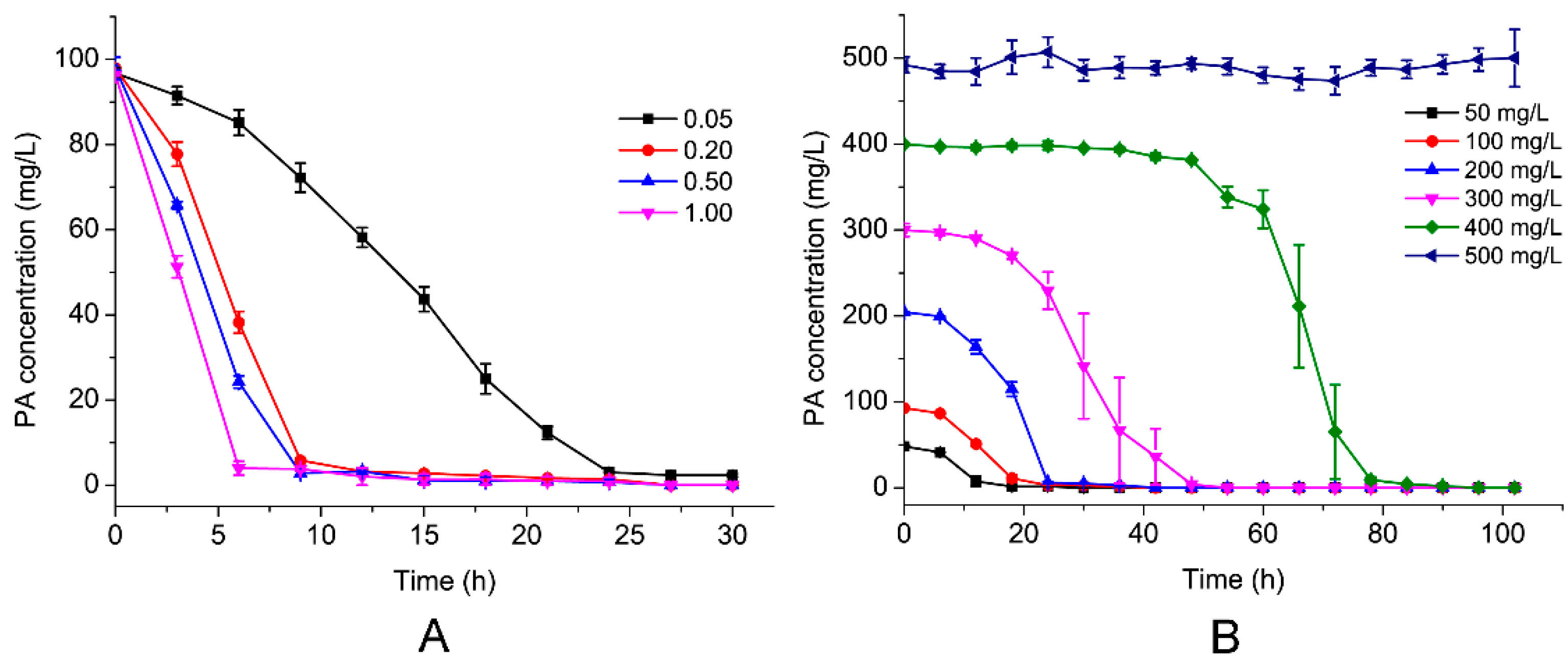
3.3. Effects of Initial PA Concentration and Inoculum of the Strain on PA Biodegradation
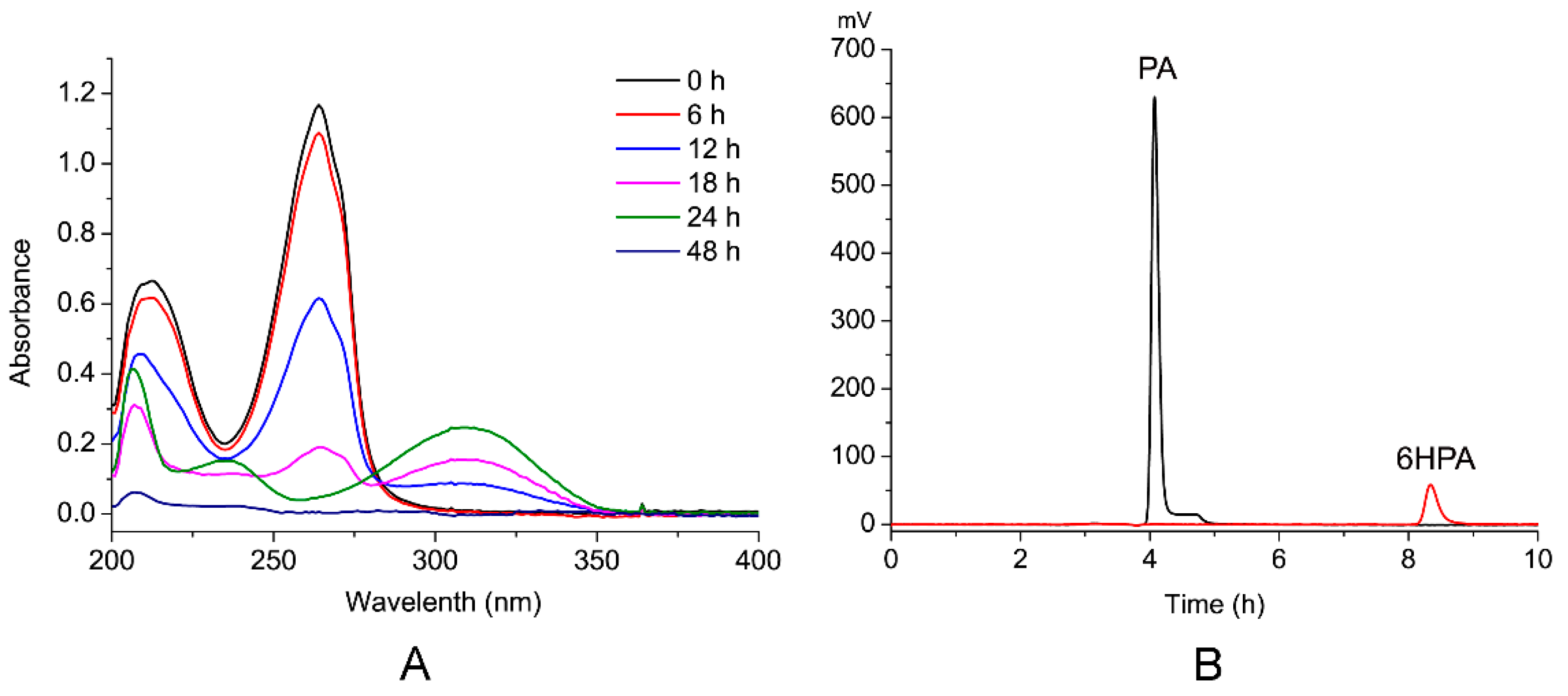
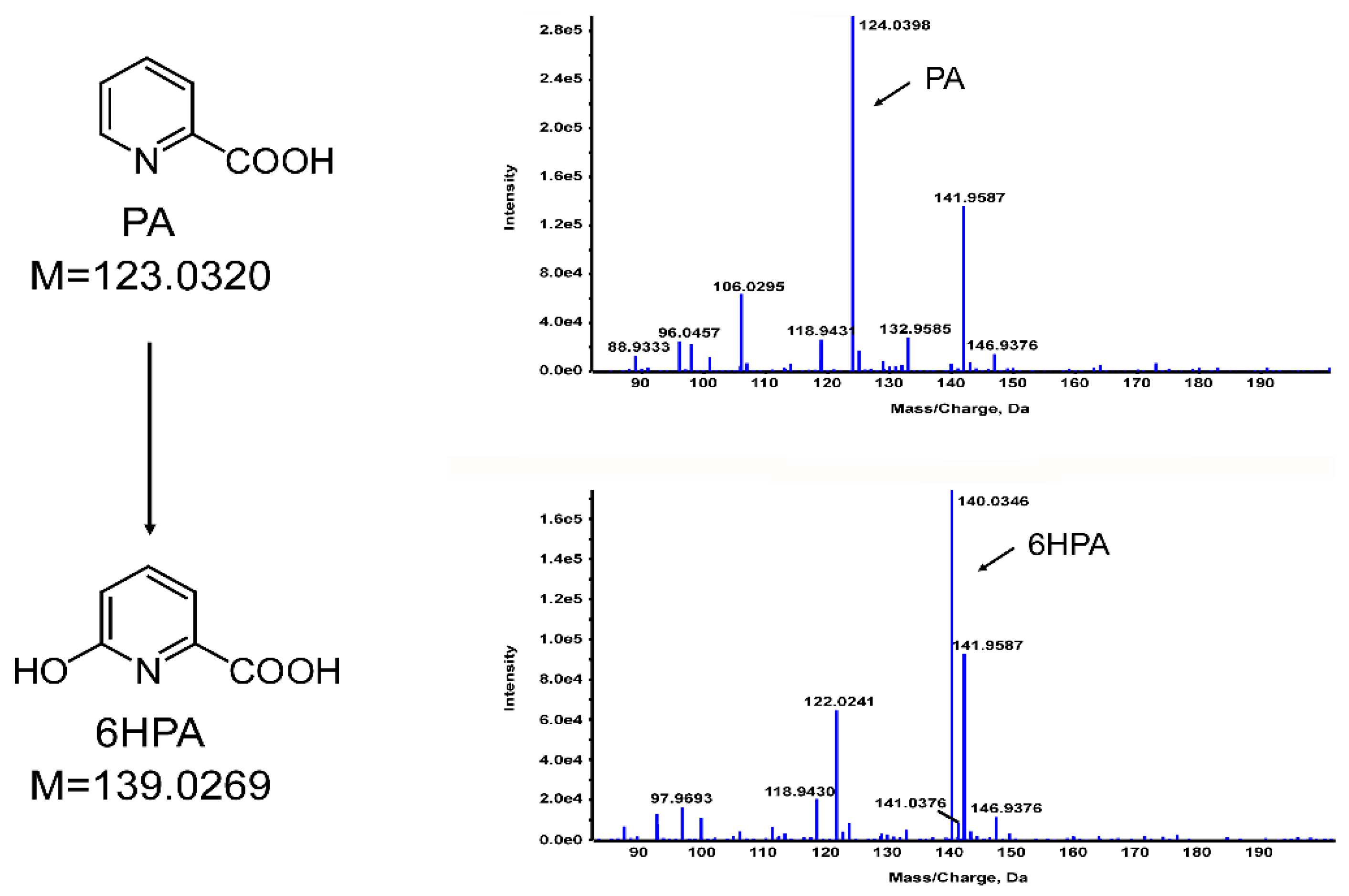
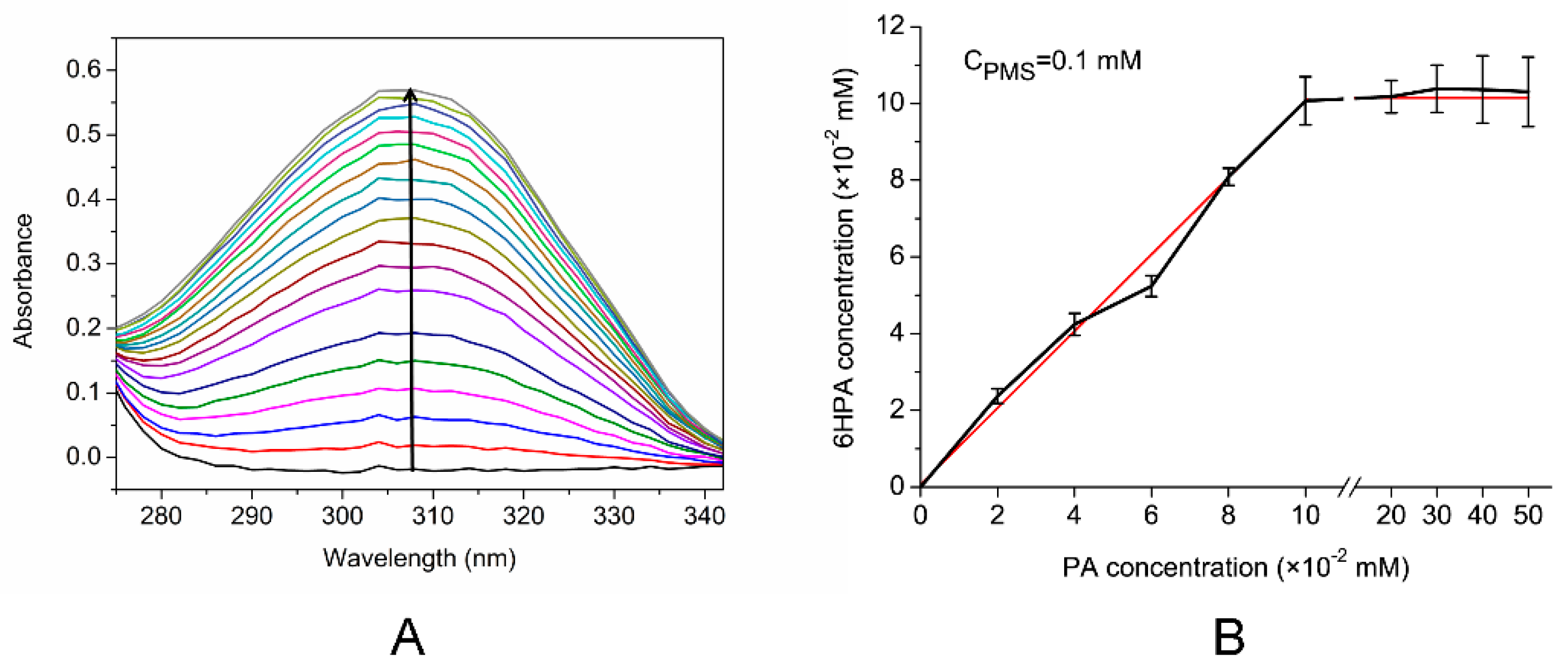
3.4. Identification of PA Metabolites
3.5. Activity of the Enzyme in the Cell Extract
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Kaiser, J.P.; Feng, Y.; Bollag, J.M. Microbial metabolism of pyridine, quinoline, acridine, and their derivatives under aerobic and anaerobic conditions. Microbiol. Rev. 1996, 60, 483–498. [Google Scholar] [PubMed]
- Yao, Y.; Tang, H.; Ren, H.; Yu, H.; Wang, L.; Zhang, W.; Behrman, E.J.; Xu, P. Iron(II)-dependent dioxygenase and N-formylamide deformylase catalyze the reactions from 5-hydroxy-2-pyridone to maleamate. Sci. Rep. 2013, 3, 3235. [Google Scholar] [CrossRef] [PubMed]
- Zheng, C.; Wang, Q.; Ning, Y.; Fan, Y.; Feng, S.; He, C.; Zhang, T.C.; Shen, Z. Isolation of a 2-picolinic acid-assimilating bacterium and its proposed degradation pathway. Bioresour. Technol. 2017, 245, 681–688. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.; Liu, W.; Wu, Z.L.; Liu, G.M.; Yin, H.; Chen, Y.R.; Hu, N.; Jia, L. Removal of pyridine from its wastewater by using a novel foam fractionation column. Chem. Eng. J. 2017, 321, 151–158. [Google Scholar] [CrossRef]
- Heyes, M.P.; Eugene, O.; Saito, K. Different kynurenine pathway enzymes limit quinolinic acid formation by various human cell types. Biochem. J. 1987, 326, 351–356. [Google Scholar] [CrossRef]
