A Novel Approach to Data Extraction on Hyperlinked Webpages
Abstract
:1. Introduction
2. Related Work
2.1. Table and Header Detection
2.2. Schema Extraction and Matching
2.3. Canonical Form Conversion
3. Methodology
- (1)
- Dataset—used for research.
- (2)
- CRF—used for classification of web table rows, and returns a row label sequence.
- (3)
- Automaton—used to classify web tables as relational or non-relational.
3.1. Dataset
3.2. HTML Input File
3.3. Conditional Random Fields
3.4. Row Class Labels
3.5. Table Schema Extraction
- Varchar: column contains a string data type.
- Numeric/integer: contains a numeric type of data.
- Date: contains a date with ‘/’ or ‘-’ separator and data values are marked as the date.
3.6. Finding the Link Table and Schema
| Algorithm 1: Table Identification and Schema Extraction Process |
| Input: ← web page |
| tables[] ← identifying relational tables(web page) // using CRF and Automaton |
| while(tables[]){ |
| tables[]← real tables using CRF and automaton |
| if(table[] have “a=href”) |
| fetch_childtable and extract_schema() |
| assign PK and FK |
| else |
| remaining table[] are non-relational |
| extract_schema(string table) // Function Definition |
| String header ←identify_header(table); //using CRF |
| datatype_row ← finding max probability of data type in one column’s cell. |
| Output: header+datatype_row |
| String datatype(string table) |
| Cell ← Identifying cells |
| if(cell is numeric) |
| cell_datatype ”numeric” |
| else if(cell is date) |
| cell_datatype ”date” |
| else if(cell is varchar) |
| cell_datatype ”varchar”Function: find_schema(tables); |
4. Results
4.1. Identification of Data Tables
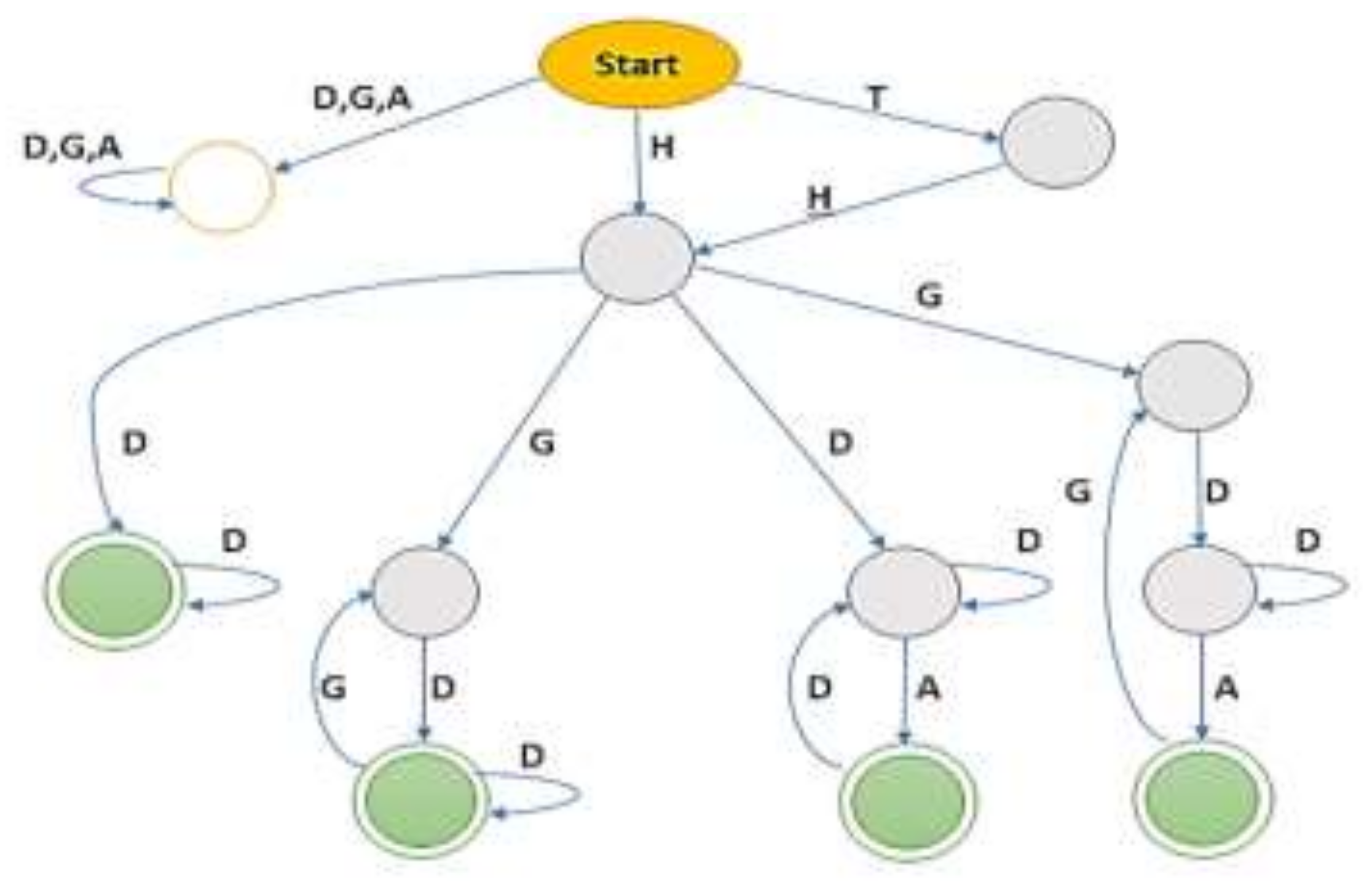
4.2. Automaton Construction
- If no path existed from one state of a specific row label to reach to another state, then it was non-deterministic, finite-state automaton (NFA), consequently.
- If all input labels were parsed on the automaton and could not reach a final state, then it could be either on an intermediate state or on the dead state.
4.3. Table Classification
