Green Production and Biotechnological Applications of Cell Wall Lytic Enzymes
Abstract
1. Introduction
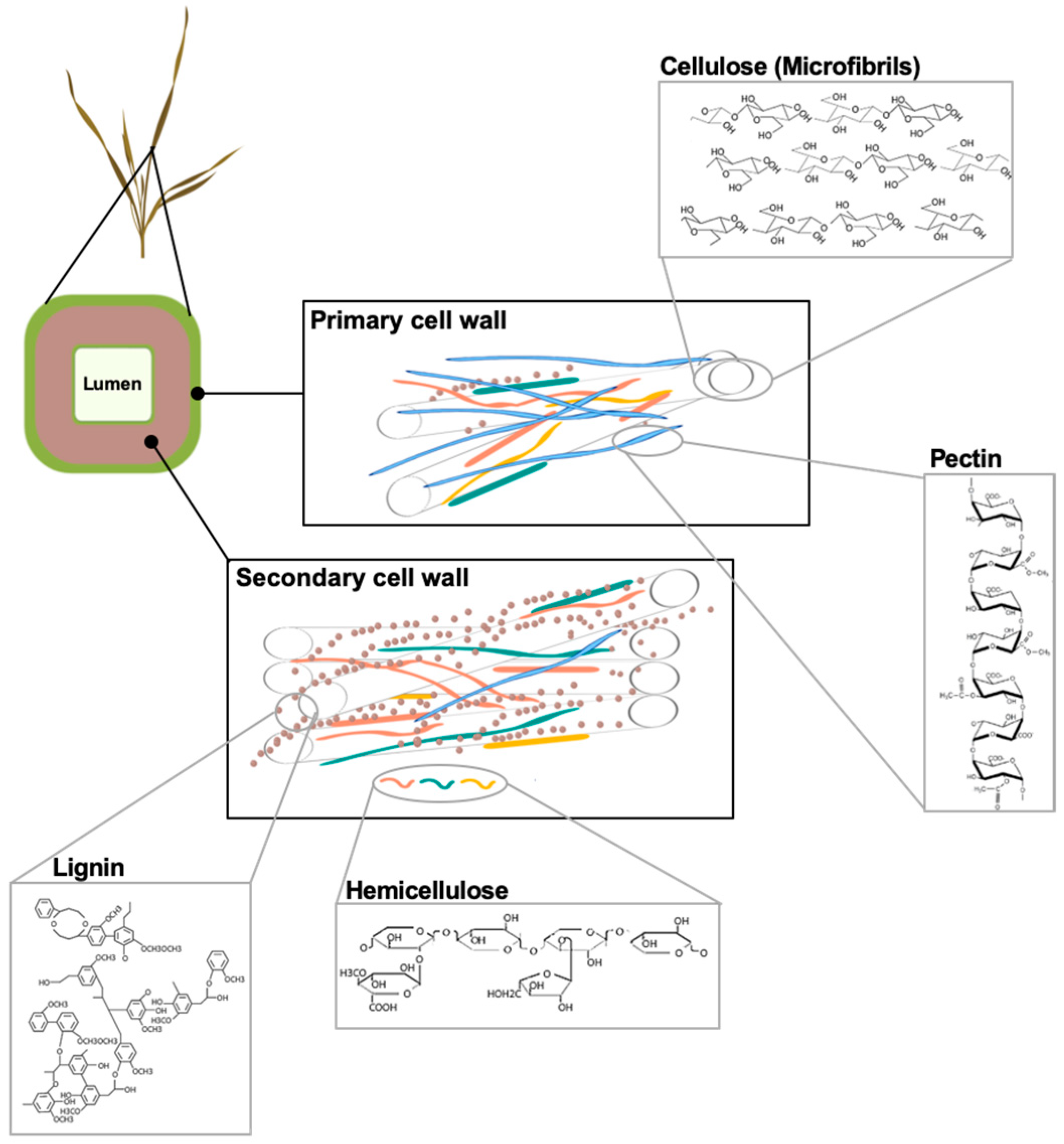
2. Lignocellulose: Structure and Functions
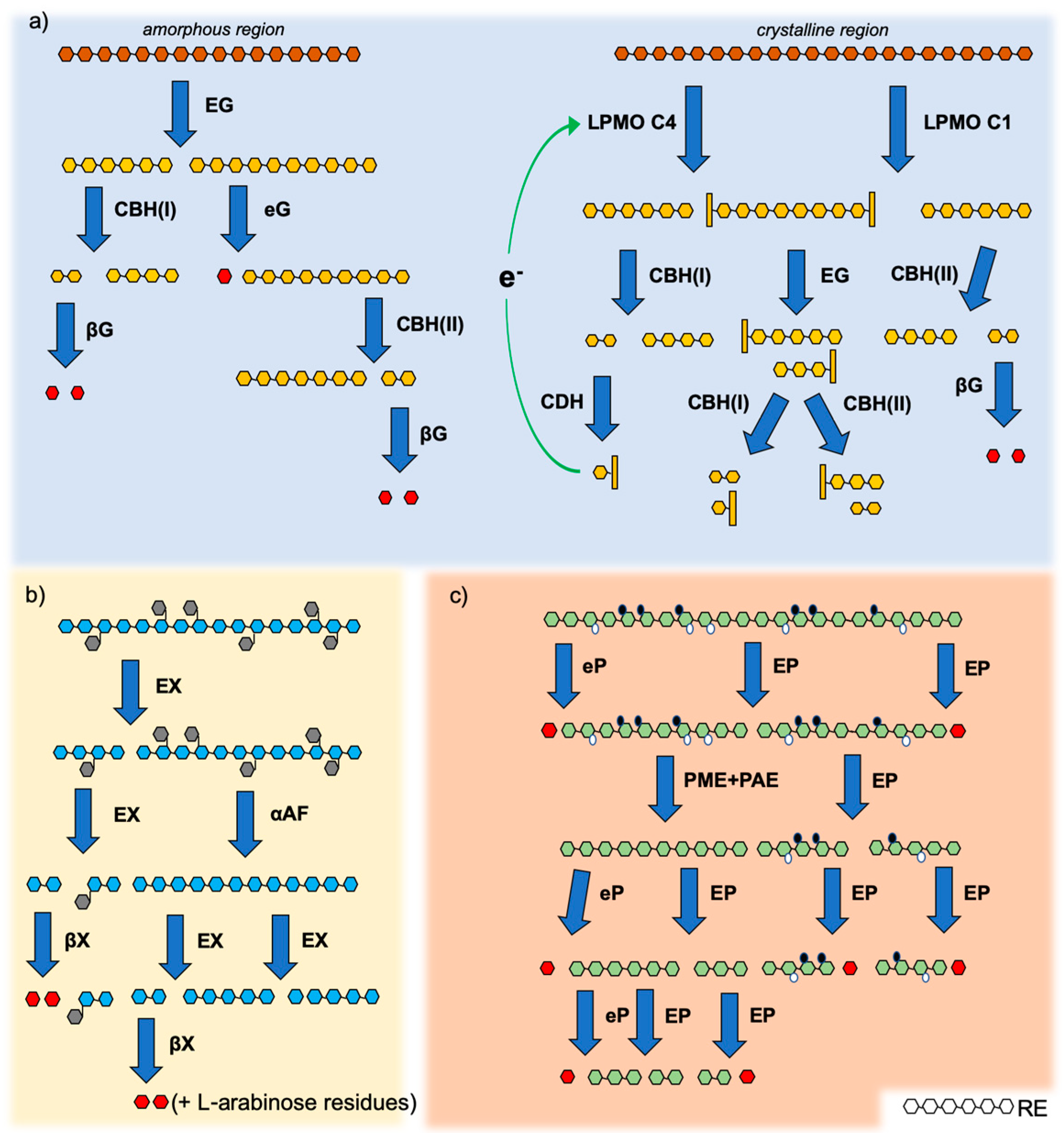
3. Degradation of Lignocellulose by CWLEs
3.1. Cellulases and Lytic Polysaccharide Mono-Oxygenases
3.2. Hemicellulases
- xylanolytic enzymes: endo-1,4-β-xylanase degrades xylan and arabinoxylan in oligomers with different degrees of polymerization and xylobiose that, in turn, is degraded by β-xylosidase in two d-xylose units;
- mannanolytic enzymes: endo-1,4-β-mannanase degrades mannan and galactomannan in oligomers with different degrees of polymerization and mannobiose that, in turn, is degraded by β-mannosidase in two d-mannose units;
- galactanolytic enzymes: endo-1,4-β-galactanase degrades galactan and type I arabinogalactan in oligomers with different degrees of polymerization and galactobiose that, in turn, is degraded by β-galactosidase in two d-galactose units;
- α-l-arabinofuranosidase hydrolyses terminal, nonreducing α-l-arabinofuranoside residues in α-l-arabinosides, which can be found in arabinoxylan and arabinogalactan;
- xyloglucanolytic enzymes: xyloglucanases are divided into two subgroups depending on their ability to cleave xyloglucan through an endo- or an exo-mode of action. Xyloglucan is mainly distributed in the primary cell walls of dicotyledonous plants [34].
