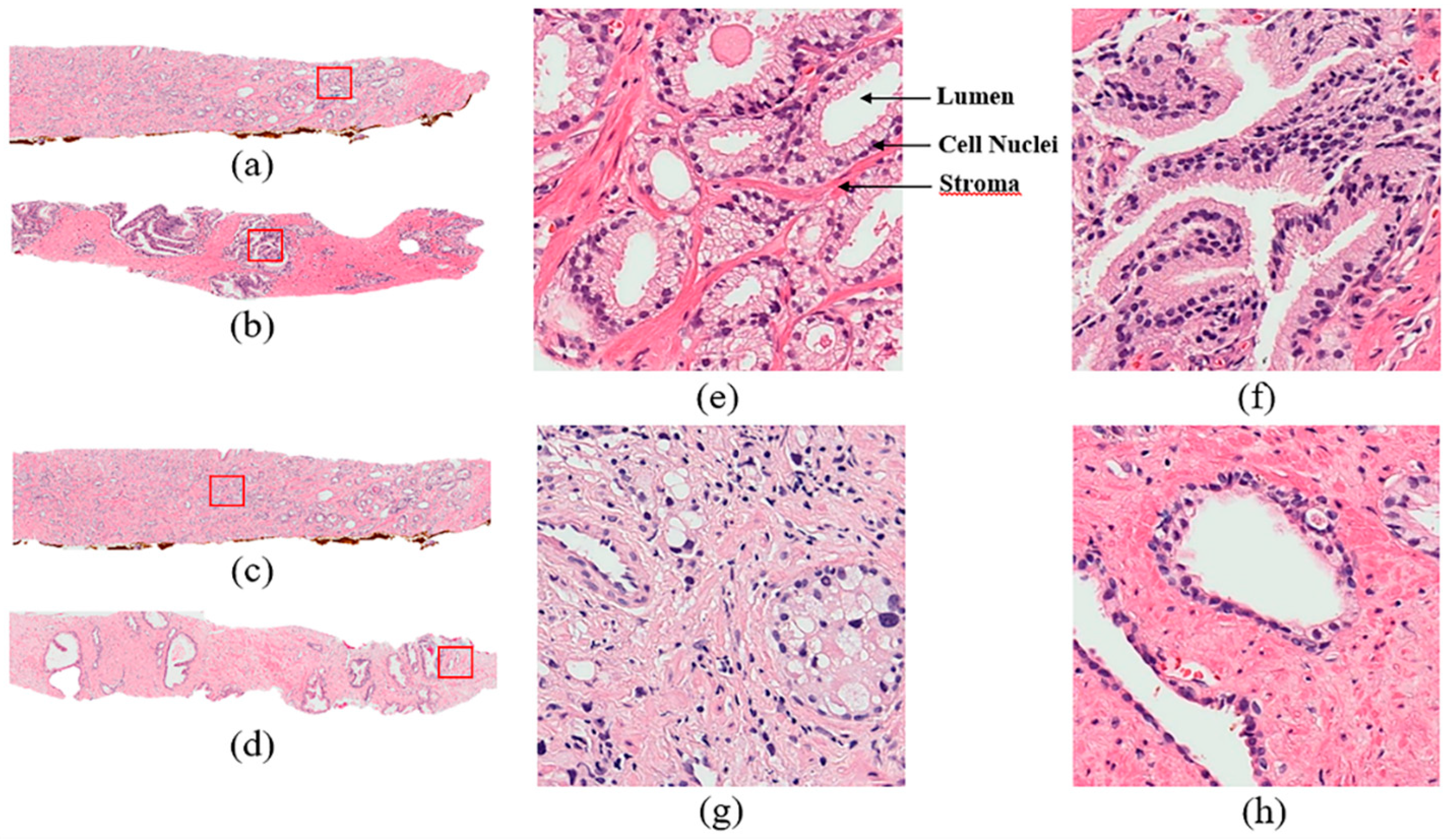
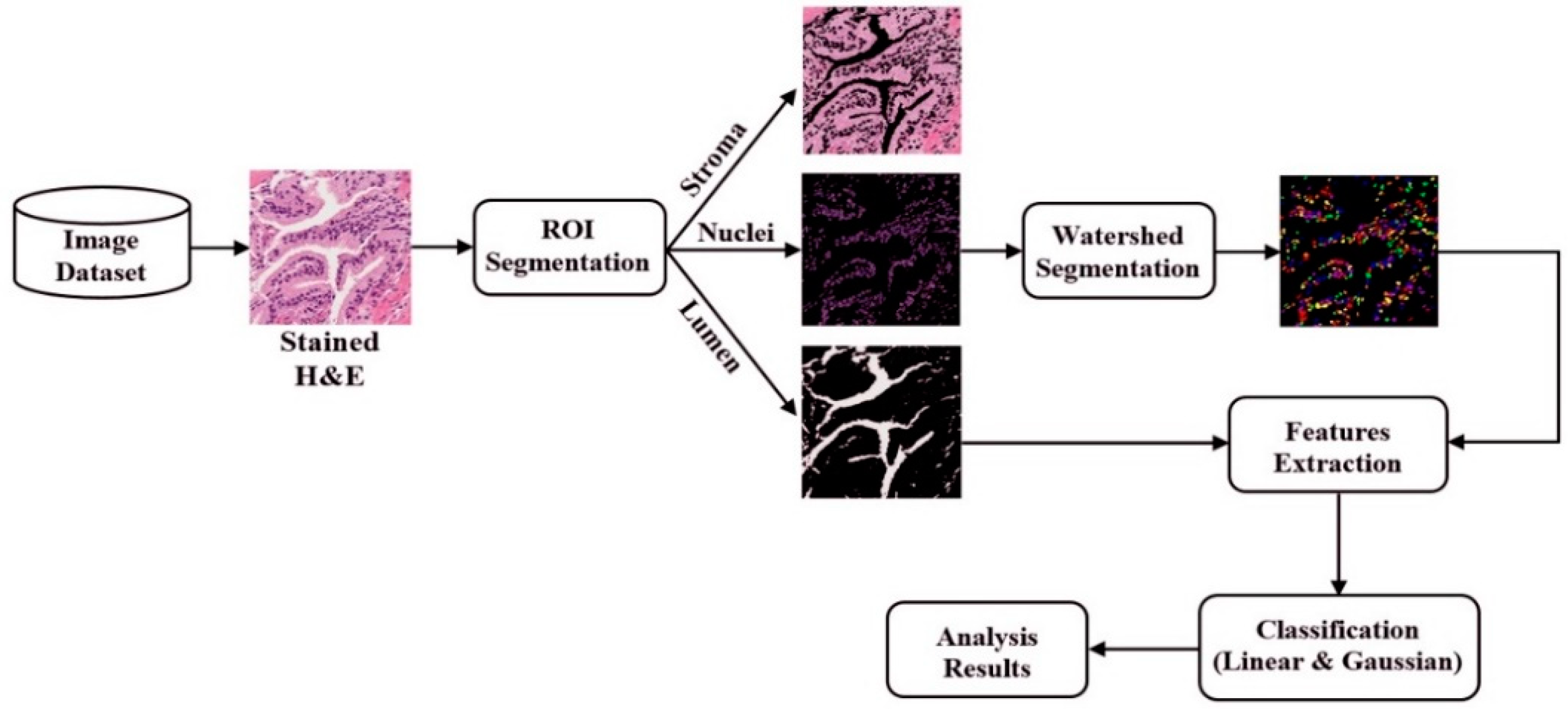
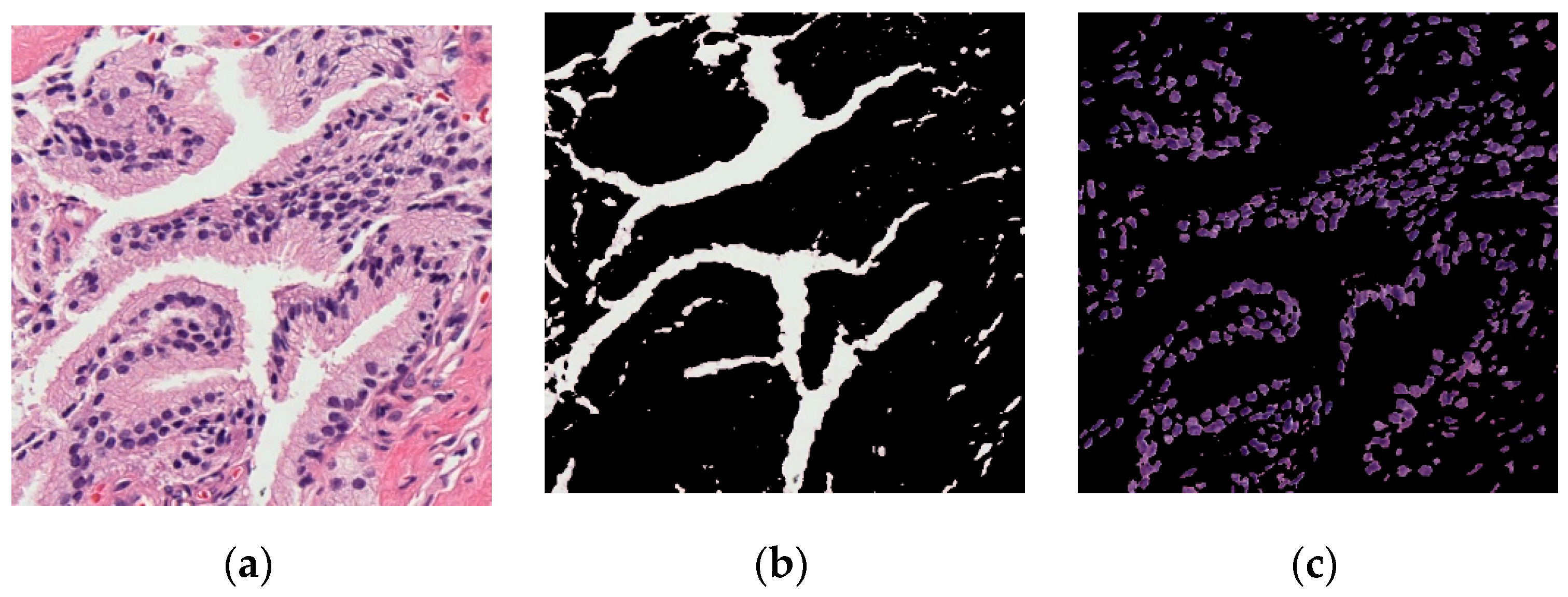
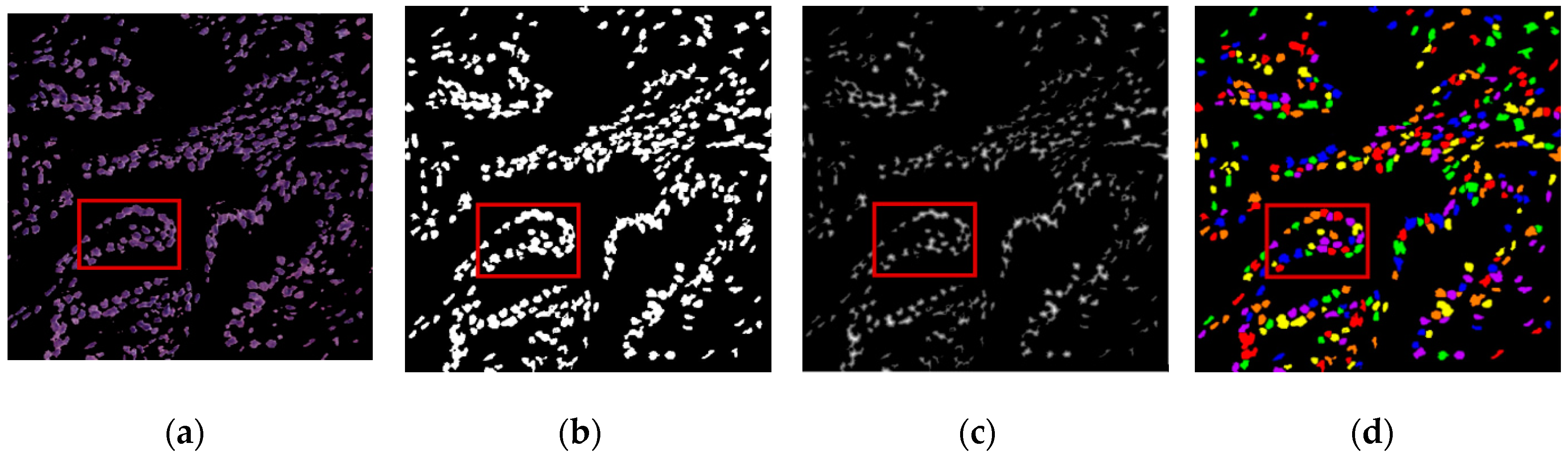
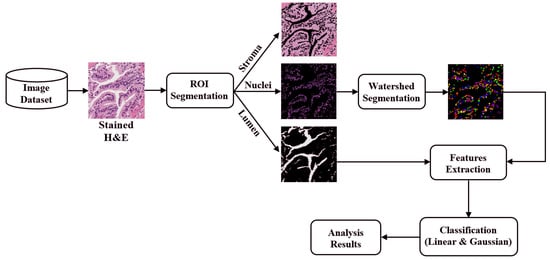
1. Introduction
Prostate adenocarcinoma, a type of prostate cancer, is the second most commonly diagnosed cancer. In the United States, the incidence of prostate cancer ranks first among all malignant tumors in men. The Gleason score is currently the most common grading system of prostate adenocarcinoma and is widely used to assess the prognosis of men with prostate cancer using samples from a prostate biopsy. There are some diagnostic protocols for cancer grading, for which microscopic evaluation of tissue specimens is required. For this, the samples need to be appropriately stained using Hematoxylin and Eosin (H&E) compounds. The cancer grade is assessed by a pathologist based on the morphological features of lumen and cell nucleus observed in the tissue. Cancer diagnosis and grading based on digital pathology have become increasingly complex due to the increase in cancer occurrence and specific treatment options for patients [
1].