- Bryleva, E.Y.; Brundin, L. Kynurenine pathway metabolites and suicidality. Neuropharmacology 2017, 112, 324–330. [Google Scholar] [CrossRef] [PubMed]
- Esquive, D.G.; Ramirez-Ortega, D.; Pineda, B.; Castro, N.; Rios, C.; de la Cruz, V.P. Kynurenine pathway metabolites and enzymes involved in redox reactions. Neuropharmacology 2017, 112, 331–345. [Google Scholar]
- Asano, Y.; Yamamoto, Y.; Yamada, H. Catechol 2, 3-dioxygenase-catalyzed synthesis of picolinic acids from catechols. Biosci. Biotechnol. Biochem. 1994, 58, 2054–2056. [Google Scholar] [CrossRef]
- Nishino, S.F.; Spain, J.C. Degradation of nitrobenzene by a Pseudomonas pseudoalcaligenes. Appl. Environ. Microbiol. 1993, 59, 2520–2525. [Google Scholar] [PubMed]
- Tian, Y.Q.; Li, W.H. A new process of synthesis of chromium-2-picolinate by chromic anhydride oxidation. Chem. Eng. 2005, 19, 53–54. (In Chinese) [Google Scholar]
- Kutanovas, S.; Karvelis, L.; Vaitekūnas, J.; Stankevičiūtė, J.; Gasparavičiūtė, R.; Meškys, R. Isolation and characterization of novel pyridine dicarboxylic acid-degrading microorganisms. Chemija 2016, 30, 74–83. [Google Scholar]
- Qiu, J.G.; Zhang, J.J.; Zhang, Y.T.; Wang, Y.H.; Tong, L.; Hong, Q.; He, J. Biodegradation of picolinic acid by a newly isolated bacterium Alcaligenes faecalis strain JQ135. Curr. Microbiol. 2017, 74, 508–514. [Google Scholar] [CrossRef] [PubMed]
- Siegmund, I.; Koenig, K.; Andreesen, J.R. Molybdenum involvement in aerobic degradation of picolinic acid by Arthrobacter picolinophilus. FEMS Microbiol. Lett. 1990, 67, 281–284. [Google Scholar] [CrossRef]
