Gradient-Guided Convolutional Neural Network for MRI Image Super-Resolution
Abstract
1. Introduction
- We design a gradient-guided residual network for solving the single contrast MRI image super-resolution problem. The proposed network exploits the mutual relation of the super-resolution and the image gradient priors. Thus, the network employs image gradient information for image super-resolution intentionally.
- With a suitable model, image gradient is exploited for MR image super-resolution to supply the clues regarding the high-frequency details. Under the guidance of gradient, the forward super-resolution process reconstructs HR image explicitly, thereby leading a more accurate HR image.
- The experimental results of three public databases show that the gradient-guided CNN outperforms the conventional feed-forward architecture CNNs in MRI image super-resolution. The proposed approach provides a flexible model of employing image prior for CNN-based super-resolution.
2. Related Works
2.1. CNN-Based MRI Super-Resolution
2.2. High-Frequency Details Recovery
- The image gradient is employed as a regularization item in the loss function. In a correctly restored image, the edges and texture (related to the image gradients) should be accurate. The regularization term, which is induced by additional sources of information, helps recover high-frequency details. , where is defined asin which denotes the gradient detector, and is the gradient magnitude of image x.
- The alternative approach to incorporating image gradient in the SR process is to concatenate the gradient maps with the input LR image y as a joint input of the network. Thus, the mapping function is
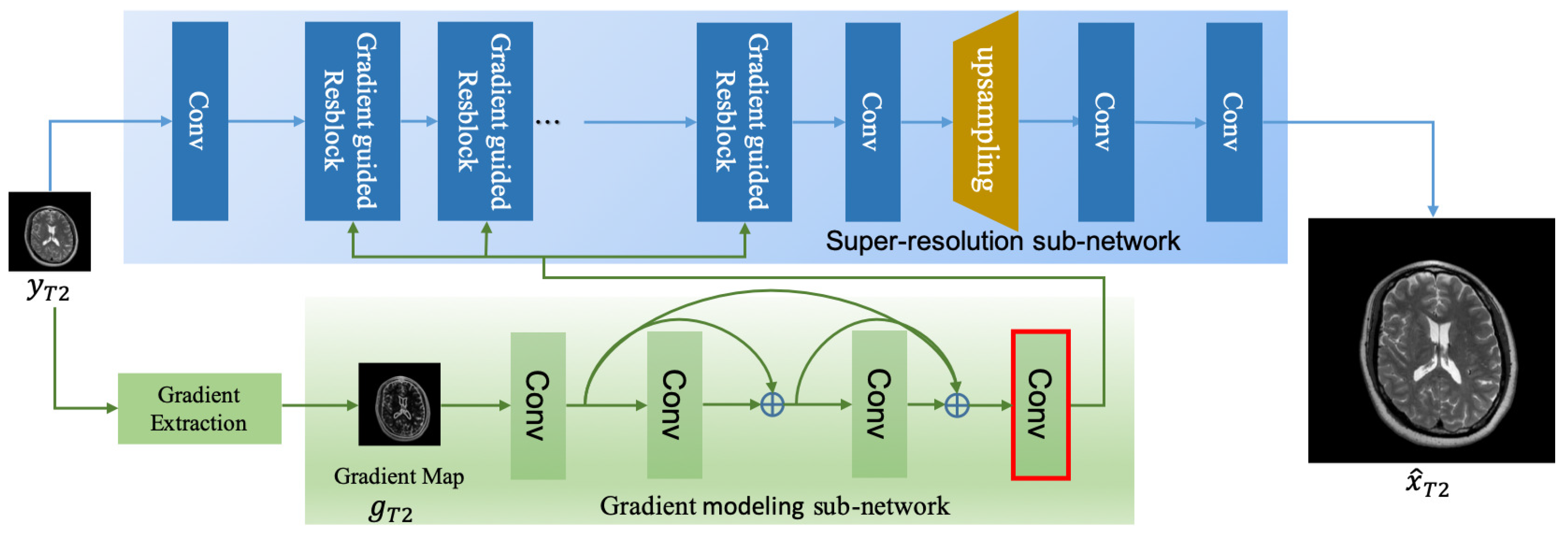
3. Proposed Methods
3.1. Gradient Modeling (GM) Subnet
3.2. Super-Resolution Subnet
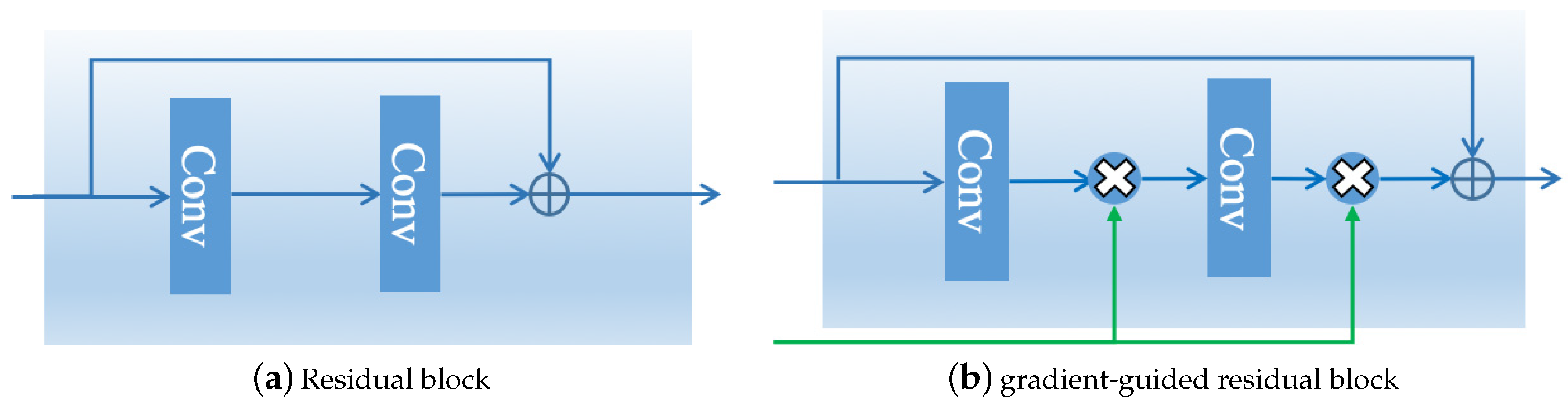
3.2.1. Gradient-Guided Resblock
3.2.2. Reconstruction Block