3.3. Pectinases
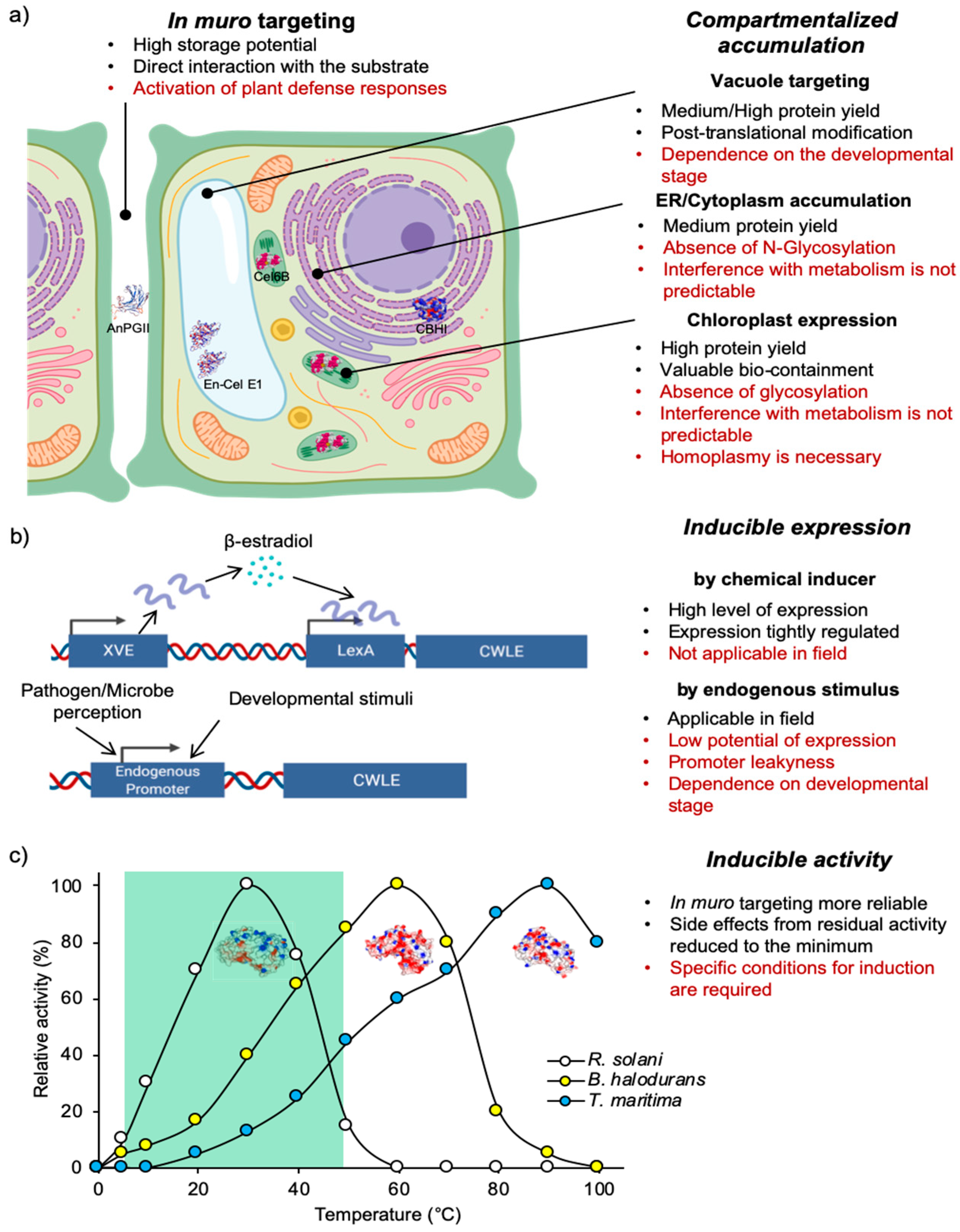
4. Production of CWLEs in Microbial and Plant Expression Systems
4.1. Heterologous Expression of CWLEs in Fungi and Bacteria
4.2. Heterologous Expression of CWLEs in Plants
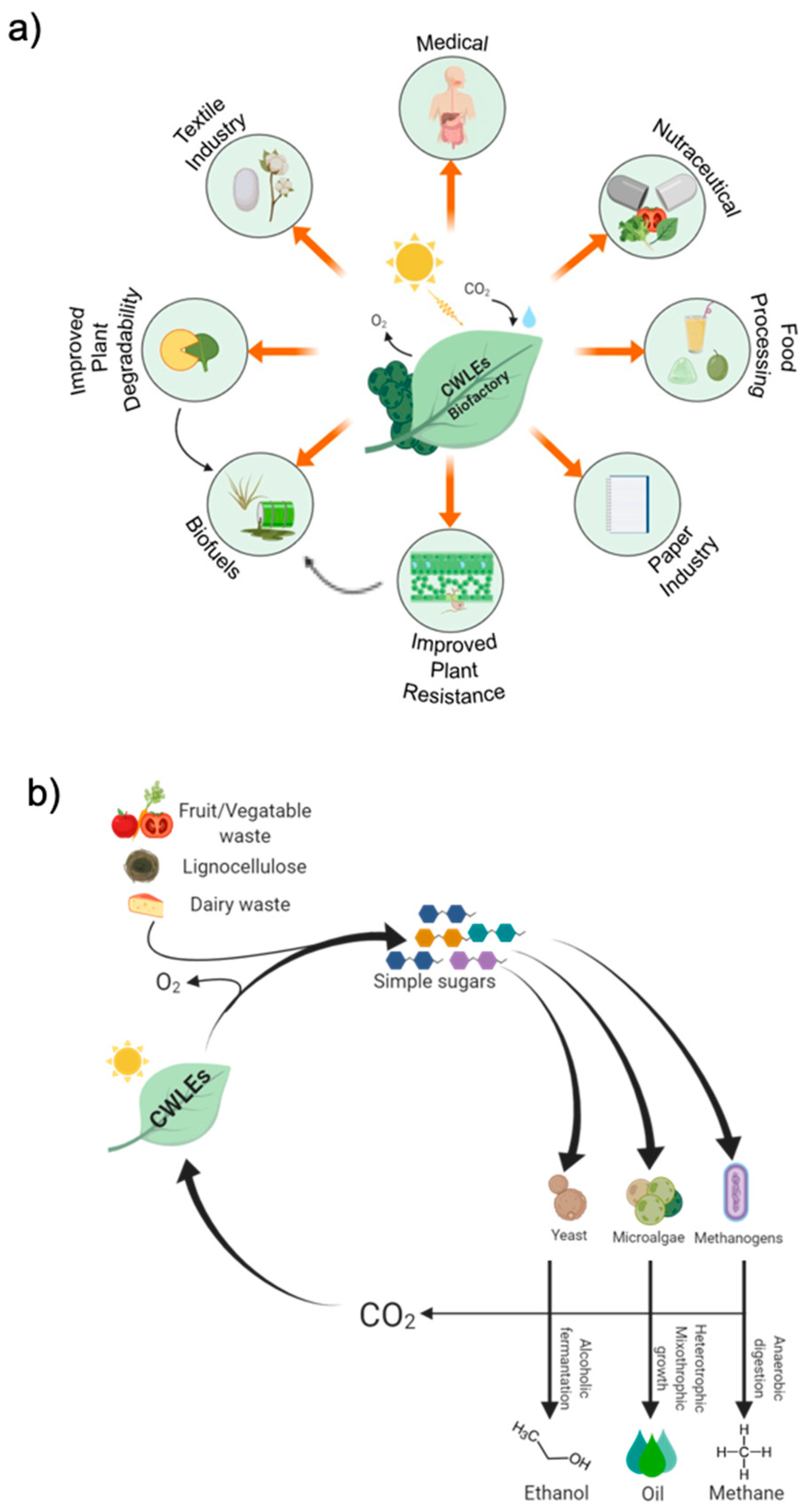
5. Bio-Applications of CWLEs
5.1. Use of CWLEs for the Production of Biofuels from Agricultural Feedstock
5.2. Use of Plant-Expressed CWLEs in Medical and Nutraceutical Fields and in Food Processing
5.3. Use of CWLEs to Improve Plant Resistance Against Pathogens
6. Conclusions
Structure Modelling and Figure Preparation
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Keegstra, K. Plant cell walls. Plant Physiol. 2010, 154, 483–486. [Google Scholar] [CrossRef] [PubMed]
- Hamann, T. Plant cell wall integrity maintenance as an essential component of biotic stress response mechanisms. Front. Plant Sci. 2012, 3, 77. [Google Scholar] [CrossRef] [PubMed]
- Bayer, E.A.; Bélaich, J.P.; Shoham, Y.; Lamed, R. The Cellulosomes: Multienzyme Machines for Degradation of Plant Cell Wall Polysaccharides. Annu. Rev. Microbiol. 2004, 58, 521–554. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.K.; Sharma, S. Recent updates on different methods of pretreatment of lignocellulosic feedstocks: A review. Bioresour. Bioprocess. 2017, 4, 7. [Google Scholar] [CrossRef] [PubMed]
- Jørgensen, H.; Pinelo, M. Enzyme recycling in lignocellulosic biorefineries. Biofuels Bioprod. Biorefin. 2017, 11, 150–167. [Google Scholar] [CrossRef]
- Bajpai, P. Pretreatment of Lignocellulosic Biomass for Biofuel Production; SpringerBriefs in Molecular Science; Springer: Singapore, 2016; ISBN 978-981-10-0686-9. [Google Scholar]
- Poutanen, K.; Sundberg, M.; Korte, H.; Puls, J. Deacetylation of xylans by acetyl esterases of Trichoderma reesei. Appl. Microbiol. Biotechnol. 1990, 33, 506–510. [Google Scholar] [CrossRef]
- Vanholme, R.; Demedts, B.; Morreel, K.; Ralph, J.; Boerjan, W. Lignin biosynthesis and structure. Plant Physiol. 2010, 153, 895–905. [Google Scholar] [CrossRef]
- Lionetti, V.; Francocci, F.; Ferrari, S.; Volpi, C.; Bellincampi, D.; Galletti, R.; D’Ovidio, R.; De Lorenzo, G.; Cervone, F. Engineering the cell wall by reducing de-methyl-esterified homogalacturonan improves saccharification of plant tissues for bioconversion. Proc. Natl. Acad. Sci. USA 2010, 107, 616–621. [Google Scholar] [CrossRef]
- Ferrari, S.; Savatin, D.V.; Sicilia, F.; Gramegna, G.; Cervone, F.; De Lorenzo, G. Oligogalacturonides: Plant damage-associated molecular patterns and regulators of growth and development. Front. Plant Sci. 2013, 4, 49. [Google Scholar] [CrossRef]
- Saini, J.K.; Saini, R.; Tewari, L. Lignocellulosic agriculture wastes as biomass feedstocks for second-generation bioethanol production: Concepts and recent developments. 3 Biotech 2015, 5, 337–353. [Google Scholar] [CrossRef]
- Agrawal, K.; Chaturvedi, V.; Verma, P. Fungal laccase discovered but yet undiscovered. Bioresour. Bioprocess. 2018, 5, 4. [Google Scholar] [CrossRef]
- Prajapati, H.V. Laccase—A Wonder Molecule: A Review of its Properties and Applications. Int. J. Pure Appl. Biosci. 2018, 6, 766–773. [Google Scholar] [CrossRef]
- Patel, N.; Shahane, S.; Majumdar, R.; Mishra, U.; Shivam; Shivam, S. Mode of Action, Properties, Production, and Application of Laccase: A Review. Recent Pat. Biotechnol. 2019, 13, 19–32. [Google Scholar] [CrossRef] [PubMed]
- Horn, S.J.; Vaaje-Kolstad, G.; Westereng, B.; Eijsink, V.G. Novel enzymes for the degradation of cellulose. Biotechnol. Biofuels 2012, 5, 45. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Bo, Y.; Han, L.; He, J.; Lanczycki, C.J.; Lu, S.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; et al. CDD/SPARCLE: Functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2017, 45, D200–D203. [Google Scholar] [CrossRef]
- Zverlov, V.; Piotukh, K.; Dakhova, O.; Velikodvorskaya, G.; Borriss, R. The multidomain xylanase A of the hyperthermophilic bacterium Thermotoga neapolitana is extremely thermoresistant. Appl. Microbiol. Biotechnol. 1996, 45, 245–247. [Google Scholar] [CrossRef]
- Park, J.I.; Kent, M.S.; Datta, S.; Holmes, B.M.; Huang, Z.; Simmons, B.A.; Sale, K.L.; Sapra, R. Enzymatic hydrolysis of cellulose by the cellobiohydrolase domain of CelB from the hyperthermophilic bacterium Caldicellulosiruptor saccharolyticus. Bioresour. Technol. 2011, 102, 5988–5994. [Google Scholar] [CrossRef]
- Dimarogona, M.; Topakas, E.; Christakopoulos, P. Cellulose degradation by oxidative enzymes. Comput. Struct. Biotechnol. J. 2012, 2, e201209015. [Google Scholar] [CrossRef]
- Singhania, R.R.; Patel, A.K.; Sukumaran, R.K.; Larroche, C.; Pandey, A. Role and significance of beta-glucosidases in the hydrolysis of cellulose for bioethanol production. Bioresour. Technol. 2013, 127, 500–507. [Google Scholar] [CrossRef]
- Zhao, Y. Mechanism of cellobiose inhibition in cellulose hydrolysis by cellobiohydrolase. Sci. China Ser. C Life Sci. 2004, 47, 18–24. [Google Scholar] [CrossRef]
- Sørensen, A.; Lübeck, M.; Lübeck, P.S.; Ahring, B.K. Fungal Beta-Glucosidases: A Bottleneck in Industrial Use of Lignocellulosic Materials. Biomolecules 2013, 3, 612–631. [Google Scholar] [CrossRef]
- Villares, A.; Moreau, C.; Bennati-Granier, C.; Garajova, S.; Foucat, L.; Falourd, X.; Saake, B.; Berrin, J.G.; Cathala, B. Lytic polysaccharide monooxygenases disrupt the cellulose fibers structure. Sci. Rep. 2017, 7, 40262. [Google Scholar] [CrossRef]
- Cannella, D.; Möllers, K.B.; Frigaard, N.U.; Jensen, P.E.; Bjerrum, M.J.; Johansen, K.S.; Felby, C. Light-driven oxidation of polysaccharides by photosynthetic pigments and a metalloenzyme. Nat. Commun. 2016, 7, 11134. [Google Scholar] [CrossRef]
- Rodrigues, K.B.; Macêdo, J.K.A.; Teixeira, T.; Barros, J.S.; Araújo, A.C.B.; Santos, F.P.; Quirino, B.F.; Brasil, B.S.A.F.; Salum, T.F.C.; Abdelnur, P.V.; et al. Recombinant expression of Thermobifida fusca E7 LPMO in Pichia pastorisand Escherichia coli and their functional characterization. Carbohydr. Res. 2017, 448, 175–181. [Google Scholar] [CrossRef]