- Zheng, C.; Zhou, J.; Wang, J.; Qu, B.; Lu, H.; Zhao, H. Aerobic degradation of 2-picolinic acid by a nitrobenzene-assimilating strain: Streptomyces sp. Z2. Bioresour. Technol. 2009, 100, 2082–2084. [Google Scholar] [CrossRef] [PubMed]
- Nie, Z.J.; Hang, B.J.; Cai, S.; Xie, X.T.; He, J.; Li, S.P. Degradation of cyhalofop-butyl (CyB) by Pseudomonas azotoformans strain QDZ-1 and cloning of a novel gene encoding CyB-hydrolyzing esterase. J. Agric. Food Chem. 2011, 59, 6040–6046. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Qiu, J.G.; Liu, B.; Zhao, L.; Zhang, Y.T.; Cheng, D.; Yan, X.; Jiang, J.D.; Hong, Q.; He, J. A novel degradation mechanism for pyridine derivatives in Alcaligenes faecalis JQ135. Appl. Environ. Microbiol. 2018, 84, e00910-18. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.G.; Jiang, W.K.; Wang, X.H.; Cheng, T.; Zhang, D.S.; Wang, H.; Qiu, J.G.; Cao, L.; Hong, Q. An amidase gene ipaH is responsible for the initial degradation step of iprodione in strain Paenarthrobacter sp. YJN-5. Appl. Environ. Microbiol. 2018, 84, e01150-18. [Google Scholar] [CrossRef] [PubMed]
- Warhurst, A.M.; Fewson, C.A. Biotransformations catalyzed by the genus Rhodococcus. Crit. Rev. Biotechnol. 1994, 14, 29–73. [Google Scholar] [CrossRef]
- Fahy, A.; McGenity, T.J.; Timmis, K.N.; Ball, A.S. Heterogeneous aerobic benzene-degrading communities in oxygen-depleted groundwaters. FEMS Microbiol. Ecol. 2006, 58, 260–270. [Google Scholar] [CrossRef] [PubMed]