4. Experiments and Results
4.1. Datasets
4.2. Implementation Details
- The original image x were convolved by Gaussian kernel with standard deviation of 1.
- The results of convolution were down-sampled with factors of and 4, respectively.
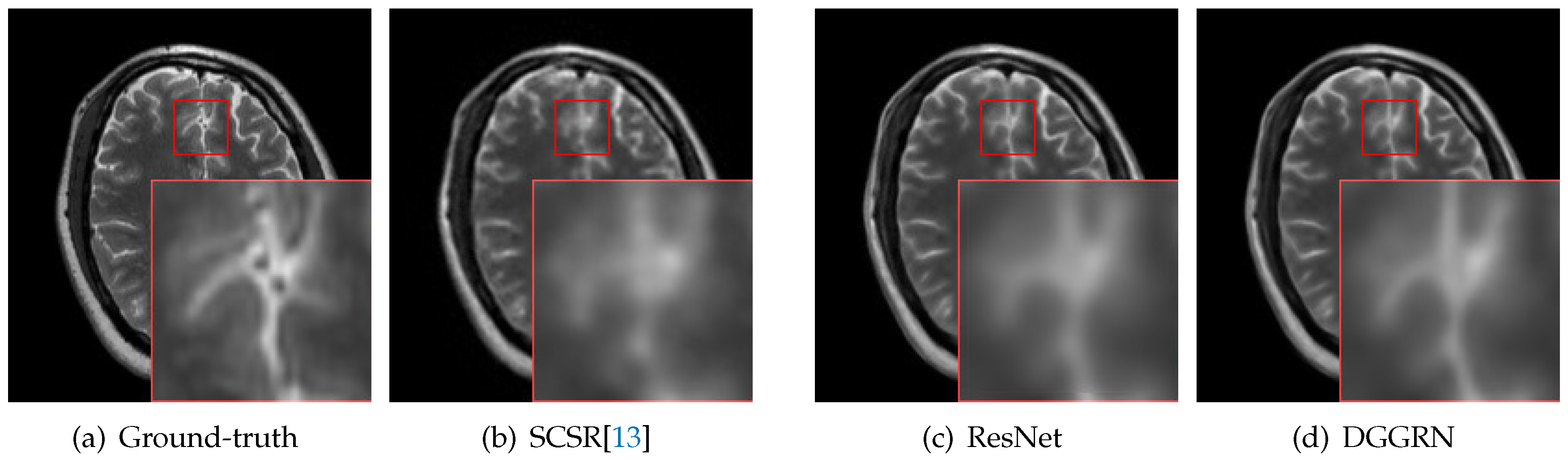
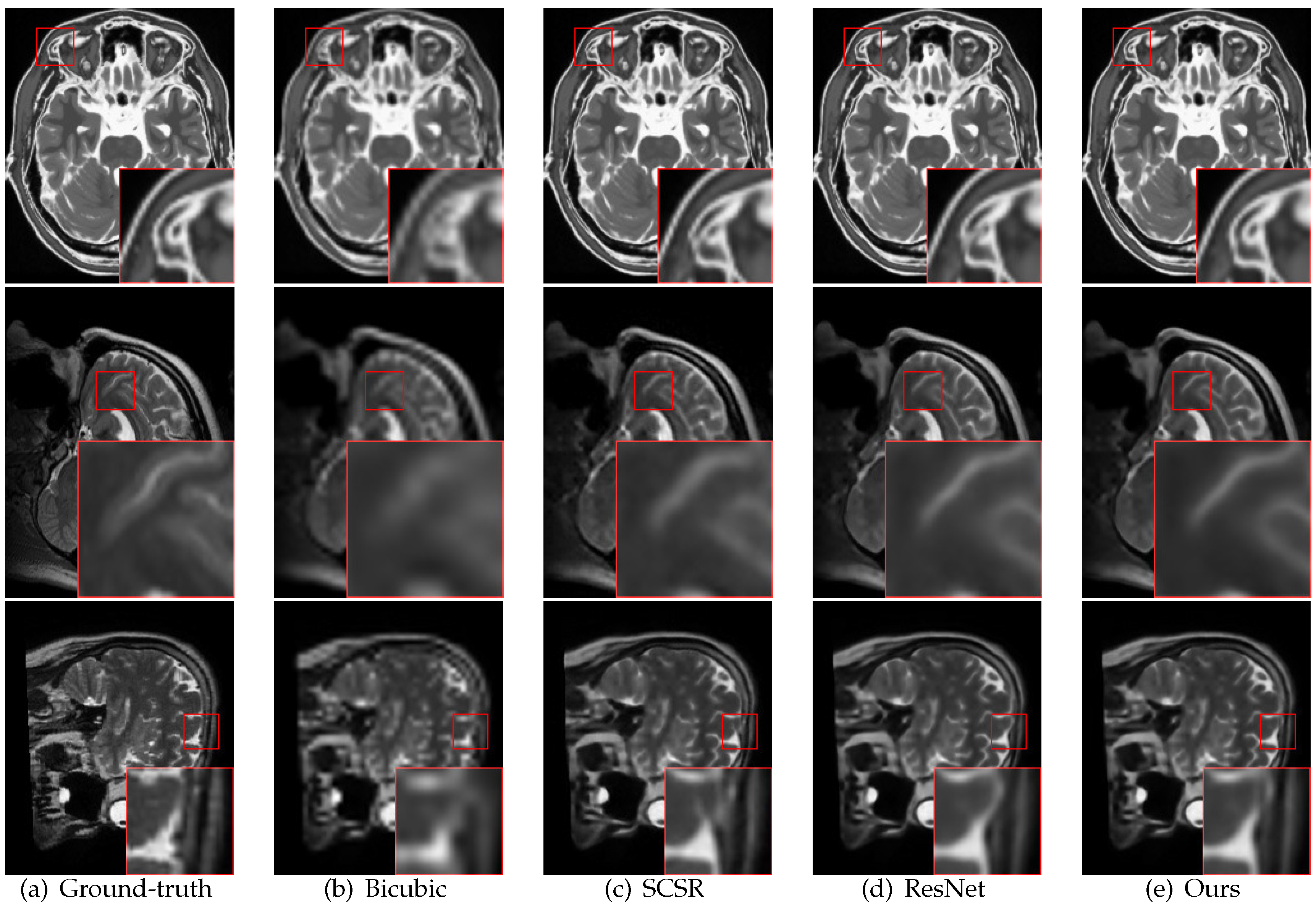
4.3. Comparison with State-of-the-Art Methods
5. Discussion
5.1. Benefits of Gradient-Guided Resblock
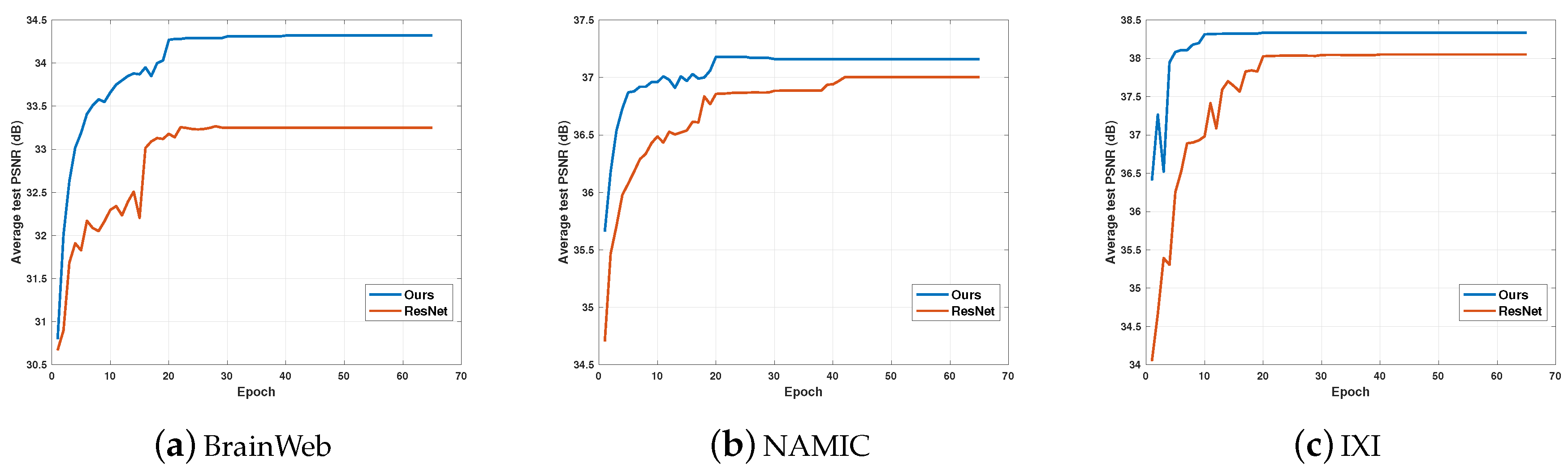
5.2. Performance and Training Epochs
5.3. Parameters and Performance
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Greenspan, H. Super-resolution in medical imaging. Comput. J. 2008, 52, 43–63. [Google Scholar] [CrossRef]
- Manjón, J.V.; Coupé, P.; Buades, A.; Fonov, V.; Collins, D.L.; Robles, M. Non-local MRI upsampling. Med. Image Anal. 2010, 14, 784–792. [Google Scholar] [CrossRef] [PubMed]
- Rueda, A.; Malpica, N.; Romero, E. Single-image super-resolution of brain MR images using overcomplete dictionaries. Med. Image Anal. 2013, 17, 113–132. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Zhang, R.; Wu, S.; Hu, J.; Xie, Y. An edge-directed interpolation method for fetal spine MR images. Biomed. Eng. Online 2013, 12, 102. [Google Scholar] [CrossRef]
- Wang, Y.H.; Qiao, J.; Li, J.B.; Fu, P.; Chu, S.C.; Roddick, J.F. Sparse representation-based MRI super-resolution reconstruction. Measurement 2014, 47, 946–953. [Google Scholar] [CrossRef]
- Jafari-Khouzani, K. MRI upsampling using feature-based nonlocal means approach. IEEE Trans. Med. Imaging 2014, 33, 1969–1985. [Google Scholar] [CrossRef]
- Lu, X.; Huang, Z.; Yuan, Y. MR image super-resolution via manifold regularized sparse learning. Neurocomputing 2015, 162, 96–104. [Google Scholar] [CrossRef]
- Zhang, D.; He, J.; Zhao, Y.; Du, M. MR image super-resolution reconstruction using sparse representation, nonlocal similarity and sparse derivative prior. Comput. Biol. Med. 2015, 58, 130–145. [Google Scholar] [CrossRef]