- True positive (TP) = the number of cases correctly identified as real.
- False positive (FP) = the number of cases incorrectly identified as real.
- True negative (TN) = the number of cases correctly identified as non-real.
- False negative (FN) = the number of cases incorrectly identified as non-real
4.4. Linked Web Tables
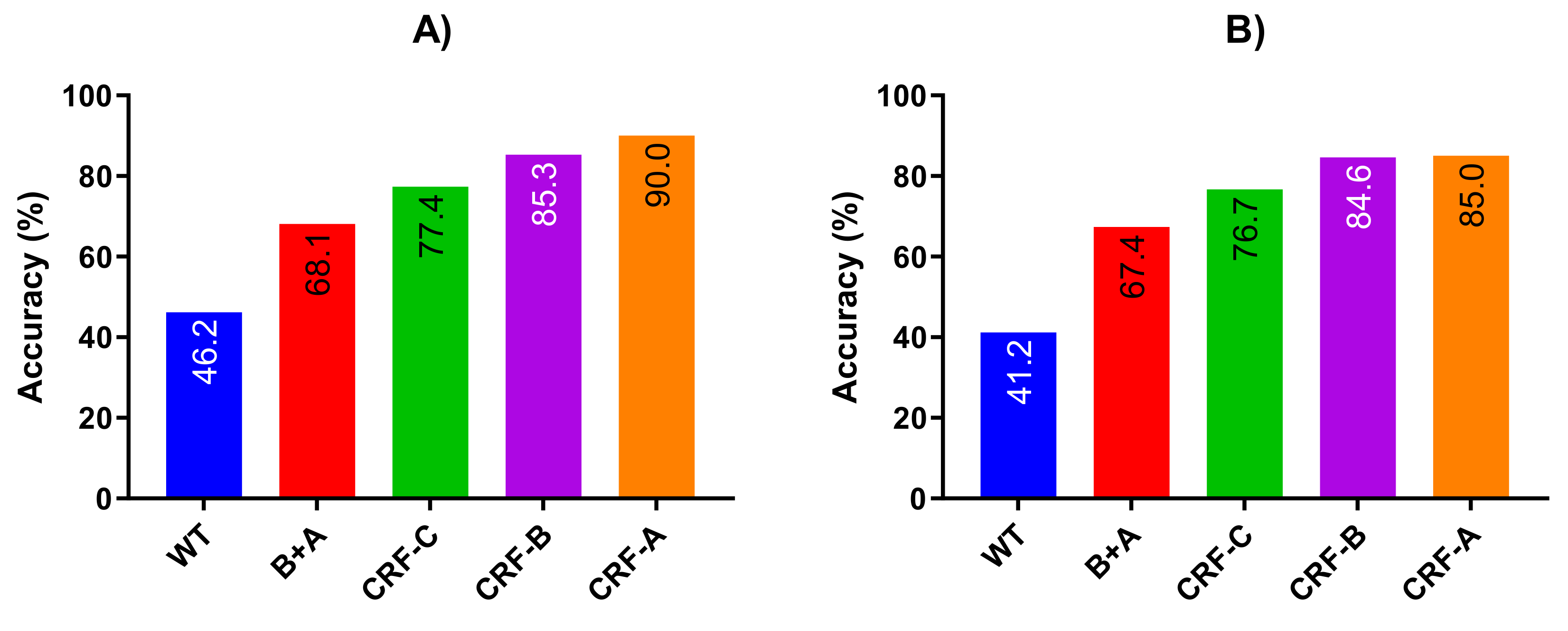
4.5. Comparison with Previous Studies
5. Conclusions and Future Work
Author Contributions
Funding
Conflicts of Interest
References
- Limaye, G.; Sarawagi, S.; Chakrabarti, S. Annotating and searching web tables using entities, types and relationships. Proc. VLDB Endow. 2010, 3, 1338–1347. [Google Scholar] [CrossRef]
- Wang, Y.; Hu, J. Detecting tables in html documents. In Proceedings of the International Workshop on Document Analysis Systems, Princeton, NJ, USA, 19–21 August 2002; pp. 249–260. [Google Scholar]
- Zanibbi, R.; Blostein, D.; Cordy, J.R. A survey of table recognition. Doc. Anal. Recognit. 2004, 7, 1–16. [Google Scholar] [CrossRef]
- Cafarella, M.J.; Halevy, A.; Wang, D.Z.; Wu, E.; Zhang, Y. Webtables: Exploring the power of tables on the web. Proc. VLDB Endow. 2008, 1, 538–549. [Google Scholar] [CrossRef]
- Cafarella, M.J.; Halevy, A.; Zhang, Y.; Wang, D.Z.; Wu, E. Uncovering the Relational Web. In Proceedings of the 11th International Workshop on Web and Databases (WebDB 2008), Vancouver, BC, Canada, 13 June 2008. [Google Scholar]
- Cafarella, M.; Halevy, A.; Lee, H.; Madhavan, J.; Yu, C.; Wang, D.Z.; Wu, E. Ten years of webtables. Proc. VLDB Endow. 2018, 11, 2140–2149. [Google Scholar] [CrossRef]
- Embley, D.W.; Krishnamoorthy, M.; Nagy, G.; Seth, S. Factoring web tables. In Proceedings of the International Conference on Industrial, Engineering and Other Applications of Applied Intelligent Systems, Syracuse, NY, USA, 28 June–1 July 2011; pp. 253–263. [Google Scholar]
- Chen, H.-H.; Tsai, S.-C.; Tsai, J.-H. Mining tables from large scale HTML texts. In Proceedings of the 18th Conference on Computational Linguistics, Saarbrücken, Germany, 31 July–4 August 2000; Volume 1, pp. 166–172. [Google Scholar]
