Reservoir Computing Based Echo State Networks for Ventricular Heart Beat Classification
Abstract
1. Introduction
2. Materials and Methods
2.1. ECG Data Sources
2.2. ECG Signal Preprocessing
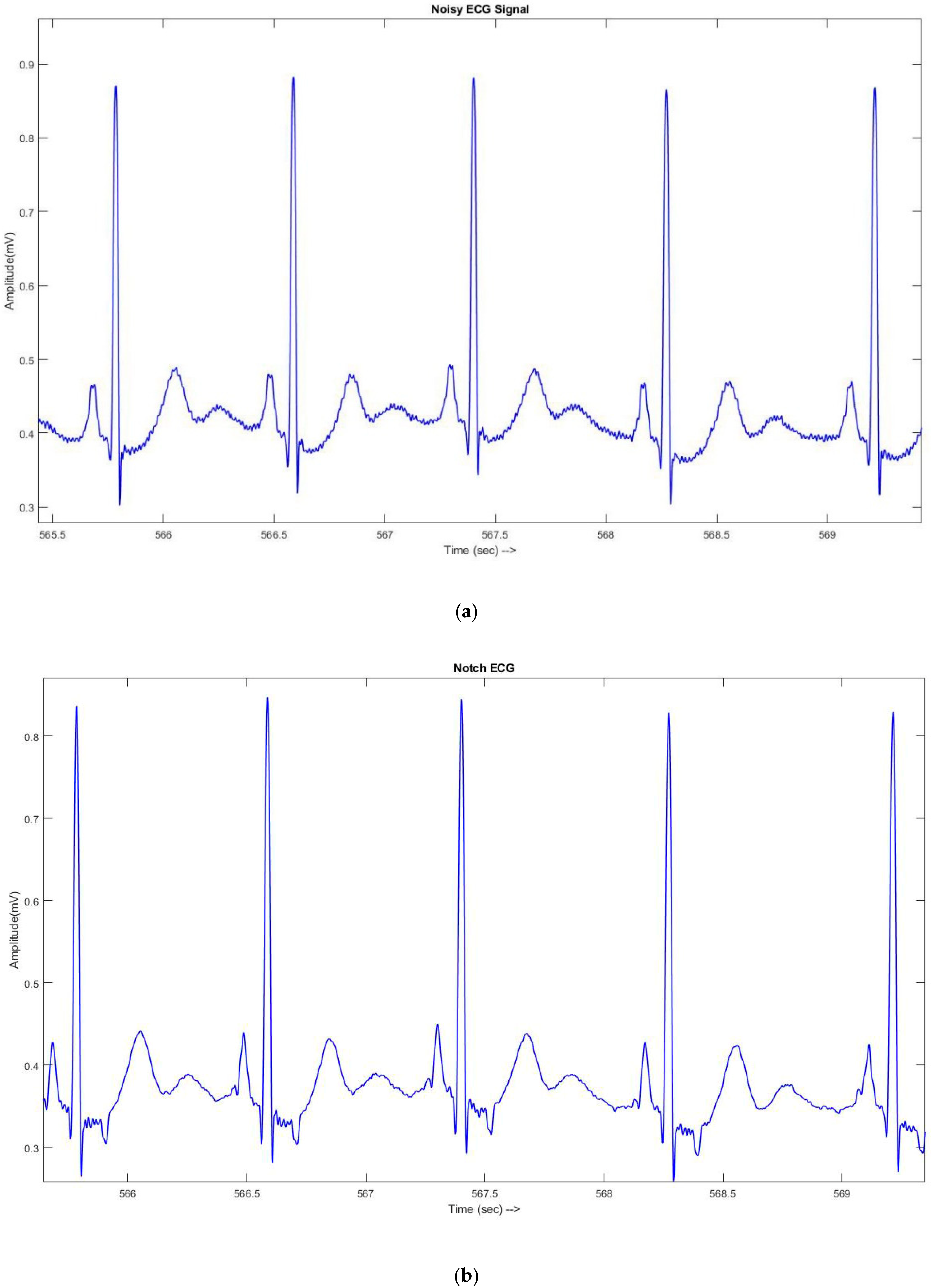
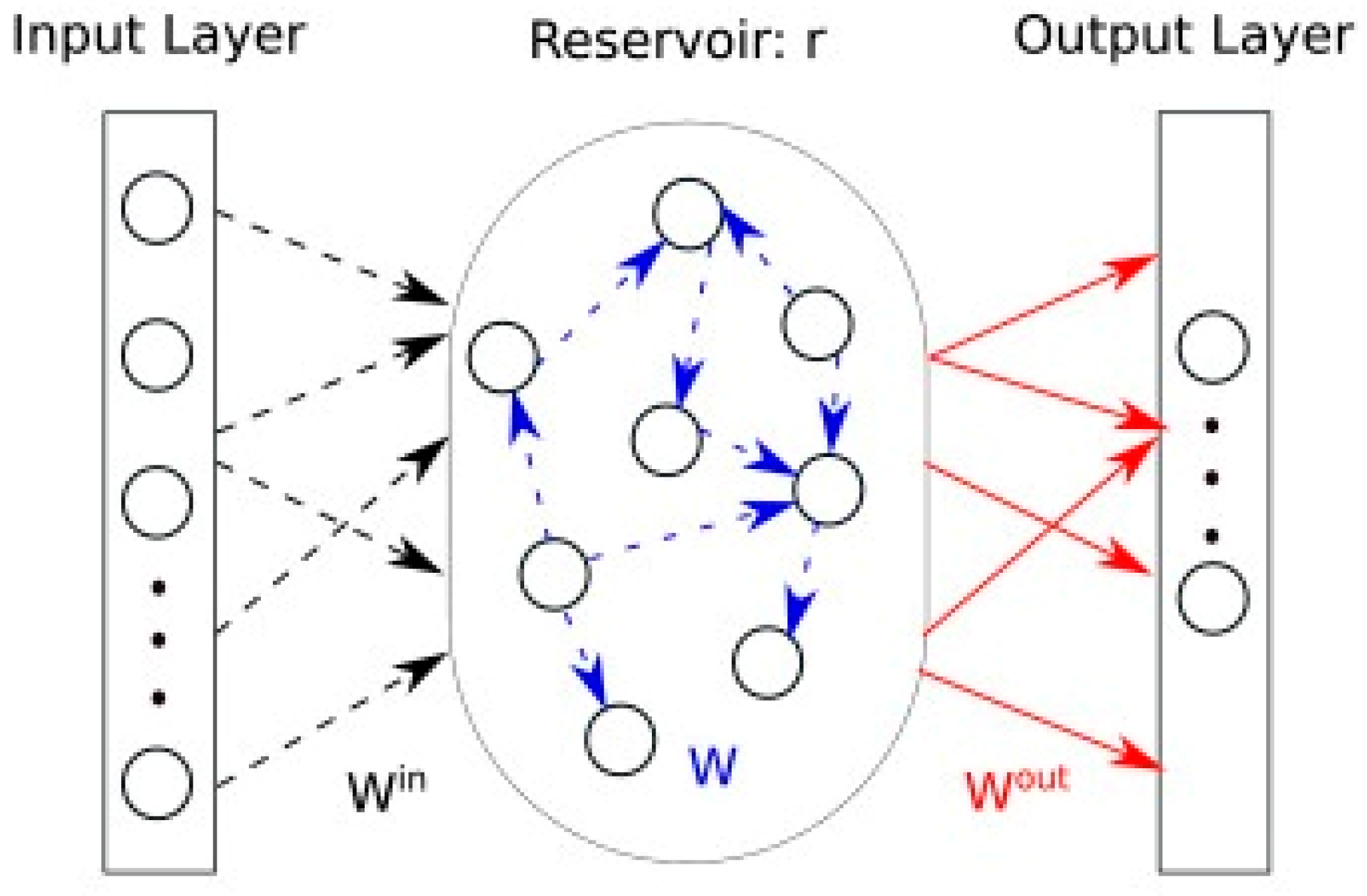
2.2.1. ECG Signal Denoising
2.2.2. High Frequency component (Peak) Detection
2.2.3. Heart Beat Segmentation
2.3. Temporal Feature Extraction
- 1.
- The Previous R-R interval is defined as the time duration between the current beat and previous beat.
- 2.
- The Subsequent R-R Interval is explained as the time duration between the current beat and subsequent beat.
- 3.
- The Standard deviation of Successive Difference (SDSD) is defined as the standard feature of physiological signals for arrhythmia classification. It is the difference between 10 consequent RR intervals [48].
- 4.
- The Average of RR-Interval is defined as the average ratio of 10 consecutive RR intervals.
- 5.