- Laurent, C.V.F.P.; Breslmayr, E.; Tunega, D.; Ludwig, R.; Oostenbrink, C. Interaction between Cellobiose Dehydrogenase and Lytic Polysaccharide Monooxygenase. Biochemistry 2019, 58, 1226–1235. [Google Scholar] [CrossRef]
- Brenelli, L.; Squina, F.M.; Felby, C.; Cannella, D. Laccase-derived lignin compounds boost cellulose oxidative enzymes AA9. Biotechnol. Biofuels 2018, 11, 10. [Google Scholar] [CrossRef]
- Gao, D.; Haarmeyer, C.; Balan, V.; Whitehead, T.A.; Dale, B.E.; Chundawat, S.P. Lignin triggers irreversible cellulase loss during pretreated lignocellulosic biomass saccharification. Biotechnol. Biofuels 2014, 7, 175. [Google Scholar] [CrossRef]
- Benedetti, M.; Vecchi, V.; Betterle, N.; Natali, A.; Bassi, R.; Dall’Osto, L. Design of a highly thermostable hemicellulose-degrading blend from Thermotoga neapolitana for the treatment of lignocellulosic biomass. J. Biotechnol. 2019, 296, 42–52. [Google Scholar] [CrossRef]
- Jönsson, L.J.; Martin, C. Pretreatment of lignocellulose: Formation of inhibitory by-products and strategies for minimizing their effects. Bioresour. Technol. 2016, 199, 103–112. [Google Scholar] [CrossRef]
- Baumann, M.J.; Borch, K.; Westh, P. Xylan oligosaccharides and cellobiohydrolase I (TrCel7A) interaction and effect on activity. Biotechnol. Biofuels 2011, 4, 45. [Google Scholar] [CrossRef]
- Momeni, M.H.; Ubhayasekera, W.; Sandgren, M.; Ståhlberg, J.; Hansson, H. Structural insights into the inhibition of cellobiohydrolase Cel7A by xylo-oligosaccharides. FEBS J. 2015, 282, 2167–2177. [Google Scholar] [CrossRef] [PubMed]
- Garcia, N.H.; Benedetti, M.; Bolzonella, D. Effects of Enzymes Addition on Biogas Production From Anaerobic Digestion of Agricultural Biomasses. Waste Biomass Valorization 2019, 10, 3711–3722. [Google Scholar] [CrossRef]
- Scheller, H.V.; Ulvskov, P. Hemicelluloses. Annu. Rev. Plant Biol. 2010, 61, 263–289. [Google Scholar] [CrossRef] [PubMed]
- Ridley, B.L.; O’Neill, M.A.; Mohnen, D. Pectins: Structure, biosynthesis, and oligogalacturonide-related signaling. Phytochemistry 2001, 57, 929–967. [Google Scholar] [CrossRef]
- Jayani, R.S.; Saxena, S.; Gupta, R. Microbial pectinolytic enzymes: A review. Process Biochem. 2005, 40, 2931–2944. [Google Scholar] [CrossRef]
- Rha, E.; Park, H.J.; Lee, C.W.; Kim, J.W.; Kim, M.O.; Chung, Y.R. Expression of exo-polygalacturonases in Botrytis cinerea. FEMS Microbiol. Lett. 2001, 201, 105–109. [Google Scholar] [CrossRef][Green Version]
- Kalunke, R.M.; Tundo, S.; Benedetti, M.; Cervone, F.; De Lorenzo, G.; D’Ovidio, R. An update on polygalacturonase-inhibiting protein (PGIP), a leucine-rich repeat protein that protects crop plants against pathogens. Front. Plant Sci. 2015, 6, 146. [Google Scholar] [CrossRef]
- Benedetti, M.; Andreani, F.; Leggio, C.; Galantini, L.; Di Matteo, A.; Pavel, N.V.; De Lorenzo, G.; Cervone, F.; Federici, L.; Sicilia, F. A Single Amino-Acid Substitution Allows Endo-Polygalacturonase of Fusarium verticillioides to Acquire Recognition by PGIP2 from Phaseolus vulgaris. PLoS ONE 2013, 8, e80610. [Google Scholar] [CrossRef]
- Kotchoni, O.S.; Shonukan, O.O.; Gachomo, W.E. Bacillus pumilus BpCRI 6, a promising candidate for cellulase production under conditions of catabolite repression. Afr. J. Biotechnol. 2003, 2, 140–146. [Google Scholar] [CrossRef][Green Version]
- Garcia-Martinez, D.V.; Shinmyo, A.; Madia, A.; Demain, A.L. Studies on cellulase production by Clostridium thermocellum. Appl. Microbiol. Biotechnol. 1980, 9, 189–197. [Google Scholar] [CrossRef]
- Li, X.; Wang, H.; Zhou, C.; Ma, Y.; Li, J.; Song, J. Cloning, expression and characterization of a pectate lyase from Paenibacillus sp. 0602 in recombinant Escherichia coli. BMC Biotechnol. 2014, 14, 18. [Google Scholar] [CrossRef] [PubMed]
- Fang, S.; Li, J.; Liu, L.; Du, G.; Chen, J. Overproduction of alkaline polygalacturonate lyase in recombinant Escherichia coli by a two-stage glycerol feeding approach. Bioresour. Technol. 2011, 102, 10671–10678. [Google Scholar] [CrossRef] [PubMed]
- Arai, T.; Matsuoka, S.; Cho, H.Y.; Yukawa, H.; Inui, M.; Wong, S.L.; Doi, R.H. Synthesis of Clostridium cellulovorans minicellulosomes by intercellular complementation. Proc. Natl. Acad. Sci. USA 2007, 104, 1456–1460. [Google Scholar] [CrossRef] [PubMed]
- Linger, J.G.; Adney, W.S.; Darzins, A. Heterologous Expression and Extracellular Secretion of Cellulolytic Enzymes by Zymomonas mobilis. Appl. Environ. Microbiol. 2010, 76, 6360–6369. [Google Scholar] [CrossRef]
- Mingardon, F.; Chanal, A.; Tardif, C.; Fierobe, H.P. The Issue of Secretion in Heterologous Expression of Clostridium cellulolyticum Cellulase-Encoding Genes in Clostridium acetobutylicum ATCC 824. Appl. Environ. Microbiol. 2011, 77, 2831–2838. [Google Scholar] [CrossRef]
- Wieczorek, A.S.; Martin, V.J. Engineering the cell surface display of cohesins for assembly of cellulosome-inspired enzyme complexes on Lactococcus lactis. Microb. Cell Factories 2010, 9, 69. [Google Scholar] [CrossRef]
- Mingardon, F.; Perret, S.; Bélaïch, A.; Tardif, C.; Bélaïch, J.P.; Fierobe, H.P. Heterologous Production, Assembly, and Secretion of a Minicellulosome by Clostridium acetobutylicum ATCC. Appl. Environ. Microbiol. 2005, 71, 1215–1222. [Google Scholar] [CrossRef]
- Lee, S.J.; Pan, J.G.; Park, S.H.; Choi, S.K. Development of a stationary phase-specific autoinducible expression system in Bacillus subtilis. J. Biotechnol. 2010, 149, 16–20. [Google Scholar] [CrossRef]
- Angov, E.; Legler, P.M.; Mease, R.M. Adjustment of codon usage frequencies by codon harmonization improves protein expression and folding. In Heterologous Gene Expression in E. coli; Evans, T.C., Xu, M.Q., Eds.; Humana Press: Totowa, NJ, USA, 2011; Volume 705, pp. 1–13. ISBN 978-1-61737-966-6. [Google Scholar]