- Shi, F.; Cheng, J.; Wang, L.; Yap, P.T.; Shen, D. LRTV: MR image super-resolution with low-rank and total variation regularizations. IEEE Trans. Med. Imaging 2015, 34, 2459–2466. [Google Scholar] [CrossRef]
- Tourbier, S.; Bresson, X.; Hagmann, P.; Thiran, J.P.; Meuli, R.; Cuadra, M.B. An efficient total variation algorithm for super-resolution in fetal brain MRI with adaptive regularization. NeuroImage 2015, 118, 584–597. [Google Scholar] [CrossRef]
- Pham, C.H.; Tor-Díez, C.; Meunier, H.; Bednarek, N.; Fablet, R.; Passat, N.; Rousseau, F. Multiscale brain MRI super-resolution using deep 3D convolutional networks. Comput. Med. Imaging Graph. 2019, 77, 101647. [Google Scholar] [CrossRef] [PubMed]
- Oktay, O.; Bai, W.; Lee, M.; Guerrero, R.; Kamnitsas, K.; Caballero, J.; de Marvao, A.; Cook, S.; O’Regan, D.; Rueckert, D. Multi-input cardiac image super-resolution using convolutional neural networks. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Athens, Greece, 17–21 October 2016; Springer: Cham, Switzerland, 2016; pp. 246–254. [Google Scholar]
- Zeng, K.; Zheng, H.; Cai, C.; Yang, Y.; Zhang, K.; Chen, Z. Simultaneous single- and multi-contrast super-resolution for brain MRI images based on a convolutional neural network. Comput. Biol. Med. 2018, 99, 133–141. [Google Scholar] [CrossRef] [PubMed]
- Alom, M.Z.; Taha, T.M.; Yakopcic, C.; Westberg, S.; Sidike, P.; Nasrin, M.S.; Hasan, M.; Van Essen, B.C.; Awwal, A.A.; Asari, V.K. A state-of-the-art survey on deep learning theory and architectures. Electronics 2019, 8, 292. [Google Scholar] [CrossRef]
- Cascio, D.; Taormina, V.; Raso, G. Deep CNN for IIF Images Classification in Autoimmune Diagnostics. Appl. Sci. 2019, 9, 1618. [Google Scholar] [CrossRef]
- Cascio, D.; Taormina, V.; Raso, G. Deep Convolutional Neural Network for HEp-2 Fluorescence Intensity Classification. Appl. Sci. 2019, 9, 408. [Google Scholar] [CrossRef]