- The Average Derivate of RR interval is defined as the average ratio of the derivate of the RR interval, where the derivation of the segmented beat is calculated by using this central difference Equation (2).
- 6.
- PCC is defined as the Pearson’s correlation coefficients between the template beat and ECG signal heartbeat.
- 7.
- PCC () is defined as the Pearson’s correlation coefficients between the derivatives value of the template beat and ECG signal segmented beat.
- 8.
- PCC () is defined as the Pearson’s correlation coefficients between the squares ratio of the template beat and ECG signal segmented beat.
- 9.
- PCC is defined as the Pearson’s correlation coefficients between the squares of the derivatives of the template beat and ECG signal segmented beat.
- 10.
- The Average of Template beat is defined as the average of the template beat.
- 11.
- The Average Derivate of Template beat is defined as the average derivatives of the template beat.
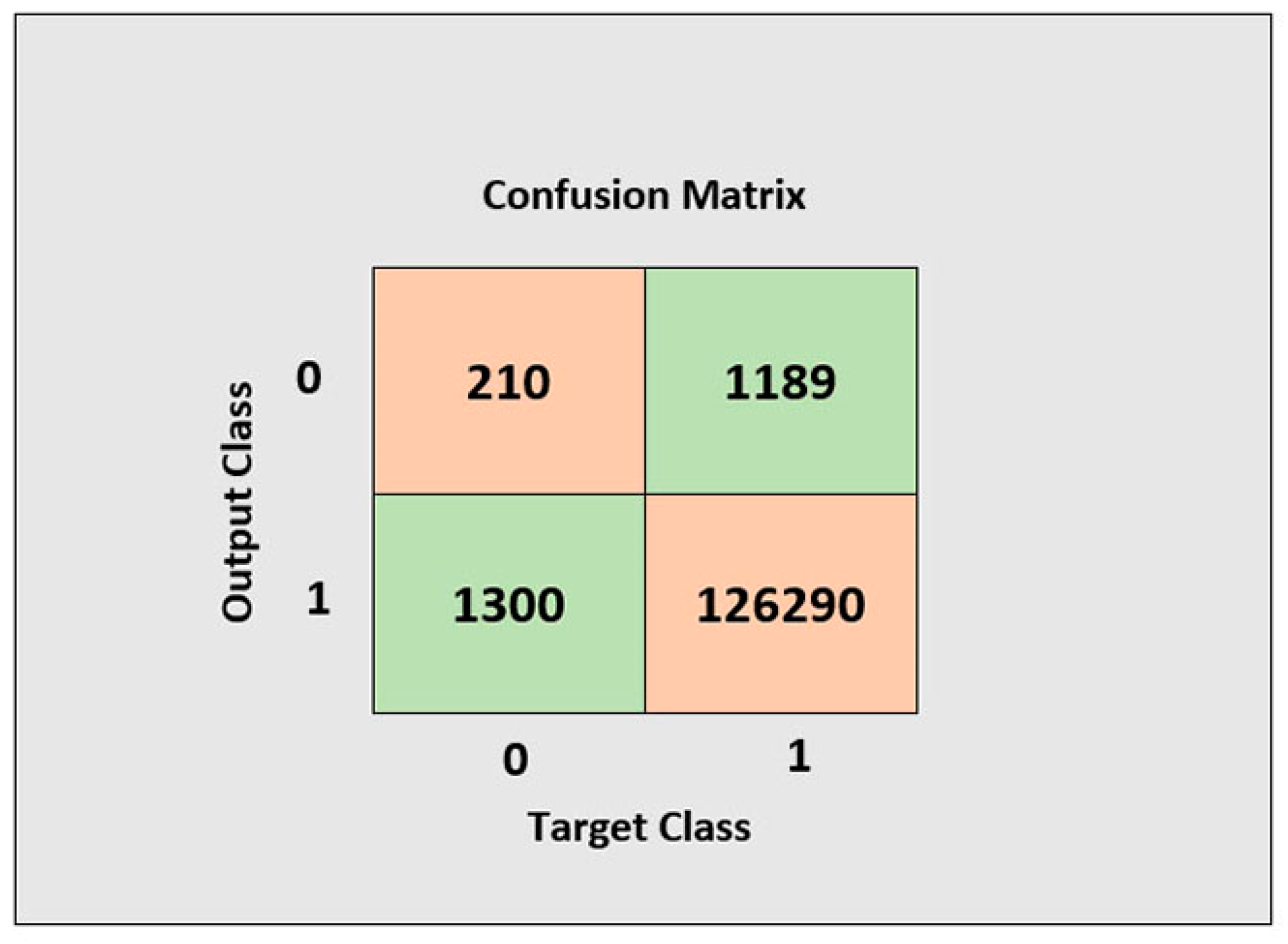
2.4. The Configuration of the Echo State Network Based Reservoir Computing for Classification
2.4.1. Learning Phase of The ESN Model
3. Results and Discussion
3.1. Performance Evaluation Parameters
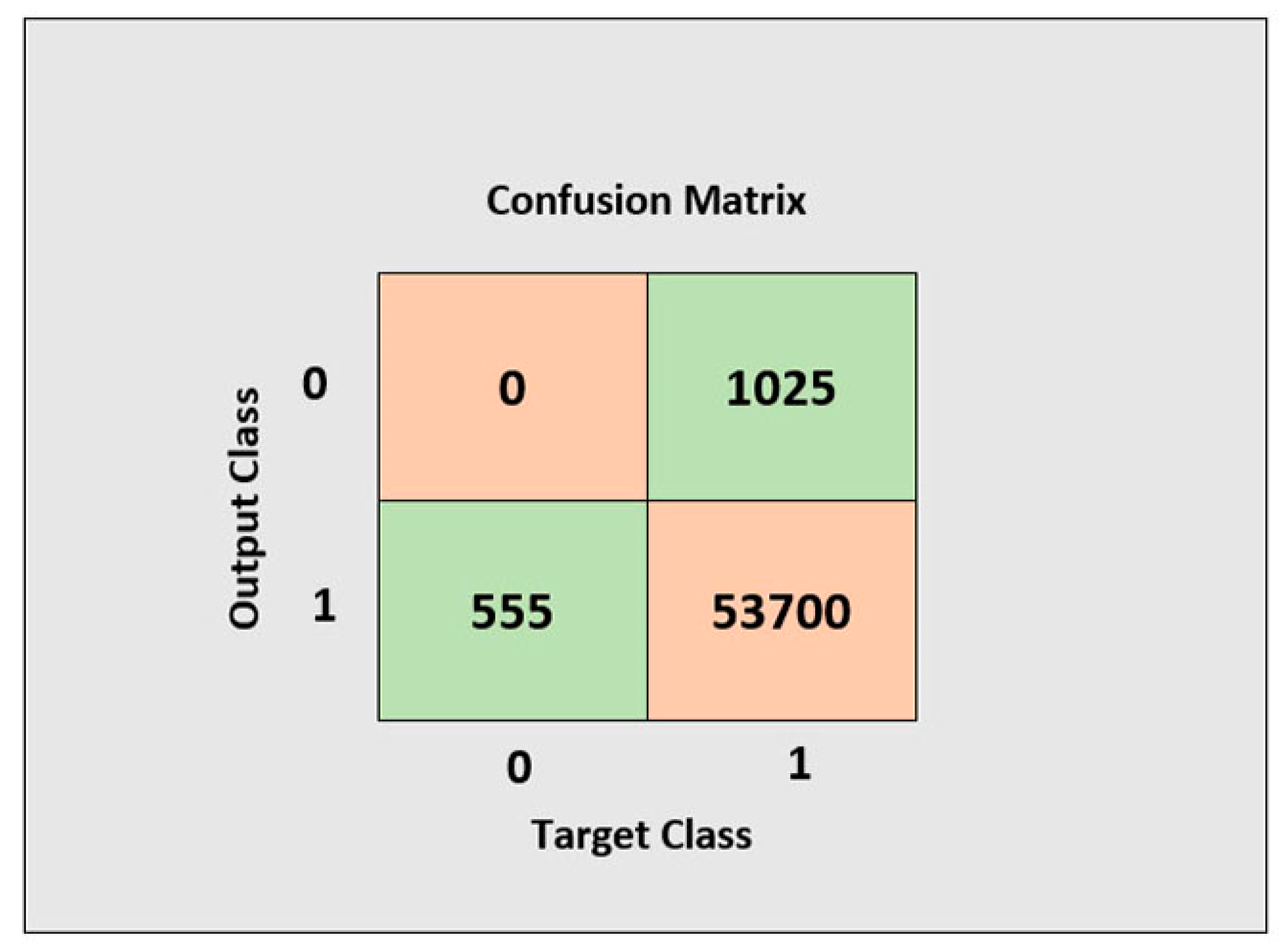
3.2. Evaluation
3.3. Comparison with State-of-the-art Methods
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Writing Group Members; Thom, T.; Haase, N.; Rosamond, W.; Howard, V.J.; Rumsfeld, J.; Manolio, T.; Zheng, Z.J.; Flegal, K.; O’Donnell, C.; et al. Heart disease and stroke statistics—2006 update: A report from the American Heart Association Statistics Committee and Stroke Statistics Subcommittee. Circulation 2006, 113, e85–e151. [Google Scholar] [PubMed]
- Stevens, G.; Mascarenhas, M.; Mathers, C. WHO brochure». Bull. World Health Organ. 2009, 87, 646. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Ge, J. Cardiovascular diseases in China: Current status and future perspectives. IJC Heart Vasc. 2015, 6, 25–31. [Google Scholar] [CrossRef] [PubMed]
- Carnevale, L.; Celesti, A.; Fazio, M.; Bramanti, P.; Villari, M. Heart disorder detection with menard algorithm on apache spark. In European Conference on Service-Oriented and Cloud Computing; Springer: Berlin/Heidelberg, Germany, 2017; pp. 229–237. [Google Scholar]
- Melillo, P.; Castaldo, R.; Sannino, G.; Orrico, A.; de Pietro, G.; Pecchia, L. Wearable technology and ECG processing for fall risk assessment, prevention and detection. In Proceedings of the 2015 37th Annual International Conference of the IEEE, Engineering in Medicine and Biology Society (EMBC), Milan, Italy, 25–29 August 2015; pp. 7740–7743. [Google Scholar]