- Chen, Z.; Cafarella, M. Automatic web spreadsheet data extraction. In Proceedings of the 3rd International Workshop on Semantic Search over the Web, Riva del Garda, Italy, 30 August 2013; p. 1. [Google Scholar]
- Fang, J.; Mitra, P.; Tang, Z.; Giles, C.L. Table header detection and classification. In Proceedings of the Twenty-Sixth AAAI Conference on Artificial Intelligence, Toronto, ON, Canada, 22–26 July 2012. [Google Scholar]
- Caragea, C.; Wu, J.; Ciobanu, A.; Williams, K.; Fernández-Ramírez, J.; Chen, H.H. Citeseer x: A scholarly big dataset. In Proceedings of the European Conference on Information Retrieval, Amsterdam, The Netherlands, 13–16 April 2014; pp. 311–322. [Google Scholar]
- Penn, G.; Hu, J.; Luo, H.; McDonald, R. Flexible web document analysis for delivery to narrow-bandwidth devices. In Proceedings of the Sixth International Conference on Document Analysis and Recognition, Seattle, WA, USA, 13 September 2001; pp. 1074–1078. [Google Scholar]
- Han, J.; Pei, J.; Kamber, M. Data Mining: Concepts and Techniques; Elsevier: Amsterdam, The Netherlands, 2011. [Google Scholar]
- Lautert, L.R.; Scheidt, M.M.; Dorneles, C.F. Web table taxonomy and formalization. ACM SIGMOD Rec. 2013, 42, 28–33. [Google Scholar] [CrossRef]
- Nagy, G. Learning the characteristics of critical cells from web tables. In Proceedings of the 21st International Conference on Pattern Recognition (ICPR2012), Tsukuba, Japan, 11–15 November 2012; pp. 1554–1557. [Google Scholar]
- Khushi, M.; Carpenter, J.E.; Balleine, R.L.; Clarke, C.L. Development of a data entry auditing protocol and quality assurance for a tissue bank database. Cell Tissue Bank. 2012, 13, 9–13. [Google Scholar] [CrossRef] [PubMed]
- Hurst, M. Layout and language: Challenges for table understanding on the web. Available online: http://wda2001.csc.liv.ac.uk//Papers/12_hurst_wda2001 (accessed on 31 October 2019).
- Nagy, G.; Padmanabhan, R.; Jandhyala, R.; Silversmith, W.; Krishnamoorthy, M. Table metadata: Headers, augmentations and aggregates. Available online: https://www.ecse.rpi.edu/~nagy/PDF_chrono/2010_Padmanabhan_Nagy_etal_DAS2010 (accessed on 31 October 2019).