- Dong, C.; Loy, C.C.; He, K.; Tang, X. Image super-resolution using deep convolutional networks. IEEE Trans. Pattern Anal. Mach. Intell. 2016, 38, 295–307. [Google Scholar] [CrossRef]
- Kim, J.; Kwon Lee, J.; Mu Lee, K. Accurate image super-resolution using very deep convolutional networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 26 June–1 July 2016; pp. 1646–1654. [Google Scholar]
- Shi, W.; Caballero, J.; Huszár, F.; Totz, J.; Aitken, A.P.; Bishop, R.; Rueckert, D.; Wang, Z. Real-time single image and video super-resolution using an efficient sub-pixel convolutional neural network. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 26 June–1 July 2016; pp. 1874–1883. [Google Scholar]
- Lim, B.; Son, S.; Kim, H.; Nah, S.; Mu Lee, K. Enhanced deep residual networks for single image super-resolution. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition Workshops, Honolulu, HI, USA, 21–26 July 2017; pp. 136–144. [Google Scholar]
- Timofte, R.; Gu, S.; Wu, J.; Van Gool, L. NTIRE 2018 challenge on single image super-resolution: methods and results. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition Workshops, Salt Lake City, UT, USA, 18–22 June 2018; pp. 852–863. [Google Scholar]
- Pham, C.H.; Ducournau, A.; Fablet, R.; Rousseau, F. Brain MRI super-resolution using deep 3D convolutional networks. In Proceedings of the 2017 IEEE 14th International Symposium on Biomedical Imaging (ISBI 2017), Melbourne, Australia, 18–21 April 2017; pp. 197–200. [Google Scholar]
- Bahrami, K.; Shi, F.; Rekik, I.; Shen, D. Convolutional neural network for reconstruction of 7T-like images from 3T MRI using appearance and anatomical features. In Deep Learning and Data Labeling for Medical Applications; Springer: Cham, Switzerland, 2016; pp. 39–47. [Google Scholar]