- Sannino, G.; de Pietro, G. An evolved ehealth monitoring system for a nuclear medicine department. In Proceedings of the Developments in E-Systems Engineering (DeSE), Dubai, United Arab Emirates, 6–8 December 2011; pp. 3–6. [Google Scholar]
- Abbasi, Q.H.; Rehman, M.U.; Qaraqe, K.; Alomainy, A. Advances in body-centric wireless communication: Applications and state-of-the-art. Institution of Engineering and Technology 2016. [Google Scholar]
- De Chazal, P.; O’Dwyer, M.; Reilly, R.B.R. Automatic classification of heartbeats using ECG morphology and heartbeat interval features. IEEE Trans. Biomed. Eng. 2004, 51, 1196–1206. [Google Scholar] [CrossRef] [PubMed]
- Tsipouras, M.G.; Fotiadis, D.I. Automatic arrhythmia detection based on time and time–frequency analysis of heart rate variability. Comput. Methods Progr. Biomed. 2004, 74, 95–108. [Google Scholar] [CrossRef]
- Alonso-Atienza, F.; Morgado, E.; Fernandez-Martinez, L.; Garcia-Alberola, A.; Rojo-Alvarez, J.L. Detection of life-threatening arrhythmias using feature selection and support vector machines. IEEE Trans. Biomed. Eng. 2014, 61, 832–840. [Google Scholar] [CrossRef]
- Zhang, Z.; Dong, J.; Luo, X.; Choi, K.S.; Wu, X. Heartbeat classification using disease-specific feature selection. Comput. Biol. Med. 2014, 46, 79–89. [Google Scholar] [CrossRef]
- Huang, H.; Liu, J.; Zhu, Q.; Wang, R.; Hu, G. A new hierarchical method for inter-patient heartbeat classification using random projections and RR intervals. Biomed. Eng. Online 2014, 13, 90. [Google Scholar] [CrossRef]
- Zidelmal, Z.; Amirou, A.; Ould-Abdeslam, D.; Merckle, J. ECG beat classification using a cost sensitive classifier. Comput. Methods Progr. Biomed. 2013, 111, 570–577. [Google Scholar] [CrossRef]
- Garcia, G.; Moreira, G.; Menotti, D.; Luz, E.J.S.R. Inter-Patient ECG Heartbeat Classification with Temporal VCG Optimized by PSO. Sci. Rep. 2017, 7, 10543. [Google Scholar] [CrossRef] [PubMed]
- Qurraie, S.S.; Afkhami, R.G.J.B.E.L. ECG arrhythmia classification using time frequency distribution techniques. International journal of Intelligent Engineering and Systems 2017, 7, 325–332. [Google Scholar]
- Ye, C.; Kumar, B.V.; Coimbra, M.T. Heartbeat classification using morphological and dynamic features of ECG signals. IEEE Trans. Biomed. Eng. 2012, 59, 2930–2941. [Google Scholar] [PubMed]
- Ye, C.; Kumar, B.V.; Coimbra, M.T. An Automatic Subject-Adaptable Heartbeat Classifier Based on Multiview Learning. IEEE J. Biomed. Health Inform. 2016, 20, 1485–1492. [Google Scholar] [CrossRef] [PubMed]