- Desvaux, M.; Hebraud, M.; Talon, R.; Henderson, I.R. Secretion and subcellular localizations of bacterial proteins: A semantic awareness issue. Trends Microbiol. 2009, 17, 139–145. [Google Scholar] [CrossRef]
- Jung, S.K.; Parisutham, V.; Jeong, S.H.; Lee, S.K. Heterologous Expression of Plant Cell Wall Degrading Enzymes for Effective Production of Cellulosic Biofuels. J. Biomed. Biotechnol. 2012, 2012, 405842. [Google Scholar] [CrossRef]
- Murashima, K.; Kosugi, A.; Doi, R.H. Solubilization of cellulosomal cellulases by fusion with cellulose-binding domain of noncellulosomal cellulase engd from Clostridium cellulovorans. Proteins Struct. Funct. Bioinform. 2003, 50, 620–628. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.Y.; Yukawa, H.; Inui, M.; Doi, R.H.; Wong, S.L. Production of Minicellulosomes from Clostridium cellulovorans in Bacillus subtilis WB800. Appl. Environ. Microbiol. 2004, 70, 5704–5707. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Le Loir, Y.; Azevedo, V.; Oliveira, S.C.; Freitas, D.A.; Miyoshi, A.; Bermúdez-Humarán, L.G.; Nouaille, S.; Ribeiro, L.A.; Leclercq, S.; Gabriel, J.E.; et al. Protein secretion in Lactococcus lactis: An efficient way to increase the overall heterologous protein production. Microb. Cell Factories 2005, 4, 2. [Google Scholar] [CrossRef] [PubMed]
- Koukiekolo, R.; Cho, H.Y.; Kosugi, A.; Inui, M.; Yukawa, H.; Doi, R.H. Degradation of Corn Fiber by Clostridium cellulovorans Cellulases and Hemicellulases and Contribution of Scaffolding Protein CbpA. Appl. Environ. Microbiol. 2005, 71, 3504–3511. [Google Scholar] [CrossRef]
- Song, J.M.; An, Y.J.; Kang, M.H.; Lee, Y.H.; Cha, S.S. Cultivation at 6–10 °C is an effective strategy to overcome the insolubility of recombinant proteins in Escherichia coli. Protein Expr. Purif. 2012, 82, 297–301. [Google Scholar] [CrossRef]
- Morello, E.; Bermúdez-Humarán, L.G.; Llull, D.; Solé, V.; Miraglio, N.; Langella, P.; Poquet, I. Lactobacillus lactis, an efficient cell factory for recombinant protein production and secretion. J. Mol. Microbiol. Biotechnol. 2008, 14, 48–58. [Google Scholar] [CrossRef]
- Pohl, S.; Harwood, C.R. Heterologous Protein Secretion by Bacillus Species. In Advances in Clinical Chemistry; Elsevier: Amsterdam, The Netherlands, 2010; Volume 73, pp. 1–25. [Google Scholar]
- Zhou, H.; Li, X.; Guo, M.; Xu, Q.; Cao, Y.; Qiao, D.; Cao, Y.; Xu, H. Secretory Expression and Characterization of an Acidic Endo-Polygalacturonase from Aspergillus niger SC323 in Saccharomyces cerevisiae. J. Microbiol. Biotechnol. 2015, 25, 999–1006. [Google Scholar] [CrossRef]
- Haan, R.D.; McBride, J.E.; La Grange, D.C.; Lynd, L.R.; Van Zyl, W.H. Functional expression of cellobiohydrolases in Saccharomyces cerevisiae towards one-step conversion of cellulose to ethanol. Enzym. Microb. Technol. 2007, 40, 1291–1299. [Google Scholar] [CrossRef]
- Haan, R.D.; Kroukamp, H.; Van Zyl, J.H.D.; Van Zyl, W.H. Cellobiohydrolase secretion by yeast: Current state and prospects for improvement. Process Biochem. 2013, 48, 1–12. [Google Scholar] [CrossRef]
- Van Zyl, W.H.; Rose, S.H.; Trollope, K.; Görgens, J.F. Fungal β-mannanases: Mannan hydrolysis, heterologous production and biotechnological applications. Process Biochem. 2010, 45, 1203–1213. [Google Scholar] [CrossRef]
- Margolles-Clark, E.; Tenkanen, M.; Nakari-Setälä, T.; Penttilä, M. Cloning of genes encoding alpha-L-arabinofuranosidase and beta-xylosidase from Trichoderma reesei by expression in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 1996, 62, 3840–3846. [Google Scholar] [PubMed]
- Cereghino, J.L.; Cregg, J.M. Heterologous protein expression in the methylotrophic yeastPichia pastoris. FEMS Microbiol. Rev. 2000, 24, 45–66. [Google Scholar] [CrossRef] [PubMed]
- Cregg, J.M.; Vedvick, T.S.; Raschke, W.C. Recent Advances in the Expression of Foreign Genes in Pichia pastoris. Nat. Biotechnol. 1993, 11, 905–910. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Cao, Y.; Ding, Y.; Lu, W.; Li, D. Cloning, functional expression and characterization of Aspergillus sulphureus β-mannanase in Pichia pastoris. J. Biotechnol. 2007, 128, 452–461. [Google Scholar] [CrossRef]
- Akcapinar, G.B.; Gul, O.; Sezerman, U. Effect of codon optimization on the expression of Trichoderma reesei endoglucanase 1 in Pichia pastoris. Biotechnol. Prog. 2011, 27, 1257–1263. [Google Scholar] [CrossRef]
- Bey, M.; Berrin, J.G.; Poidevin, L.; Sigoillot, J.C. Heterologous expression of Pycnoporus cinnabarinus cellobiose dehydrogenase in Pichia pastoris and involvement in saccharification processes. Microb. Cell Factories 2011, 10, 113. [Google Scholar] [CrossRef]
- Chen, P.; Fu, X.; Ng, T.B.; Ye, X.Y. Expression of a secretory β-glucosidase from Trichoderma reesei in Pichia pastoris and its characterization. Biotechnol. Lett. 2011, 33, 2475–2479. [Google Scholar] [CrossRef]
- Akbarzadeh, A.; Ranaei Siadat, S.O.; Motallebi, M.; Zamani, M.R.; Barshan Tashnizi, M.; Moshtaghi, S. Characterization and high level expression of acidic endoglucanase in Pichia pastoris. Appl. Biochem. Biotechnol. 2014, 172, 2253–2265. [Google Scholar] [CrossRef]
- Carrasco, M.; Rozas, J.M.; Alcaíno, J.; Cifuentes, V.; Baeza, M. Pectinase secreted by psychrotolerant fungi: Identification, molecular characterization and heterologous expression of a cold-active polygalacturonase from Tetracladium sp. Microb. Cell Factories 2019, 18, 45. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, Y.; Zhang, D.; Li, J.; Hua, Z.; Du, G.; Chen, J. Enhancement of cell viability and alkaline polygalacturonate lyase production by sorbitol co-feeding with methanol in Pichia pastoris fermentation. Bioresour. Technol. 2010, 101, 1318–1323. [Google Scholar] [CrossRef]
- Wang, H.; Li, J.; Liu, L.; Li, X.; Jia, D.; Du, G.; Chen, J.; Song, J. Increased production of alkaline polygalacturonate lyase in the recombinant Pichia pastoris by controlling cell concentration during continuous culture. Bioresour. Technol. 2012, 124, 338–346. [Google Scholar] [CrossRef] [PubMed]