- Leigh, M.B.; Prouzová, P.; Macková, M.; Macek, T.; Nagle, D.P.; Fletcher, J.S. Polychlorinated biphenyl (PCB)-degrading bacteria associated with trees in a PCB-contaminated site. Appl. Environ. Microbiol. 2006, 72, 2331–2342. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.T.; Lee, S.B.; Park, Y.H. Characterization of a pyridine-degrading branched Gram-positive bacterium isolated from the anoxic zone of an oil shale column. Appl. Microbiol. Biotechnol. 1991, 35, 824–829. [Google Scholar] [CrossRef]
- Yoon, J.H.; Kang, S.S.; Cho, Y.G.; Lee, S.T.; Kho, Y.H.; Kim, C.J.; Park, Y.H. Rhodococcus pyridinivorans sp. nov., a pyridine-degrading bacterium. Int. J. Syst. Evol. Microbiol. 2000, 50, 2173–2180. [Google Scholar] [CrossRef] [PubMed]
- Mathew, C.D.; Nagasawa, T.; Kobayashi, M.; Yamada, H. Nitrilase-catalyzed production of nicotinic acid from 3-cyanopyridine in Rhodococcus rhodochrous J1. Appl. Environ. Microbiol. 1988, 54, 1030–1032. [Google Scholar] [PubMed]
- Gong, X.W.; Ma, G.H.; Duan, Y.Q.; Zhu, D.L.; Chen, Y.K.; Zhang, K.Q.; Yang, J.K. Biodegradation and metabolic pathway of nicotine in Rhodococcus sp. Y22. World J. Microbiol. Biotechnol. 2016, 32, 188. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, G.; Bauder, R.; Speer, M.; Rommet, T.O.; Lingens, F. Microbial metabolism of quinoline and related compounds. II. Degradation of quinoline by Pseudomonas fluorescens 3, Pseudomonas putida 86 and Rhodococcus spec. B1. Biol. Chem. Hoppe Seyler 1989, 370, 1183–1190. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, T.; Nagasawa, T. Enzymatic functionalization of aromatic N-heterocycles: Hydroxylation and carboxylation. J. Biosci. Bioeng. 2000, 89, 111–118. [Google Scholar] [CrossRef]