- Mar, T.; Zaunseder, S.; Martínez, J.P.; Llamedo, M.; Poll, R. Optimization of ECG classification by means of feature selection. IEEE Trans. Biomed. Eng. 2011, 58, 2168–2177. [Google Scholar] [CrossRef] [PubMed]
- Elhaj, F.A.; Salim, N.; Harris, A.R.; Swee, T.T.; Ahmed, T. Arrhythmia recognition and classification using combined linear and nonlinear features of ECG signals. Comput. Methods Progr. Biomed. 2016, 127, 52–63. [Google Scholar] [CrossRef] [PubMed]
- Raj, S.; Ray, K.C.; Shankar, O. Cardiac arrhythmia beat classification using DOST and PSO tuned SVM. Comput. Methods Progr. Biomed. 2016, 136, 163–177. [Google Scholar] [CrossRef]
- Castillo, O.; Melin, P.; Ramírez, E.; Soria, J. Hybrid intelligent system for cardiac arrhythmia classification with Fuzzy K-Nearest Neighbors and neural networks combined with a fuzzy system. Expert Syst. Appl. 2012, 39, 2947–2955. [Google Scholar] [CrossRef]
- Saini, I.; Singh, D.; Khosla, A. QRS detection using K-Nearest Neighbor algorithm (KNN) and evaluation on standard ECG databases. J. Adv. Res. 2013, 4, 331–344. [Google Scholar] [CrossRef]
- Dokur, Z.; Ölmez, T. ECG beat classification by a novel hybrid neural network. Comput. Methods Progr. Biomed. 2001, 66, 167–181. [Google Scholar] [CrossRef]
- Martis, R.J.; Acharya, U.R.; Min, L.C. ECG beat classification using PCA, LDA, ICA and discrete wavelet transform. Biomed. Signal Process. Control 2013, 8, 437–448. [Google Scholar] [CrossRef]
- Inan, O.T.; Giovangrandi, L.; Kovacs, G.T. Robust neural-network-based classification of premature ventricular contractions using wavelet transform and timing interval features. IEEE Trans. Biomed. Eng. 2006, 53, 2507–2515. [Google Scholar] [CrossRef] [PubMed]
- Javadi, M.; Ebrahimpour, R.; Sajedin, A.; Faridi, S.; Zakernejad, S. Improving ECG classification accuracy using an ensemble of neural network modules. PLoS ONE 2011, 6, e24386. [Google Scholar] [CrossRef] [PubMed]
- Afkhami, R.G.; Azarnia, G.; Tinati, M.A. Cardiac arrhythmia classification using statistical and mixture modeling features of ECG signals. Pattern Recognit. Lett. 2016, 70, 45–51. [Google Scholar] [CrossRef]
- Wu, Z.; Ding, X.; Zhang, G. A novel method for classification of ECG arrhythmias using deep belief networks. J. Comput. Intell. Appl. 2016, 15, 1650021. [Google Scholar] [CrossRef]
- Acharya, U.R.; Oh, S.L.; Hagiwara, Y.; Tan, J.H.; Adam, M.; Gertych, A.; San Tan, R. A deep convolutional neural network model to classify heartbeats. Comput. Biol. Med. 2017, 89, 389–396. [Google Scholar] [CrossRef] [PubMed]