In South Korea, the incidence of prostate cancer is increasing significantly. Prostate cancer (PCa) is the fifth most common cancer among males in Korea and the expected cancer deaths in 2018 were 82,155 [
2]. The detection of prostate cancer has always been a major issue for pathologists and medical practitioners, for both diagnosis and treatment. Usually, the cancer detection process in histopathology consists of categorizing stained microscopic biopsy images into malignant and benign.
The Gleason grade grouping system defines Gleason scores ≤ 6 as grade 1, score
as grade 2, score
as grade 3, score
as grade 4, and score
as grade 5. The Gleason score is obtained by adding the primary (most common) and secondary (second most common) scores from H&E stained tissue microscopic images. This system was developed by Dr. Donald F Gleason, who was a Pathologist in Minnesota, and members of the Veterans Administration Cooperative Urological Research Group (VACURG) [
3]. This system was tested on a large number of patients, including long-term follow-ups and is considered an outstanding success.
In recent years, an excellent and important addition to microscopy and digital imaging has been developed for microscopes that are used to convert stained tissue slides into whole slide digital images. This allows for more efficient computer-based viewing and analysis of histopathology. Early diagnosis and treatment are required, to avoid the enlargement of cancer cells in the prostate gland and control the spreading of more aggressive tumors to other parts of the body.
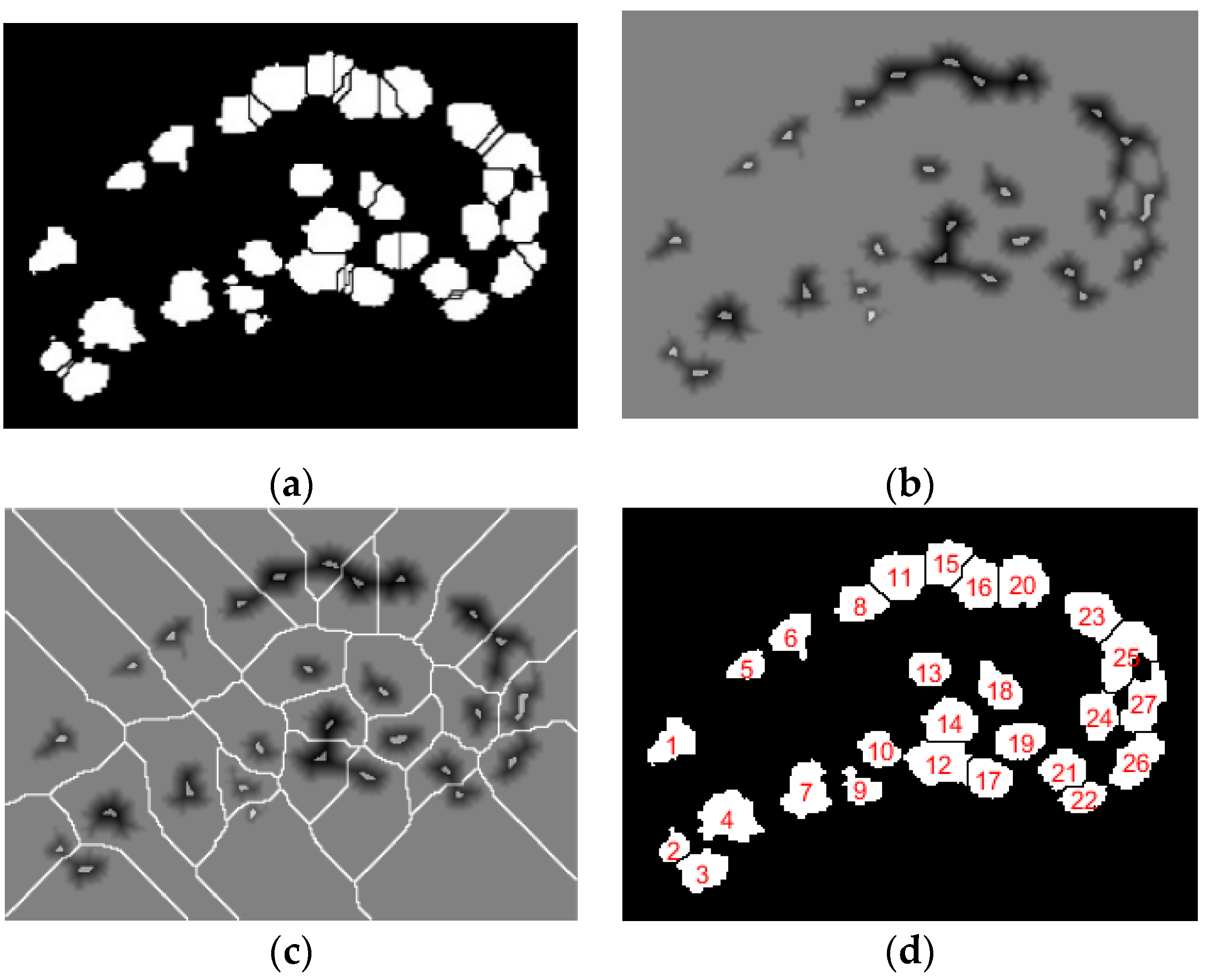
The digital pathology field has grown dramatically over recent years, largely due to technological advancements in image processing and machine learning algorithms, and increases in computational power. As part of this field, many methods have been proposed for automatic histopathological image analysis and classification. In this paper, color segmentation, based on k-means clustering method, is proposed for microscopic biopsy tissue image processing, and the watershed algorithm has been implemented to separate touching cell nuclei in tissue images.