- Wang, S.; Su, Z.; Ying, L.; Peng, X.; Zhu, S.; Liang, F.; Feng, D.; Liang, D. Accelerating magnetic resonance imaging via deep learning. In Proceedings of the 2016 IEEE 13th International Symposium on Biomedical Imaging (ISBI), Prague, Czech Republic, 13–16 April 2016; pp. 514–517. [Google Scholar]
- Schlemper, J.; Caballero, J.; Hajnal, J.V.; Price, A.; Rueckert, D. A deep cascade of convolutional neural networks for MR image reconstruction. In Proceedings of the International Conference on Information Processing in Medical Imaging, Boone, NC, USA, 25–30 June 2017; Springer: Cham, Switzerland; pp. 647–658. [Google Scholar]
- McDonagh, S.; Hou, B.; Alansary, A.; Oktay, O.; Kamnitsas, K.; Rutherford, M.; Hajnal, J.V.; Kainz, B. Context-sensitive super-resolution for fast fetal magnetic resonance imaging. In Molecular Imaging, Reconstruction and Analysis of Moving Body Organs, and Stroke Imaging and Treatment; Springer: Cham, Switzerland, 2017; pp. 116–126. [Google Scholar]
- Hu, C.; Qu, X.; Guo, D.; Bao, L.; Chen, Z. Wavelet-based edge correlation incorporated iterative reconstruction for undersampled MRI. Magn. Reson. Imaging 2011, 29, 907–915. [Google Scholar] [CrossRef]
- Mai, Z.; Rajan, J.; Verhoye, M.; Sijbers, J. Robust edge-directed interpolation of magnetic resonance images. Phys. Med. Biol. 2011, 56, 7287–7303. [Google Scholar] [CrossRef]
- Wei, Z.; Ma, K.K. Contrast-guided image interpolation. IEEE Trans. Image Process. 2013, 22, 4271–4285. [Google Scholar]
- Zheng, H.; Zeng, K.; Guo, D.; Ying, J.; Yang, Y.; Peng, X.; Huang, F.; Chen, Z.; Qu, X. Multi-Contrast Brain MRI Image Super-Resolution With Gradient-Guided Edge Enhancement. IEEE Access 2018, 6, 57856–57867. [Google Scholar] [CrossRef]
- Sun, L.; Fan, Z.; Huang, Y.; Ding, X.; Paisley, J. A Deep Information Sharing Network for Multi-contrast Compressed Sensing MRI Reconstruction. arXiv 2018, arXiv:1804.03596. [Google Scholar] [CrossRef] [PubMed]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 26 June–1 July 2016; pp. 770–778. [Google Scholar]