- Kiranyaz, S.; Ince, T.; Gabbouj, M. Real-time patient-specific ECG classification by 1-D convolutional neural networks. IEEE Trans. Biomed. Eng. 2016, 63, 664–675. [Google Scholar] [CrossRef]
- Krasteva, V.; Jekova, I. QRS template matching for recognition of ventricular ectopic beats. Ann. Biomed. Eng. 2007, 35, 2065–2076. [Google Scholar] [CrossRef]
- Jaeger, H.J.B. The “Echo State” Approach to Analysing and Training Recurrent Neural Networks-with An Erratum Note; German National Research Center for Information Technology GMD Technical Report; German National Research Center: Bonn, Germany, 2001; Volume 148, p. 13. [Google Scholar]
- Lukoševičius, M.; Jaeger, H. Reservoir computing approaches to recurrent neural network training. Comput. Sci. Rev. 2009, 3, 127–149. [Google Scholar] [CrossRef]
- Greenwald, S.D.; Patil, R.S.; Mark, R.G. Improved detection and classification of arrhythmias in noise-corrupted electrocardiograms using contextual information. In Proceedings of the Proceedings Computers in Cardiology, Chicago, IL, USA, 23–26 September 1990; pp. 461–464. [Google Scholar]
- Goldberger, A.L.; Amaral, L.A.; Glass, L.; Hausdorff, J.M.; Ivanov, P.C.; Mark, R.G.; Mietus, J.E.; Moody, G.B.; Peng, C.-K.; Stanley, H.E.J.C. PhysioBank, PhysioToolkit, and PhysioNet: Components of a new research resource for complex physiologic signals. Circulation 2000, 101, e215–e220. [Google Scholar] [CrossRef]
- ANSI/AAMI EC57:1998. Testing and Reporting Performance Results of Cardiac Rhythm and ST Segment Measurement Algorithms; AAMI: Brisbane, Australia, 1998. [Google Scholar]
- Friesen, G.M.; Jannett, T.C.; Jadallah, M.A.; Yates, S.L.; Quint, S.R.; Nagle, H.T. A comparison of the noise sensitivity of nine QRS detection algorithms. IEEE Trans. Biomed. Eng. 1990, 37, 85–98. [Google Scholar] [CrossRef] [PubMed]
- Chandrakar, B.; Yadav, O.P.; Chandra, V.K. A survey of noise removal techniques for ECG signals. Int. J. Adv. Res. Comput. Commun. Eng. 2013, 2, 1354–1357. [Google Scholar]
- Cuomo, S.; De Pietro, G.; Farina, R.; Galletti, A.; Sannino, G. A revised scheme for real time ecg signal denoising based on recursive filtering. Biomed. Signal Process. Control 2016, 27, 134–144. [Google Scholar] [CrossRef]
- Farina, R.; Dobricic, S.; Storto, A.; Masina, S.; Cuomo, S. A revised scheme to compute horizontal covariances in an oceanographic 3D-VAR assimilation system. J. Comput. Phys. 2015, 284, 631–647. [Google Scholar] [CrossRef]