This approach can be implemented in different ways; however, the marker selection approach has been carried out in this study to control over-segmentation. Diagnosing prostate cancer from a biopsy tissue image under a microscope is difficult for the pathologists and doctors. Therefore, machine learning and deep learning techniques are developed for computerized classification and cancer grading. In this study, a machine learning classification method is proposed in order to classify Gleason grade groups of prostate cancer. From a perspective of computer engineering, since the regular procedure of diagnosing prostate cancer and grading is difficult and time consuming; therefore, automated computerized methods are in high demand and are essential for medical image analysis.
2. Literature Review
Tabesh et al. [
4] extracted features that describe color, texture, and morphology from 367 and 268 H&E image patches, which were acquired from tissue microarray (TMA) datasets. These features were used for support vector machine (SVM) classification. They achieved an accuracy of 96.7% and 81% for predicting benign vs. malignant and low-grade vs. high-grade classifications, respectively, using 5-fold cross-validation.
Doyle et al. [
5] proposed a cascade approach to the multi-class grading problem. They used cascade binary classification to maximize inter- and intra-class accuracy rather than the conventional one-shot classification and one-versus-all approaches to multi-class classification. In the proposed cascade approach, each division is classified separately and independently.
Nir et al. [
6] proposed some novel features based on intra- and inter-nuclei properties for classification. They trained their classifier on 333 tissue microarray (TMA) cores annotated by six pathologists for different Gleason grades and used SVM classification to achieve an accuracy of 88.5% and 73.8% for cancer detection (benign vs. malignant) and low vs. high grade (Grade 3 vs. Grade 4, 5), respectively.
Doyle et al. [
7] extracted nearly 600 image texture features to perform pixel-wise Bayesian classification at each image scale to obtain the corresponding likelihood scene. The authors achieved an accuracy of 88.0% for distinguishing between benign and malignant samples.
Rundo et al. [
8] proposed Fuzzy C-Means (FCM) clustering algorithm for prostate multispectral MRI morphologic data processing and segmentation. The authors used co-registered T1w and T2w MR image series and achieved an average dice similarity coefficient 90.77
7.75, with respect to 81.90
6.49 and 82.55
4.93 by processing T2w and T1w imaging alone, respectively.