- Mallick, S.; Chakraborty, J.; Dutta, T.K. Role of oxygenases in guiding diverse metabolic pathways in the bacterial degradation of low-molecular-weight polycyclic aromatic hydrocarbons: A review. Crit. Rev. Microbiol. 2011, 37, 64–90. [Google Scholar] [CrossRef] [PubMed]
- Ghosal, D.; Ghosh, S.; Dutta, T.K.; Ahn, Y. Current state of knowledge in microbial degradation of polycyclic aromatic hydrocarbons (PAHs): A review. Front. Microbiol. 2016, 7, 1369. [Google Scholar] [CrossRef] [PubMed]
- Watson, G.K.; Cain, R.B. Microbial metabolism of the pyridine ring. Metabolic pathways of pyridine biodegradation by soil bacteria. Biochem. J. 1975, 146, 157–172. [Google Scholar] [CrossRef] [PubMed]
- Orpin, C.G.; Knight, M.; Evans, W.C. The bacterial oxidation of picolinamide, a photolytic product of diquat. Biochem. J. 1972, 127, 819–831. [Google Scholar] [CrossRef] [PubMed]
- Qiu, J.G.; Ma, Y.; Wen, Y.Z.; Chen, L.S.; Wu, L.F.; Liu, W.P. Functional identification of two novel genes from Pseudomonas sp. strain HZN6 involved in the catabolism of nicotine. Appl. Environ. Microbiol. 2012, 78, 2154–2160. [Google Scholar] [CrossRef] [PubMed]
- Shukla, O.P. 8-Hydroxycoumarin: An intermediate in the microbial transformation of quinoline. Curr. Sci. 1984, 53, 1145–1147. [Google Scholar]
- Yasuda, M.; Sakamoto, T.; Sashida, R.; Ueda, M.; Morimoto, Y.; Nagasawa, T. Microbial hydroxyIation of 3-cyanopyridine to 3-cyano-6-hydroxypyridine. Biosci. Biotechnol. Biochem. 1995, 59, 572–575. [Google Scholar] [CrossRef]