- Zhou, D.; Wang, R.; Lu, J.; Zhang, Q. Depth Image Super Resolution Based on Edge-Guided Method. Appl. Sci. 2018, 8, 298. [Google Scholar] [CrossRef]
- Xie, J.; Feris, R.S.; Sun, M.T. Edge-guided single depth image super resolution. IEEE Trans. Image Process. 2016, 25, 428–438. [Google Scholar] [CrossRef] [PubMed]
- Timofte, R.; Rothe, R.; Van Gool, L. Seven Ways to Improve Example-Based Single Image Super Resolution. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 26 June–1 July 2016; pp. 1865–1873. [Google Scholar]
- Zheng, H.; Qu, X.; Bai, Z.; Liu, Y.; Guo, D.; Dong, J.; Peng, X.; Chen, Z. Multi-contrast brain magnetic resonance image super-resolution using the local weight similarity. BMC Med. Imaging 2017, 17, 6. [Google Scholar] [CrossRef] [PubMed]
- Cocosco, C.A.; Kollokian, V.; Kwan, R.K.; Evans, A.C. BrainWeb: Online Interface to a 3D MRI Simulated Brain Database. NeuroImage 1997, 5, 425. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2015; Springer: Cham, Switzerland, 2015; pp. 234–241. [Google Scholar]
- Abadi, M.; Barham, P.; Chen, J.; Chen, Z.; Davis, A.; Dean, J.; Devin, M.; Ghemawat, S.; Irving, G.; Isard, M.; et al. Tensorflow: A system for large-scale machine learning. In Proceedings of the 12th {USENIX} Symposium on Operating Systems Design and Implementation ({OSDI} 16), Savannah, GA, USA, 2–4 November 2016; pp. 265–283. [Google Scholar]
- Kingma, D.P.; Ba, J. Adam: A method for stochastic optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar]

| Dataset | Upscaling Factor | Bicubic | LRTV | NMU | SCSR | ResNet | ReCNN | Ours |
|---|---|---|---|---|---|---|---|---|
| BrainWeb | 2 | 21.51 | 24.81 | 27.50 | 32.99 | 33.25 | 32.86 | 34.32 |
| 0.827 | 0.904 | 0.952 | 0.984 | 0.985 | 0.973 | 0.987 | ||
| 3 | 18.3 | 21.67 | 21.54 | 26.07 | 27.71 | 26.10 | 27.62 | |
| 0.664 | 0.820 | 0.811 | 0.922 | 0.938 | 0.925 | 0.942 | ||