Jiao et al. [
9] used combined deep learning and SVM methods for breast masses classification. The methods were applied to the Digital Database for Screening Mammography (DDSM) dataset and achieved high accuracy under two objective evaluation measures. The authors used nearly 600 images, out of these, 50% were benign and 50% were malignant. The classification accuracy achieved in this paper was 96.7% for distinguishing between benign and malignant samples.
Hu et al. [
10] presented a novel mass detection system for digital mammograms, which integrated a visual saliency model with deep learning techniques. The authors used combined deep learning and SVM methods for image and feature classification, respectively. They achieved an average accuracy of 91.5% in mass detection between cancer and benign datasets.
Naik et al. [
11] presented a method for automated histopathology images. They have demonstrated the utility of glandular and nuclear segmentation algorithm in accurate extraction of various morphological and nuclear features for automated grading of prostate cancer, breast cancer, and distinguishing between cancerous and benign breast histology specimen. The authors used a SVM classifier for classification of prostate images containing 16 Gleason grade 3 images, 11 grade 4 images, and 17 benign epithelial images of biopsy tissue. They achieved an accuracy of 95.19% for grade 3 vs. grade 4, 86.35% for grade 3 vs. benign, and 92.90% for grade 4 vs. benign.
Nguyen et al. [
12] introduced a novel approach to grade prostate malignancy using digitized histopathological specimens of the prostate tissue. They have extracted tissue structural features from the gland morphology and co-occurrence texture features from 82 regions of interest (ROI) with 620 × 550 pixels to classify a tissue pattern into three major categories: benign, grade 3 carcinoma, and grade 4 carcinoma. The authors proposed a hierarchical (binary) classification scheme and obtained 85.6% accuracy in classifying an input tissue pattern into one of the three classes.
Albashish et al. [
13] proposed some texture features, namely Haralick, Histogram of Oriented Gradient (HOG), and run-length matrix, which have been extracted from nuclei and lumen images individually. They used a total of 149 images with 4140 × 3096 pixels, and the dataset was randomly divided into 50% for training and 50% for testing. An ensemble machine learning classification system was proposed, and achieved an accuracy of 88.9% for Grade 3 vs. Grade 4, 92.4% for benign vs. Grade 4, and 97.85% for benign vs. Grade 3. These accuracies were averaged over 50 simulation runs and statistical significance.
Diamond et al. [
14] used morphological and texture features to classify the sub-region of 100 × 100 pixels and subjected each to image-processing techniques. They classified a tissue image into either stroma or prostatic carcinoma. In addition, the authors used lumen area to discriminate benign tissue from the other two classes. As a result, 79.3% of sub-regions were correctly classified.
Ding et al. [
15] introduced an automated image analysis framework capable of efficiently segmenting microglial cells from histology images and analyzing their morphology. Their experiments show that the proposed framework is accurate and scalable for large datasets. They extracted three types of features for SVM classification, namely Mono-fractal, Multi-fractal, and Gabor features.
Yang et al. [
16] used image processing and machine learning algorithms to analyze the smear images captured by the developed image-based cytometer. A low-cost, portable image-based cytometer was built for image acquisition from Giemsa stained blood smear. The authors selected 50 images manually for the training set, out of these, 25 images were parasites and 25 images were non-parasites. The selected images were then segmented separately to extract the features for Support Vector Machine (SVM) classification, and they used linear kernel classifier to train and test these features.
4. Results and Discussion
Quantitative analysis was performed on each cancerous image based on the four prostate cancer tissue groups (Grade 3, Grade 4, Grade 5, and Benign). We implemented the proposed method using MATLAB R2018a. We performed data analysis to analyze the components of the nuclei, which were segmented from prostate tissue images.