- Yu, H.; Tang, H.Z.; Xu, P. Green strategy from waste to value-added-chemical production: Efficient biosynthesis of 6-hydroxy-3-succinoyl-pyridine by an engineered biocatalyst. Sci. Rep. 2014, 4, 5397. [Google Scholar] [CrossRef] [PubMed]
- Hannemann, F.; Bichet, A.; Ewen, K.M.; Bernhardt, R. Cytochrome P450 systems-biological variations of electron transport chains. BBA-Gen Subj. 2007, 1770, 330–344. [Google Scholar] [CrossRef] [PubMed]
- Jiménez, J.I.; Canales, Á.; Jiménez-Barbero, J.; Ginalski, K.; Rychlewski, L.; García, J.L.; Díaz, E. Deciphering the genetic determinants for aerobic nicotinic acid degradation: The nic cluster from Pseudomonas putida KT2440. Proc. Natl. Acad. Sci. USA 2008, 105, 11329–11334. [Google Scholar] [CrossRef] [PubMed]

© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Ji, J.; Xu, S.; Wang, H.; Shen, B.; He, J.; Qiu, J.; Chen, Q. Biodegradation of Picolinic Acid by Rhodococcus sp. PA18. Appl. Sci. 2019, 9, 1006. https://doi.org/10.3390/app9051006
Zhang Y, Ji J, Xu S, Wang H, Shen B, He J, Qiu J, Chen Q. Biodegradation of Picolinic Acid by Rhodococcus sp. PA18. Applied Sciences. 2019; 9(5):1006. https://doi.org/10.3390/app9051006
Chicago/Turabian StyleZhang, Yanting, Junbin Ji, Siqiong Xu, Hongmei Wang, Biao Shen, Jian He, Jiguo Qiu, and Qing Chen. 2019. "Biodegradation of Picolinic Acid by Rhodococcus sp. PA18" Applied Sciences 9, no. 5: 1006. https://doi.org/10.3390/app9051006
APA StyleZhang, Y., Ji, J., Xu, S., Wang, H., Shen, B., He, J., Qiu, J., & Chen, Q. (2019). Biodegradation of Picolinic Acid by Rhodococcus sp. PA18. Applied Sciences, 9(5), 1006. https://doi.org/10.3390/app9051006