- Blazeck, J.; Reed, B.; Garg, R.; Gerstner, R.; Pan, A.; Agarwala, V.; Alper, H.S. Generalizing a hybrid synthetic promoter approach in Yarrowia lipolytica. Appl. Microbiol. Biotechnol. 2013, 97, 3037–3052. [Google Scholar] [CrossRef] [PubMed]
- Yamada, R.; Taniguchi, N.; Tanaka, T.; Ogino, C.; Fukuda, H.; Kondo, A. Direct ethanol production from cellulosic materials using a diploid strain of Saccharomyces cerevisiae with optimized cellulase expression. Biotechnol. Biofuels 2011, 4, 8. [Google Scholar] [CrossRef] [PubMed]
- Mellitzer, A.; Weis, R.; Glieder, A.; Flicker, K. Expression of lignocellulolytic enzymes in Pichia pastoris. Microb. Cell Factories 2012, 11, 61. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Jain, K.K.; Bhardwaj, K.N.; Chakraborty, S.; Kuhad, R.C. Multiple genes in a single host: Cost-effective production of bacterial laccase (cotA), pectate lyase (pel), and endoxylanase (xyl) by simultaneous expression and cloning in single vector in E. coli. PLoS ONE 2015, 10, e0144379. [Google Scholar] [CrossRef] [PubMed]
- Mazzoli, R.; Lamberti, C.; Pessione, E. Engineering new metabolic capabilities in bacteria: Lessons from recombinant cellulolytic strategies. Trends Biotechnol. 2012, 30, 111–119. [Google Scholar] [CrossRef] [PubMed]
- Tsai, S.L.; Oh, J.; Singh, S.; Chen, R.; Chen, W. Functional Assembly of Minicellulosomes on the Saccharomyces cerevisiae Cell Surface for Cellulose Hydrolysis and Ethanol Production. Appl. Environ. Microbiol. 2009, 75, 6087–6093. [Google Scholar] [CrossRef]
- Kricka, W.; Fitzpatrick, J.; Bond, U. Metabolic engineering of yeasts by heterologous enzyme production for degradation of cellulose and hemicellulose from biomass: A perspective. Front. Microbiol. 2014, 5, 174. [Google Scholar] [CrossRef]
- Li, Q.; Song, J.; Peng, S.; Wang, J.P.; Qu, G.Z.; Sederoff, R.R.; Chiang, V.L. Plant biotechnology for lignocellulosic biofuel production. Plant Biotechnol. J. 2014, 12, 1174–1192. [Google Scholar] [CrossRef]
- Kubicek, C.P.; Starr, T.L.; Glass, N.L. Plant Cell Wall–Degrading Enzymes and Their Secretion in Plant-Pathogenic Fungi. Annu. Rev. Phytopathol. 2014, 52, 427–451. [Google Scholar] [CrossRef]
- Lagaert, S.; Beliën, T.; Volckaert, G. Plant cell walls: Protecting the barrier from degradation by microbial enzymes. Semin. Cell Dev. Biol. 2009, 20, 1064–1073. [Google Scholar] [CrossRef] [PubMed]
- Jolie, R.P.; Duvetter, T.; Van Loey, A.M.; Hendrickx, M.E. Pectin methylesterase and its proteinaceous inhibitor: A review. Carbohydr. Res. 2010, 345, 2583–2595. [Google Scholar] [CrossRef] [PubMed]
- Benedetti, M.; Leggio, C.; Federici, L.; De Lorenzo, G.; Pavel, N.V.; Cervone, F. Structural Resolution of the Complex between a Fungal Polygalacturonase and a Plant Polygalacturonase-Inhibiting Protein by Small-Angle X-Ray Scattering. Plant Physiol. 2011, 157, 599–607. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Benedetti, M.; Verrascina, I.; Pontiggia, D.; Locci, F.; Mattei, B.; De Lorenzo, G.; Cervone, F. Four Arabidopsis berberine bridge enzyme-like proteins are specific oxidases that inactivate the elicitor-active oligogalacturonides. Plant J. 2018, 94, 260–273. [Google Scholar] [CrossRef] [PubMed]
- Locci, F.; Benedetti, M.; Pontiggia, D.; Citterico, M.; Caprari, C.; Mattei, B.; Cervone, F.; De Lorenzo, G. An Arabidopsis berberine bridge enzyme-like protein specifically oxidizes cellulose oligomers and plays a role in immunity. Plant J. 2019, 98, 540–554. [Google Scholar] [CrossRef]
- Boudart, G.; Charpentier, M.; Lafitte, C.; Martinez, Y.; Jauneau, A.; Gaulin, E.; Esquerré-Tugayé, M.T.; Dumas, B. Elicitor activity of a fungal endopolygalacturonase in tobacco requires a functional catalytic site and cell wall localization. Plant Physiol. 2003, 131, 93–101. [Google Scholar] [CrossRef][Green Version]
- Benedetti, M.; Pontiggia, D.; Raggi, S.; Cheng, Z.; Scaloni, F.; Ferrari, S.; Ausubel, F.M.; Cervone, F.; De Lorenzo, G. Plant immunity triggered by engineered in vivo release of oligogalacturonides, damage-associated molecular patterns. Proc. Natl. Acad. Sci. USA 2015, 112, 5533–5538. [Google Scholar] [CrossRef]
- Souza, C.D.A.; Li, S.; Lin, A.Z.; Boutrot, F.; Grossmann, G.; Zipfel, C.; Somerville, S.C. Cellulose-Derived Oligomers Act as Damage-Associated Molecular Patterns and Trigger Defense-Like Responses. Plant Physiol. 2017, 173, 2383–2398. [Google Scholar] [CrossRef]
- Poinssot, B.; Vandelle, E.; Bentejac, M.; Adrian, M.; Levis, C.; Brygoo, Y.; Garin, J.; Sicilia, F.; Coutos-Thévenot, P.; Pugin, A. The Endopolygalacturonase 1 from Botrytis cinerea Activates Grapevine Defense Reactions Unrelated to Its Enzymatic Activity. Mol. Plant Microbe Interact. 2003, 16, 553–564. [Google Scholar] [CrossRef]
- Ma, Y.; Han, C.; Chen, J.; Li, H.; He, K.; Liu, A.; Li, D. Fungal cellulase is an elicitor but its enzymatic activity is not required for its elicitor activity: Fungal cellulase as an elicitor. Mol. Plant Pathol. 2015, 16, 14–26. [Google Scholar] [CrossRef]
- Choi, H.W.; Klessig, D.F. DAMPs, MAMPs, and NAMPs in plant innate immunity. BMC Plant Biol. 2016, 16, 232. [Google Scholar] [CrossRef] [PubMed]
- Zipfel, C. Plant pattern-recognition receptors. Trends Immunol. 2014, 35, 345–351. [Google Scholar] [CrossRef] [PubMed]
- Bézier, A.; Aziz, A.; Joubert, J.M.; Heyraud, A.; Baillieul, F.; Gauthier, A.; Poinssot, B.; Pugin, A. Elicitor and resistance-inducing activities of-1,4 cellodextrins in grapevine, comparison with β-1, 3 glucans and α-1, 4 oligogalacturonides. J. Exp. Bot. 2007, 58, 1463–1472. [Google Scholar]
- Claverie, J.; Balacey, S.; Lemaître-Guillier, C.; Brulé, D.; Chiltz, A.; Granet, L.; Noirot, E.; Daire, X.; Darblade, B.; Héloir, M.C.; et al. The Cell Wall-Derived Xyloglucan Is a New DAMP Triggering Plant Immunity in Vitis vinifera and Arabidopsis thaliana. Front. Plant Sci. 2018, 9, 1725. [Google Scholar] [CrossRef]