- Yakout, M.; Ganjam, K.; Chakrabarti, K.; Chaudhuri, S. Infogather: Entity augmentation and attribute discovery by holistic matching with web tables. In Proceedings of the 2012 ACM SIGMOD International Conference on Management of Data, New York, NY, USA, 20–24 May 2012; pp. 97–108. [Google Scholar]
- Di Mauro, N.; Basile, T.M.; Ferilli, S.; Esposito, F. Optimizing probabilistic models for relational sequence learning. In Proceedings of the International Symposium on Methodologies for Intelligent Systems, Warsaw, Poland, 28–30 June 2011; pp. 240–249. [Google Scholar]
- Esposito, F.; di Mauro, N.; Basile, T.; Ferilli, S. Multi-dimensional relational sequence mining. Fundam. Inform. 2008, 89, 23–43. [Google Scholar]
- Koller, D.; Friedam, N; Džeroski, S; Sutton, C.; McCallum, A.; Pfeffer, A.; Neville, J. Introduction to Statistical Relational Learning; MIT Press: Cambridge, MA, USA, 2007. [Google Scholar]
- Shaukat, K.; Masood, N.; Mehreen, S. Population of Data in Extracted Web Table Schema; LAP Lambert Academic Publishing: Saarbrücken, Germany, 2017. [Google Scholar]
- Shaukat, K.; Masood, N.; Mehreen, S.; Haider, F.; Bakar, A.; Shaukat, U. Population of data in web-tables schema. In Proceedings of the 2016 19th International Multi-Topic Conference (INMIC), Islamabad, Pakistan, 5–6 December 2016; pp. 1–6. [Google Scholar]
- Adelfio, M.D.; Samet, H. Schema extraction for tabular data on the web. Proc. VLDB Endow. 2013, 6, 421–432. [Google Scholar] [CrossRef]
- Babu, S.; Motwani, R.; Munagala, K.; Nishizawa, I.; Widom, J. Adaptive ordering of pipelined stream filters. In Proceedings of the 2004 ACM SIGMOD international conference on Management of data, Paris, France, 13–18 June 2004; pp. 407–418. [Google Scholar]
- Lafferty, J.; McCallum, A.; Pereira, F.C. Conditional Random Fields: Probabilistic Models for Segmenting and Labeling Sequence Data. 2001. Available online: https://dl.acm.org/citation.cfm?id=655813 (accessed on 31 October 2019).