| 4 | 16.37 | 19.36 | 19.33 | 21.31 | 22.11 | 21.03 | 23.11 | |
| 0.525 | 0.697 | 0.682 | 0.776 | 0.803 | 0.771 | 0.849 | ||
| NAMIC | 2 | 28.70 | 31.98 | 33.95 | 36.86 | 37.00 | 36.64 | 37.21 |
| 0.850 | 0.910 | 0.889 | 0.922 | 0.928 | 0.920 | 0.939 | ||
| 3 | 24.93 | 29.42 | 29.34 | 31.49 | 31.52 | 31.10 | 31.97 | |
| 0.721 | 0.870 | 0.772 | 0.826 | 0.821 | 0.822 | 0.864 | ||
| 4 | 22.81 | 26.54 | 26.76 | 28.33 | 28.45 | 27.97 | 29.05 | |
| 0.613 | 0.769 | 0.642 | 0.712 | 0.717 | 0.706 | 0.737 | ||
| IXI | 2 | 28.56 | - | - | 37.86 | 38.08 | 37.31 | 38.28 |
| 0.915 | - | - | 0.982 | 0.983 | 0.970 | 0.983 | ||
| 3 | 24.68 | - | - | 31.68 | 31.79 | 31.45 | 32.06 | |
| 0.853 | - | - | 0.942 | 0.944 | 0.939 | 0.946 | ||
| 4 | 22.44 | - | - | 28.15 | 28.42 | 27.97 | 28.77 | |
| 0.723 | - | - | 0.888 | 0.893 | 0.874 | 0.895 |
| Block Number | 2 | 4 | 6 | 8 | 10 | 12 |
|---|---|---|---|---|---|---|
| ResNet | 36.83 | 36.96 | 37.06 | 37.00 | 14.20 | 14.20 |
| Ours | 37.00 | 37.10 | 37.18 | 37.21 | 37.16 | 37.23 |
| K | 32 | 64 | 128 |
|---|---|---|---|
| BrainWeb | 32.85 | 34.32 | 34.40 |
| NAMIC | 37.13 | 37.21 | 37.23 |
| IXI | 38.16 | 38.28 | 38.38 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Du, X.; He, Y. Gradient-Guided Convolutional Neural Network for MRI Image Super-Resolution. Appl. Sci. 2019, 9, 4874. https://doi.org/10.3390/app9224874
Du X, He Y. Gradient-Guided Convolutional Neural Network for MRI Image Super-Resolution. Applied Sciences. 2019; 9(22):4874. https://doi.org/10.3390/app9224874
Chicago/Turabian StyleDu, Xiaofeng, and Yifan He. 2019. "Gradient-Guided Convolutional Neural Network for MRI Image Super-Resolution" Applied Sciences 9, no. 22: 4874. https://doi.org/10.3390/app9224874
APA StyleDu, X., & He, Y. (2019). Gradient-Guided Convolutional Neural Network for MRI Image Super-Resolution. Applied Sciences, 9(22), 4874. https://doi.org/10.3390/app9224874