- Capodicasa, C.; Vairo, D.; Zabotina, O.; McCartney, L.; Caprari, C.; Mattei, B.; Manfredini, C.; Aracri, B.; Benen, J.; Knox, J.P.; et al. Targeted Modification of Homogalacturonan by Transgenic Expression of a Fungal Polygalacturonase Alters Plant Growth. Plant Physiol. 2004, 135, 1294–1304. [Google Scholar] [CrossRef]
- Klose, H.; Günl, M.; Usadel, B.; Fischer, R.; Commandeur, U. Cell wall modification in tobacco by differential targeting of recombinant endoglucanase from Trichoderma reesei. BMC Plant Biol. 2015, 15, 54. [Google Scholar] [CrossRef]
- Petersén, K.; Bock, R. High-level expression of a suite of thermostable cell wall-degrading enzymes from the chloroplast genome. Plant Mol. Biol. 2011, 76, 311–321. [Google Scholar] [CrossRef]
- Castiglia, D.; Sannino, L.; Marcolongo, L.; Ionata, E.; Tamburino, R.; De Stradis, A.; Cobucci-Ponzano, B.; Moracci, M.; La Cara, F.; Scotti, N. High-level expression of thermostable cellulolytic enzymes in tobacco transplastomic plants and their use in hydrolysis of an industrially pretreated Arundo donax L. biomass. Biotechnol. Biofuels 2016, 9, 154. [Google Scholar] [CrossRef]
- Dai, Z.; Hooker, B.S.; Anderson, D.B.; Thomas, S.R. Improved plant-based production of E1 endoglucanase using potato: Expression optimization and tissue targeting. Mol. Breed. 2000, 6, 277–285. [Google Scholar] [CrossRef]
- Harrison, M.D.; Geijskes, J.; Coleman, H.D.; Shand, K.; Kinkema, M.; Palupe, A.; Hassall, R.; Sainz, M.; Lloyd, R.; Miles, S.; et al. Accumulation of recombinant cellobiohydrolase and endoglucanase in the leaves of mature transgenic sugar cane: Cellulolytic enzyme production in sugar cane leaf. Plant Biotechnol. J. 2011, 9, 884–896. [Google Scholar] [CrossRef]
- Dai, Z.; Hooker, B.S.; Quesenberry, R.D.; Gao, J. Expression of Trichoderma reeseiexo-cellobiohydrolase I in transgenic tobacco leaves and calli. Appl. Biochem. Biotechnol. 1999, 79, 689–700. [Google Scholar] [CrossRef]
- Ziegelhoffer, T.; Will, J.; Austin-Phillips, S. Expression of bacterial cellulase genes in transgenic alfalfa (Medicago sativa L.), potato (Solanum tuberosum L.) and tobacco (Nicotiana tabacum L.). Mol. Breed. 1999, 5, 309–318. [Google Scholar] [CrossRef]
- Greene, E.R.; Himmel, M.E.; Beckham, G.T.; Tan, Z. Glycosylation of cellulases. In Advances in Carbohydrate Chemistry and Biochemistry; Elsevier: Amsterdam, The Netherlands, 2015; Volume 72, pp. 63–112. ISBN 978-0-12-802141-5. [Google Scholar]
- Daniell, H.; Muthukumar, B.; Lee, S.B. Marker free transgenic plants: Engineering the chloroplast genome without the use of antibiotic selection. Curr. Genet. 2001, 39, 109–116. [Google Scholar] [CrossRef] [PubMed]
- Neuhaus, J.M.; Sticher, L.; Meins, F.; Boller, T. A short C-terminal sequence is necessary and sufficient for the targeting of chitinases to the plant vacuole. Proc. Natl. Acad. Sci. USA 1991, 88, 10362–10366. [Google Scholar] [CrossRef]
- Pereira, C.; Pereira, S.; Pissarra, J. Delivering of Proteins to the Plant Vacuole—An Update. Int. J. Mol. Sci. 2014, 15, 7611–7623. [Google Scholar] [CrossRef]
- Marin Viegas, V.S.; Ocampo, C.G.; Petruccelli, S. Vacuolar deposition of recombinant proteins in plant vegetative organs as a strategy to increase yields. Bioengineered 2017, 8, 203–211. [Google Scholar] [CrossRef]
- Harrison, M.D.; Geijskes, R.J.; Lloyd, R.; Miles, S.; Palupe, A.; Sainz, M.B.; Dale, J.L. Recombinant Cellulase Accumulation in the Leaves of Mature, Vegetatively Propagated Transgenic Sugarcane. Mol. Biotechnol. 2014, 56, 795–802. [Google Scholar] [CrossRef]
- Oraby, H.; Venkatesh, B.; Dale, B.; Ahmad, R.; Ransom, C.; Oehmke, J.; Sticklen, M. Enhanced conversion of plant biomass into glucose using transgenic rice-produced endoglucanase for cellulosic ethanol. Transgenic Res. 2007, 16, 739–749. [Google Scholar] [CrossRef]
- Ziegelhoffer, T.; Raasch, J.A.; Austin-Phillips, S. Dramatic effects of truncation and sub-cellular targeting on the accumulation of recombinant microbial cellulase in tobacco. Mol. Breed. 2001, 8, 147–158. [Google Scholar] [CrossRef]
- Biswas, G.C.G.; Ransom, C.; Sticklen, M. Expression of biologically active Acidothermus cellulolyticus endoglucanase in transgenic maize plants. Plant Sci. 2006, 171, 617–623. [Google Scholar] [CrossRef]
- Brunecky, R.; Selig, M.J.; Vinzant, T.B.; Himmel, M.E.; Lee, D.; Blaylock, M.J.; Decker, S.R. In planta expression of A. cellulolyticus Cel5A endocellulase reduces cell wall recalcitrance in tobacco and maize. Biotechnol. Biofuels 2011, 4, 1. [Google Scholar] [CrossRef] [PubMed]
- Klose, H.; Günl, M.; Usadel, B.; Fischer, R.; Commandeur, U. Ethanol inducible expression of a mesophilic cellulase avoids adverse effects on plant development. Biotechnol. Biofuels 2013, 6, 53. [Google Scholar] [CrossRef] [PubMed]
- Zuo, J.; Niu, Q.W.; Chua, N.H. An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J. 2000, 24, 265–273. [Google Scholar] [CrossRef] [PubMed]
- Potenza, C.; Aleman, L.; Sengupta-Gopalan, C. Targeting transgene expression in research, agricultural, and environmental applications: Promoters used in plant transformation. Vitr. Cell. Dev. Biol. Plant 2004, 40, 1–22. [Google Scholar] [CrossRef]
- Tomassetti, S.; Pontiggia, D.; Verrascina, I.; Reca, I.B.; Francocci, F.; Salvi, G.; Cervone, F.; Ferrari, S. Controlled expression of pectic enzymes in Arabidopsis thaliana enhances biomass conversion without adverse effects on growth. Phytochemistry 2015, 112, 221–230. [Google Scholar] [CrossRef]
- Mir, B.A.; Mewalal, R.; Mizrachi, E.; Myburg, A.A.; Cowan, D.A. Recombinant hyperthermophilic enzyme expression in plants: A novel approach for lignocellulose digestion. Trends Biotechnol. 2014, 32, 281–289. [Google Scholar] [CrossRef]
- Mir, B.A.; Myburg, A.A.; Mizrachi, E.; Cowan, D.A. In planta expression of hyperthermophilic enzymes as a strategy for accelerated lignocellulosic digestion. Sci. Rep. 2017, 7, 11462. [Google Scholar] [CrossRef]