- De Chazal, P.; Reilly, R.B. A patient-adapting heartbeat classifier using ECG morphology and heartbeat interval features. IEEE Trans. Biomed. Eng. 2006, 53, 2535–2543. [Google Scholar] [CrossRef] [PubMed]
- Pratt, W.K. Digital Image Processing: PIKS Scientific Inside; Wiley-Interscience: Hoboken, NJ, USA, 2007. [Google Scholar]
- IEEE Acoustics, Speech, and Signal Processing Society. Digital Signal Processing Committee. In Programs for digital signal processing; IEEE: Piscataway, NJ, USA, 1979. [Google Scholar]
- Pan, J.; Tompkins, W.J. A real-time QRS detection algorithm. IEEE Trans. Biomed. Eng. 1985, 32, 230–236. [Google Scholar] [CrossRef] [PubMed]
- Acharya, U.R.; Fujita, H.; Oh, S.L.; Raghavendra, U.; Tan, J.H.; Adam, M.; Gertych, A.; Hagiwara, Y. Automated identification of shockable and non-shockable life-threatening ventricular arrhythmias using convolutional neural network. Future Gen. Comput. Syst. 2018, 79, 952–959. [Google Scholar] [CrossRef]
- Nong, W. A novel algorithm for ventricular arrhythmia classification using a fuzzy logic approach. Aust. Phys. Eng. Sci. Med. 2016, 39, 903–912. [Google Scholar]
- Jain, A.K.; Duin, R.P.; Mao, J. Statistical pattern recognition: A review. IEEE Trans.Pattern Anal. Mach. Intell. 2000, 22, 4–37. [Google Scholar] [CrossRef]
- Kim, Y.J.; Heo, J.; Park, K.S.; Kim, S. Proposition of novel classification approach and features for improved real-time arrhythmia monitoring. Comput. Biol. Med. 2016, 75, 190–202. [Google Scholar] [CrossRef]
- Maass, W.; Natschläger, T.; Markram, H. Real-time computing without stable states: A new framework for neural computation based on perturbations. Neural Comput. 2002, 14, 2531–2560. [Google Scholar] [CrossRef] [PubMed]
- Yamazaki, T.; Tanaka, S. The cerebellum as a liquid state machine. Neural Netw. 2007, 20, 290–297. [Google Scholar] [CrossRef] [PubMed]
- Brunner, D.; Soriano, M.C.; Mirasso, C.R.; Fischer, I. Parallel photonic information processing at gigabyte per second data rates using transient states. Nat. Commun. 2013, 4, 1364. [Google Scholar] [CrossRef] [PubMed]
- Albert, A. Regression and the Moore-Penrose Pseudoinverse; Elsevier: Amsterdam, The Netherlands, 1972. [Google Scholar]
- Tikhonov, A.; Goncharsky, A.; Stepanov, V.; Yagola, A.G. Numerical methods for the solution of ill-posed problems (Mathematics and its Applications). 1995. [Google Scholar]