- Condon, A.; Deshpande, A.; Hellerstein, L.; Wu, N. Flow algorithms for two pipelined filter ordering problems. In Proceedings of the Twenty-Fifth ACM SIGMOD-SIGACT-SIGART Symposium on Principles of Database Systems, Chicago, IL, USA, 26–28 June 2006; pp. 193–202. [Google Scholar]
- Kodialam, M.S. The throughput of sequential testing. In Proceedings of the International Conference on Integer Programming and Combinatorial Optimization, Utrecht, The Netherlands, 13–15 June 2001; pp. 280–292. [Google Scholar]
- Srivastava, U.; Munagala, K.; Widom, J.; Motwani, R. Query optimization over web services. In Proceedings of the 32nd international conference on Very large data bases, Seoul, Korea, 12–15 September 2006; pp. 355–366. [Google Scholar]
- Sekhavat, Y.A.; di Paolo, F.; Barbosa, D.; Merialdo, P. Knowledge Base Augmentation using Tabular Data. In Proceedings of the LDOW, Seoul, Korea, 8 April 2014. [Google Scholar]
- DiFranzo, D.; Ding, L.; Graves, A.; Michaelis, J.R.; Li, X.; McGuinness, D.L.; Hendler, J. Data-gov wiki: Towards linking government data. In Proceedings of the 2010 AAAI Spring Symposium Series, Palo Alto, CA, USA, 22–24 March 2010. [Google Scholar]
- Venetis, P.; Halevy, A.; Madhavan, J.; Paşca, M.; Shen, W.; Wu, F.; Wu, C. Recovering semantics of tables on the web. Proc. VLDB Endow. 2011, 4, 528–553. [Google Scholar] [CrossRef]
- Embley, D.W.; Seth, S.; Nagy, G. Transforming web tables to a RELATIONAL database. In Proceedings of the 2014 22nd International Conference on Pattern Recognition, Stockholm, Sweden, 24–28 August 2014; pp. 2781–2786. [Google Scholar]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Khushi, M.; Carpenter, J.E.; Balleine, R.L.; Clarke, C.L. Electronic biorepository application system: Web-based software to manage receipt, peer review, and approval of researcher applications to a biobank. Biopreserv. Biobank. 2012, 10, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Hassan, M.U.; Shaukat, K.; Niu, D.; Mahreen, S.; Ma, Y.; Haider, F.; Zhao, X. An Overview of Schema Extraction and Matching Techniques. In Proceedings of the 2018 2nd IEEE Advanced Information Management, Communicates, Electronic and Automation Control Conference (IMCEC), Xi’an, China, 25–27 May 2018; pp. 1290–1294. [Google Scholar]
- Cafarella, M.J.; Halevy, A.; Khoussainova, N. Data integration for the relational web. Proc. VLDB Endow. 2009, 2, 1090–1101. [Google Scholar] [CrossRef]
- Khushi, M. Benchmarking Database Performance for Genomic Data. J. Cell. Biochem. 2018, 6, 877–883. [Google Scholar] [CrossRef] [PubMed]



| Form Name | A | B | Total Fields (X = A × B) | Fields with Errors (Y) | Error Rate* (Y/X) × 100 (%) | |
|---|---|---|---|---|---|---|
| X/Y | % age | |||||
| Breast pathology | 48 | 733 | 35,184 | 188 | 0.0053 | 0.53 |
| Specimen accession | 28 | 667 | 18,676 | 63 | 0.0034 | 0.34 |
| Patient consent | 11 | 379 | 4169 | 11 | 0.0026 | 0.26 |
| Demographics | 20 | 348 | 6960 | 9 | 0.0013 | 0.13 |
| Clinical follow-up | 15 | 1115 | 16,725 | 2 | 0.0001 | 0.01 |
| Total | 122 | 3242 | 81,714 | 273 | 0.0033 | 0.33 |