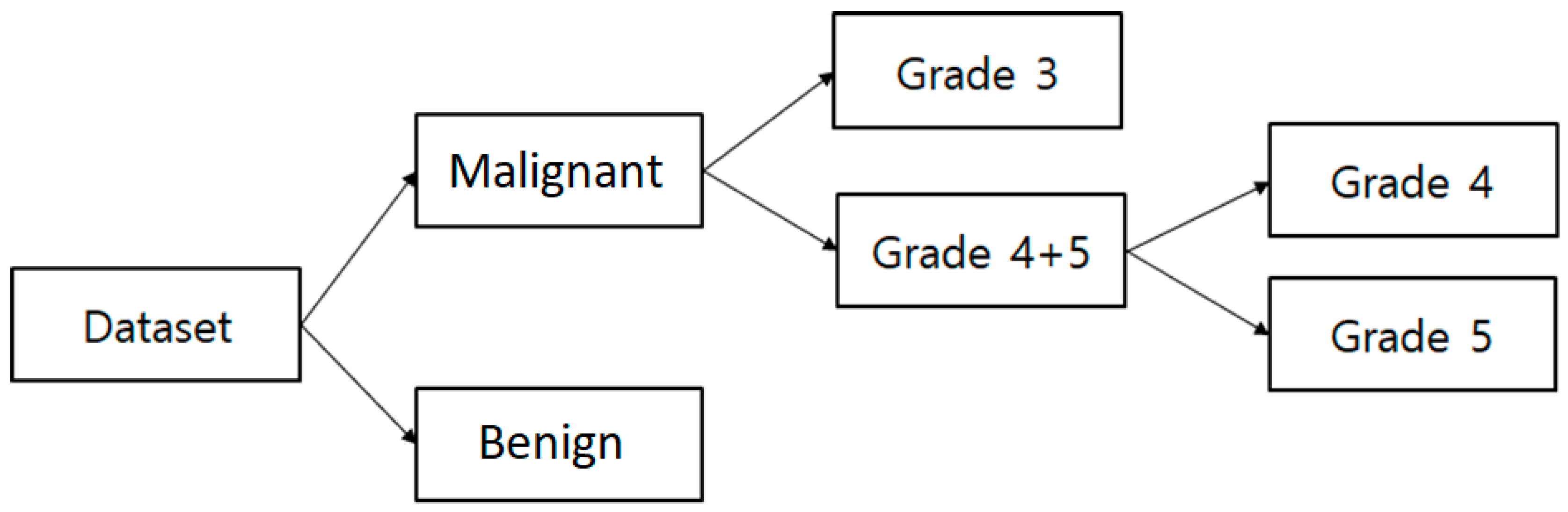
In this paper, 400 images were used in total. Of these, 240 were used for training and 160 were used for testing. The number of images considered for each group was 100, and these were classified as malignant vs. benign, Grade 3 vs. Grade 4+5, and Grade 4 vs. Grade 5. Each image was 24-bits/pixel with a size of 512 × 512 pixels. All of the possible results are shown in
Table 2,
Table 3 and
Table 4, where we show the confusion matrices of SVM binary classification for training and testing separately.
Table 2,
Table 3 and
Table 4 show the confusion matrices used to evaluate the performance of machine learning algorithms and the classifiers on a set of train and test data. We have shown these confusion matrix tables to get a better idea about the errors of a classification model. Each one of these tables is divided into two parts to show the correctly classified and misclassified data with respect to the training and testing process respectively.
In
Table 5, we used four types of performance metrics, namely, accuracy, sensitivity, specificity, and Matthews’s correlation coefficient (MCC). These metrics were calculated using our confusion matrices, i.e., true positive (TP), true negative (TN), false positive (FP), and false negative (FN). We multiplied the accuracy by 100% to normalize it with respect to the other measurements. The four types of performance metrics used in
Table 5 are explained as follow,
Accuracy is measure of the proportion of correctly classified samples.
Sensitivity is a measure of the proportion of positive correctly classified samples.
Specificity is a measure of the proportion of negative correctly classified samples.
Matthew’s correlation coefficient (MCC) is the eminence of binary class classification. It is a correlation coefficient between target and predictions.