- Kutlu, Y.; Kuntalp, D. Feature extraction for ECG heartbeats using higher order statistics of WPD coefficients. Comput. Methods Progr. Biomed. 2012, 105, 257–267. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H. System for Cardiac Arrhythmia Detection and Characterization. U.S. Google Patent 8,233,972, 31 July 2012. [Google Scholar]
- Teijeiro, T.; Félix, P.; Presedo, J.; Castro, D. Heartbeat classification using abstract features from the abductive interpretation of the ECG. IEEE J. Biomed. Health Inform. 2018, 22, 409–420. [Google Scholar] [CrossRef]
- Llamedo, M.; Martínez, J.P. Heartbeat classification using feature selection driven by database generalization criteria. IEEE Trans. Biomed. Eng. 2011, 58, 616–625. [Google Scholar] [CrossRef]
- Llamedo, M.; Martínez, J.P. An automatic patient-adapted ECG heartbeat classifier allowing expert assistance. IEEE Trans. Biomed. Eng. 2012, 59, 2312–2320. [Google Scholar] [CrossRef]







| Acronyms | |
|---|---|
| MIT-BIH-AR | Massachusetts Institute of Technology-Beth Israel arrhythmia |
| MIT-BIH-SVDM | Massachusetts Institute of Technology-Beth Israel Hospital Supraventricular Arrhythmia database |
| AHA | American Heart Association |
| ECG | Electrocardiogram |
| RC | Reservoir computing |
| MIoT | Medical Internet of Things |
| ANDSI/AAMI | American Nation Standards Institute/Association for the Advancement of Medical Instrumentation |
| SVB | Supraventricular Beat |
| VB | Ventricular Beat |
| Dataset | ECG Rec | Patients | Leads | SVB | VB |
|---|---|---|---|---|---|
| AHA | 155 | - | 2 | 317,612 | 32,403 |
| MIT-BIH-SVDB | 78 | - | 2 | 174,317 | 9953 |
| MIT-BIH-AR | 48 | 47 | 2 | 92,754 | 7803 |
| Leads | Acc (%) | Se (%) | Er |
|---|---|---|---|
| ECG 1 | 98 | 98.98 | 0.02 |
| ECG 2 | 97 | 98.97 | 0.03 |
| AHA | 96 | 97.95 | 0.04 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mastoi, Q.-u.-a.; Wah, T.Y.; Gopal Raj, R. Reservoir Computing Based Echo State Networks for Ventricular Heart Beat Classification. Appl. Sci. 2019, 9, 702. https://doi.org/10.3390/app9040702
Mastoi Q-u-a, Wah TY, Gopal Raj R. Reservoir Computing Based Echo State Networks for Ventricular Heart Beat Classification. Applied Sciences. 2019; 9(4):702. https://doi.org/10.3390/app9040702
Chicago/Turabian StyleMastoi, Qurat-ul-ain, Teh Ying Wah, and Ram Gopal Raj. 2019. "Reservoir Computing Based Echo State Networks for Ventricular Heart Beat Classification" Applied Sciences 9, no. 4: 702. https://doi.org/10.3390/app9040702
APA StyleMastoi, Q.-u.-a., Wah, T. Y., & Gopal Raj, R. (2019). Reservoir Computing Based Echo State Networks for Ventricular Heart Beat Classification. Applied Sciences, 9(4), 702. https://doi.org/10.3390/app9040702