| Sr# | Paper Title | Published In | Table Detection | Worked On | Corpus Size | Schema Extraction | Linked >Tables |
|---|---|---|---|---|---|---|---|
| 1 | Mining tables from large scale HTML | Computational Linguistics, 2000 | Yes | HTML | - | No | No |
| 2 | Detecting tables in HTML documents | Springer, 2002 | Yes | HTML | 1393 | No | No |
| 3 | Uncovering the relational web | Web DB, 2008 | Yes | HTML | 14.1B | Yes | No |
| 4 | Factoring web tables | Springer, 2011 | No | Spread Sheets | 200 | No | No |
| 5 | Table header detection and classification | AAAI, 2012 | Yes | 200 | No | No | |
| 6 | Schema extraction from tabular data | VLDB, 2013 | Yes | HTML, Spread Sheets | 7883, 14669 | No | No |
| 7 | Web table taxonomy and formalization | SIGMOD, 2013 | Yes | HTML | 30000 | No | No |
| 8 | Ten years of webtables | Proc. VLDB. 2018 | Yes | HTML | - | Yes | No |
| Dataset Characteristics | |
|---|---|
| Number of Pages | 15,000 |
| Number of tables | 30,000 |
| Number of pages with tables | 12,900 |
| Number of rows | 210,000 |
| Number of Complex Tables | 18,000 |
| Number of Non-Real Tables | 9000 |
| Number of Simple Tables | 3000 |
| Number of Real Tables having Links | 6000 |
| Label | Functionality |
|---|---|
| T | Title of the table mostly table name describing the whole domain of the table. |
| H | Columns names as cell values define the domain of subsequent data rows beneath. |
| G | Cluster subsequent rows in a group. |
| D | Data Tuples/Rows define the header |
| A | Aggregate/total of above rows. |
| Sr# | Sequence | Status |
|---|---|---|
| 1 | THD+ | Accepted |
| 2 | HD+ | Accepted |
| 3 | H((D) + A)+ | Accepted |
| 4 | TH((D) + A)+ | Accepted |
| 5 | H(GD+)+ | Accepted |
| 6 | TH(GD+)+ | Accepted |
| 7 | DD * | Rejected |
| 8 | DAA * | Rejected |
| Actual Class\Predicted Class | Real Tables | Non-Real Tables | Total |
|---|---|---|---|
| Real Tables | 14,280 (68%) | 6720 (32%) | 21,000 |
| Non-Real Tables | 60 (0.7%) | 8940 (99.3%) | 9000 |
| Total | 14,340 | 15,660 | 30,000 |
| Actual Class\Predicted Class | Links Having Real Tables | Links Having Simple Text | Total |
|---|---|---|---|
| Links having Real Tables | 2235 | 1365 | 3600 |
| Links having Simple Text | 90 | 2310 | 2400 |
| Total | 2325 | 3675 | 6000 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shaukat, K.; Masood, N.; Khushi, M. A Novel Approach to Data Extraction on Hyperlinked Webpages. Appl. Sci. 2019, 9, 5102. https://doi.org/10.3390/app9235102
Shaukat K, Masood N, Khushi M. A Novel Approach to Data Extraction on Hyperlinked Webpages. Applied Sciences. 2019; 9(23):5102. https://doi.org/10.3390/app9235102
Chicago/Turabian StyleShaukat, Kamran, Nayyer Masood, and Matloob Khushi. 2019. "A Novel Approach to Data Extraction on Hyperlinked Webpages" Applied Sciences 9, no. 23: 5102. https://doi.org/10.3390/app9235102
APA StyleShaukat, K., Masood, N., & Khushi, M. (2019). A Novel Approach to Data Extraction on Hyperlinked Webpages. Applied Sciences, 9(23), 5102. https://doi.org/10.3390/app9235102