- Peng, X.; Qiao, W.; Mi, S.; Jia, X.; Su, H.; Han, Y. Characterization of hemicellulase and cellulase from the extremely thermophilic bacterium Caldicellulosiruptor owensensis and their potential application for bioconversion of lignocellulosic biomass without pretreatment. Biotechnol. Biofuels 2015, 8, 131. [Google Scholar] [CrossRef]
- Sarmiento, F.; Peralta, R.; Blamey, J.M. Cold and Hot Extremozymes: Industrial Relevance and Current Trends. Front. Bioeng. Biotechnol. 2015, 3, 148. [Google Scholar] [CrossRef]
- York, W.S.; Qin, Q.; Rose, J.K.C. Proteinaceous inhibitors of endo-β-glucanases. Biochim. Biophys. Acta BBA Proteins Proteom. 2004, 1696, 223–233. [Google Scholar] [CrossRef]
- Juge, N. Plant protein inhibitors of cell wall degrading enzymes. Trends Plant Sci. 2006, 11, 359–367. [Google Scholar] [CrossRef] [PubMed]
- Souza, T.V.; Araujo, J.N.; da Silva, V.M.; Liberato, M.V.; Pimentel, A.C.; Alvarez, T.M.; Squina, F.M.; Garcia, W. Chemical stability of a cold-active cellulase with high tolerance toward surfactants and chaotropic agent. Biotechnol. Rep. 2016, 9, 1–8. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Li, Y.; Sun, Z.; Ge, X.; Zhang, J. Effects of lignin and surfactant on adsorption and hydrolysis of cellulases on cellulose. Biotechnol. Biofuels 2016, 9, 20. [Google Scholar] [CrossRef] [PubMed]
- Benedetti, M.; Vecchi, V.; Barera, S.; Dall’Osto, L. Biomass from microalgae: The potential of domestication towards sustainable biofactories. Microb. Cell Factories 2018, 17, 173. [Google Scholar] [CrossRef]
- Imam, S.H.; Buchanan, M.J.; Shin, H.C.; Snell, W.J. The Chlamydomonas cell wall: Characterization of the wall framework. J. Cell Biol. 1985, 101, 1599–1607. [Google Scholar] [CrossRef]
- Blifernez-Klassen, O.; Klassen, V.; Doebbe, A.; Kersting, K.; Grimm, P.; Wobbe, L.; Kruse, O. Cellulose degradation and assimilation by the unicellular phototrophic eukaryote Chlamydomonas reinhardtii. Nat. Commun. 2012, 3, 1214. [Google Scholar] [CrossRef]
- Rasala, B.A.; Lee, P.A.; Shen, Z.; Briggs, S.P.; Mendez, M.; Mayfield, S.P. Robust Expression and Secretion of Xylanase1 in Chlamydomonas reinhardtii by Fusion to a Selection Gene and Processing with the FMDV 2A Peptide. PLoS ONE 2012, 7, e43349. [Google Scholar] [CrossRef]
- Kumar, P.; Barrett, D.M.; Delwiche, M.J.; Stroeve, P. Methods for Pretreatment of Lignocellulosic Biomass for Efficient Hydrolysis and Biofuel Production. Ind. Eng. Chem. Res. 2009, 48, 3713–3729. [Google Scholar] [CrossRef]
- Brodeur, G.; Yau, E.; Badal, K.; Collier, J.; Ramachandran, K.B.; Ramakrishnan, S. Chemical and Physicochemical Pretreatment of Lignocellulosic Biomass: A Review. Enzym. Res. 2011, 2011, 787532. [Google Scholar] [CrossRef]
- Azhar, S.H.M.; Abdulla, R.; Jambo, S.A.; Marbawi, H.; Gansau, J.A.; Faik, A.A.M.; Rodrigues, K.F. Yeasts in sustainable bioethanol production: A review. Biochem. Biophys. Rep. 2017, 10, 52–61. [Google Scholar]
- de Farias Silva, C.E.; Bertucco, A. Bioethanol from microalgae and cyanobacteria: A review and technological outlook. Process Biochem. 2016, 51, 1833–1842. [Google Scholar] [CrossRef]
- Bogen, C.; Al-Dilaimi, A.; Albersmeier, A.; Wichmann, J.; Grundmann, M.; Rupp, O.; Lauersen, K.J.; Blifernez-Klassen, O.; Kalinowski, J.; Goesmann, A.; et al. Reconstruction of the lipid metabolism for the microalga Monoraphidium neglectum from its genome sequence reveals characteristics suitable for biofuel production. BMC Genom. 2013, 14, 926. [Google Scholar] [CrossRef] [PubMed]
- Dall’Osto, L.; Cazzaniga, S.; Guardini, Z.; Barera, S.; Benedetti, M.; Mannino, G.; Maffei, M.E.; Bassi, R. Combined resistance to oxidative stress and reduced antenna size enhance light-to-biomass conversion efficiency in Chlorella vulgaris cultures. Biotechnol. Biofuels 2019, 12, 221. [Google Scholar] [CrossRef] [PubMed]
- Prathima, A.; Karthikeyan, S. Characteristics of micro-algal biofuel from Botryococcus braunii. Energy Sources Part A Recover. Util. Environ. Eff. 2017, 39, 206–212. [Google Scholar] [CrossRef]
- Rodolfi, L.; Zittelli, G.C.; Bassi, N.; Padovani, G.; Biondi, N.; Bonini, G.; Tredici, M.R. Microalgae for oil: Strain selection, induction of lipid synthesis and outdoor mass cultivation in a low-cost photobioreactor. Biotechnol. Bioeng. 2009, 102, 100–112. [Google Scholar] [CrossRef]
- Perez-Garcia, O.; Bashan, Y. Microalgal Heterotrophic and Mixotrophic Culturing for Bio-Refining: From Metabolic Routes to Techno-Economics; Springer Science and Business Media LLC: Berlin/Heidelberg, Germany, 2015; pp. 61–131. [Google Scholar]
- Milano, J.; Ong, H.C.; Masjuki, H.; Chong, W.T.; Lam, M.K.; Loh, P.K.; Vellayan, V. Microalgae biofuels as an alternative to fossil fuel for power generation. Renew. Sustain. Energy Rev. 2016, 58, 180–197. [Google Scholar] [CrossRef]
- Kainthola, J.; Kalamdhad, A.S.; Goud, V.V. A review on enhanced biogas production from anaerobic digestion of lignocellulosic biomass by different enhancement techniques. Process Biochem. 2019, 84, 81–90. [Google Scholar] [CrossRef]
- Pillai, C.; Paul, W.; Sharma, C.P. Chitin and chitosan polymers: Chemistry, solubility and fiber formation. Prog. Polym. Sci. 2009, 34, 641–678. [Google Scholar] [CrossRef]
- Rinaudo, M. Chitin and chitosan: Properties and applications. Prog. Polym. Sci. 2006, 31, 603–632. [Google Scholar] [CrossRef]
- Zhang, J.; Xia, W.; Liu, P.; Cheng, Q.; Tahi, T.; Gu, W.; Li, B.; Tahirou, T. Chitosan Modification and Pharmaceutical/Biomedical Applications. Mar. Drugs 2010, 8, 1962–1987. [Google Scholar] [CrossRef]
- Fong, D.; Hoemann, C.D. Chitosan immunomodulatory properties: Perspectives on the impact of structural properties and dosage. Future Sci. 2018, 4. [Google Scholar] [CrossRef]
- Abdel-Tawwab, M.; Razek, N.A.; Abdel-Rahman, A.M. Immunostimulatory effect of dietary chitosan nanoparticles on the performance of Nile tilapia, Oreochromis niloticus (L.). Fish Shellfish Immunol. 2019, 88, 254–258. [Google Scholar] [CrossRef]
- Kengen, S.; Kengen, S.; Stams, A.J.; De Vos, W.M. Sugar metabolism of hyperthermophiles. FEMS Microbiol. Rev. 1996, 18, 119–137. [Google Scholar] [CrossRef]
- Shin, K.C.; Nam, H.K.; Oh, D.K. Hydrolysis of flavanone glycosides by β-Glucosidase from Pyrococcus furiosus and its application to the production of flavanone aglycones from citrus extracts. J. Agric. Food Chem. 2013, 61, 11532–11540. [Google Scholar] [CrossRef]
- Jadaun, J.S. Pectinase: A Useful Tool in Fruit Processing Industries. Nutr. Food Sci. Int. J. 2018, 5. [Google Scholar] [CrossRef]
- Zerva, I.; Remmas, N.; Ntougias, S. Biocatalyst Potential of Cellulose-Degrading Microorganisms Isolated from Orange Juice Processing Waste. Beverages 2019, 5, 21. [Google Scholar] [CrossRef]
- Sharma, R.; Sharma, P.C.; Rana, J.C.; Joshi, V.K. Improving the olive oil yield and quality through enzyme-assisted mechanical extraction, antioxidants and packaging. J. Food Process. Preserv. 2015, 39, 157–166. [Google Scholar] [CrossRef]
- Butt, M.S.; Tahir-Nadeem, M.; Ahmad, Z.; Sultan, M.T. Xylanases and their applications in baking industry. Food Technol. Biotechnol. 2008, 46, 22–31. [Google Scholar]
- Nadeem, M.T.; Butt, M.S.; Anjum, F.M.; Asgher, M. Improving bread quality by carboxymethyl cellulase application. Int. J. Agric. Biol. 2009, 11, 727–730. [Google Scholar]
- Savatin, D.V.; Ferrari, S.; Sicilia, F.; De Lorenzo, G. Oligogalacturonide-Auxin Antagonism Does Not Require Posttranscriptional Gene Silencing or Stabilization of Auxin Response Repressors in Arabidopsis. Plant Physiol. 2011, 157, 1163–1174. [Google Scholar] [CrossRef]
- Huot, B.; Yao, J.; Montgomery, B.L.; He, S.Y. Growth-defense tradeoffs in plants: A balancing act to optimize fitness. Mol. Plant 2014, 7, 1267–1287. [Google Scholar] [CrossRef] [PubMed]
- Couto, D.; Zipfel, C. Regulation of pattern recognition receptor signalling in plants. Nat. Rev. Immunol. 2016, 16, 537–552. [Google Scholar] [CrossRef] [PubMed]
- Bacete, L.; Mélida, H.; Miedes, E.; Molina, A. Plant cell wall-mediated immunity: Cell wall changes trigger disease resistance responses. Plant J. 2018, 93, 614–636. [Google Scholar] [CrossRef] [PubMed]
- Boller, T.; Felix, G. A Renaissance of Elicitors: Perception of Microbe-Associated Molecular Patterns and Danger Signals by Pattern-Recognition Receptors. Annu. Rev. Plant Biol. 2009, 60, 379–406. [Google Scholar] [CrossRef] [PubMed]
- Galletti, R.; Ferrari, S.; De Lorenzo, G. Arabidopsis MPK3 and MPK6 play different roles in basal and oligogalacturonide- or flagellin-induced resistance against Botrytis cinerea. Plant Physiol. 2011, 157, 804–814. [Google Scholar] [CrossRef]
- Cervone, F.; De Lorenzo, G.; Ferrari, S.; Benedetti, M.; Pontiggia, D. Fusion Protein and Transgenic Plant Expressing Said Protein. U.S. Patent 10385347B2, 20 August 2019. [Google Scholar]
- Borras-Hidalgo, O.; Caprari, C.; Hernandez-Estevez, I.; De Lorenzo, G.; Cervone, F. A gene for plant protection: Expression of a bean polygalacturonase inhibitor in tobacco confers a strong resistance against Rhizoctonia solani and two oomycetes. Front. Plant Sci. 2012, 3, 268. [Google Scholar] [CrossRef]
- Janni, M.; Sella, L.; Favaron, F.; Blechl, A.E.; De Lorenzo, G.; D’Ovidio, R. The expression of a bean PGIP in transgenic wheat confers increased resistance to the fungal pathogen Bipolaris sorokiniana. Mol. Plant Microbe Interact. 2008, 21, 171–177. [Google Scholar] [CrossRef]
- Mohammadzadeh, R.; Zamani, M.; Motallebi, M.; Norouzi, P.; Jourabchi, E.; Benedetti, M.; Lorenzo, G.D. Agrobacterium tumefaciens-mediated introduction of polygalacturonase inhibiting protein 2 gene (PvPGIP2) from Phaseolus vulgaris into sugar beet (Beta vulgaris L.). Aust. J. Crop Sci. 2012, 6, 1290–1297. [Google Scholar]
- Brunner, K.; Zeilinger, S.; Ciliento, R.; Woo, S.L.; Lorito, M.; Kubicek, C.P.; Mach, R.L. Improvement of the Fungal Biocontrol Agent Trichoderma atroviride to Enhance both Antagonism and Induction of Plant Systemic Disease Resistance. Appl. Environ. Microbiol. 2005, 71, 3959–3965. [Google Scholar] [CrossRef]
| SUBSTRATE | CWLE | AA |
|---|---|---|
| CELLULOSE | Lytic Polysaccharide Mono-Oxygenase | 9, 10, 15, 16. |
| Cellobiose dehydrogenase | 3. | |
| endo-1,4-β-glucanase | GH | |
| 5, 6, 7, 8, 9, 10, 12, 44, 45, 48, 51, 74, 124. | ||
| Exoglucanase | 1, 5, 9. | |
| Cellobiohydrolase | 6, 7, 9, 48. | |
| β-glucosidase | 1, 3, 5, 9, 30, 116. | |
| HEMICELLULOSE | ||
| Xylan | endo-1,4-β-xylanase | 5, 8, 11, 30, 43, 51, 98. |
| β-xylosidase | 1, 2, 3, 30, 39, 43, 51, 52, 54, 116, 120. | |
| Mannan | endo-1,4-β-mannanase | 26, 45, 113, 134. |
| β-mannosidase | 1, 2, 5, 164. | |
| Galactan | endo-1,4-β-galactanase | 53. |
| β-galactosidase | 1, 2, 35, 42, 59, 147, 165. | |
| Xyloglucan | Endoxyloglucanase | 5, 9, 12, 16, 44, 45. |
| Exoxyloglucanase | 3, 74. | |
| Arabinose branched | l-α-Arabinofuranosidase | 2, 3, 43, 51, 54, 62. |
| PECTIN | ||
| Homogalacturonan | endo-1,4-α-polygalacturonase | 28. |
| Exopolygalacturonase | 28. | |
| Pectate lyase | PL | |
| 1, 2, 3, 9, 10. | ||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Benedetti, M.; Locci, F.; Gramegna, G.; Sestili, F.; Savatin, D.V. Green Production and Biotechnological Applications of Cell Wall Lytic Enzymes. Appl. Sci. 2019, 9, 5012. https://doi.org/10.3390/app9235012
Benedetti M, Locci F, Gramegna G, Sestili F, Savatin DV. Green Production and Biotechnological Applications of Cell Wall Lytic Enzymes. Applied Sciences. 2019; 9(23):5012. https://doi.org/10.3390/app9235012
Chicago/Turabian StyleBenedetti, Manuel, Federica Locci, Giovanna Gramegna, Francesco Sestili, and Daniel V. Savatin. 2019. "Green Production and Biotechnological Applications of Cell Wall Lytic Enzymes" Applied Sciences 9, no. 23: 5012. https://doi.org/10.3390/app9235012
APA StyleBenedetti, M., Locci, F., Gramegna, G., Sestili, F., & Savatin, D. V. (2019). Green Production and Biotechnological Applications of Cell Wall Lytic Enzymes. Applied Sciences, 9(23), 5012. https://doi.org/10.3390/app9235012