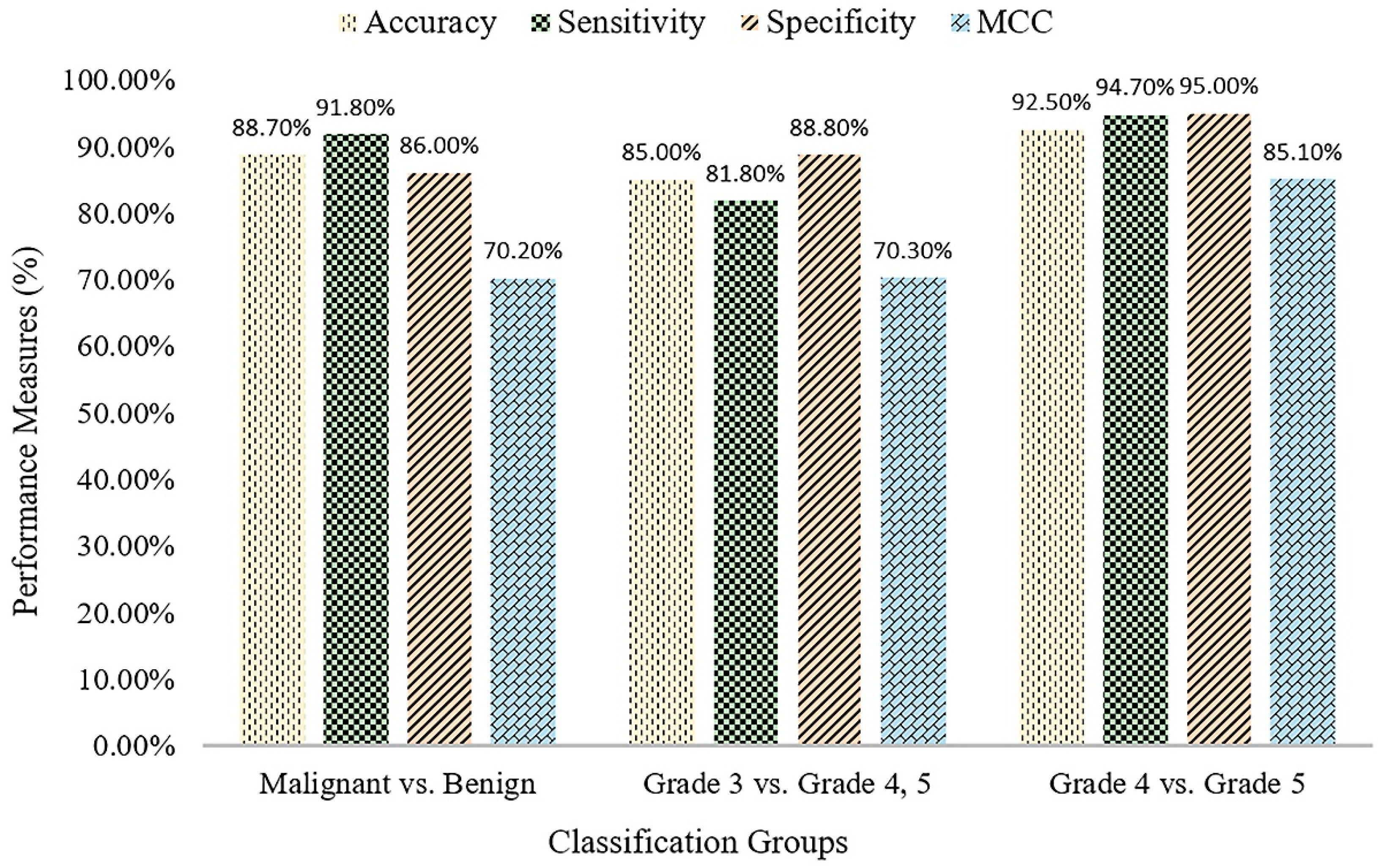
Table 5 shows the classification results of the proposed method for three different groups. The SVM binary classification accuracy, sensitivity, specificity, and MCC for malignant vs. benign are 88.7%, 91.8%, 86.0%, and 70.2%, respectively. For Grade 3 vs. Grade 4+5, the classification accuracy, sensitivity, specificity, and MCC are 85.0%, 81.8%, 88.8%, and 70.3, respectively. For Grade 4 vs. Grade 5, the classification accuracy, sensitivity, specificity, and MCC are 92.5%, 94.7%, 95.0%, and 85.1, respectively.
For the purpose of validation, we also performed prostate cancer grading classification using multilayer perceptron (MLP) technique in Weka, shown in
Table 6. MLP is a class of feed-forward artificial neural network, which consists of at least three layers of node: an input layer, hidden layer, and an output layer. Each node is a neuron except input nodes and uses a non-linear activation function. MLP utilizes a supervised learning technique like SVM. From the results shown in
Table 5 and
Table 6, we can see that the proposed SVM binary classification works significantly better than MLP, and the highest accuracy obtained was 92.5%, for Grade 4 vs. Grade 5. First, classification was performed to detect cancer in all of the samples in the dataset. The second and third classification was performed within the cancer group for low- and high-grade cancer detection. In
Figure 7, the bar graph shows the comparison results for the three different binary divisions that are used for SVM classification.
To predict, automatically, prostate cancer gradings, we used machine learning and deep learning algorithms such as SVM and MLP, respectively. To do so, we first applied image segmentation as a preprocessing step. Secondly, we converted the images from RGB to binary to carry out watershed segmentation. Thirdly, we calculated a set of morphological features based on the segmented nucleus and lumen tissue images. Finally, the SVM and MLP classification was performed based on the significant features selected.
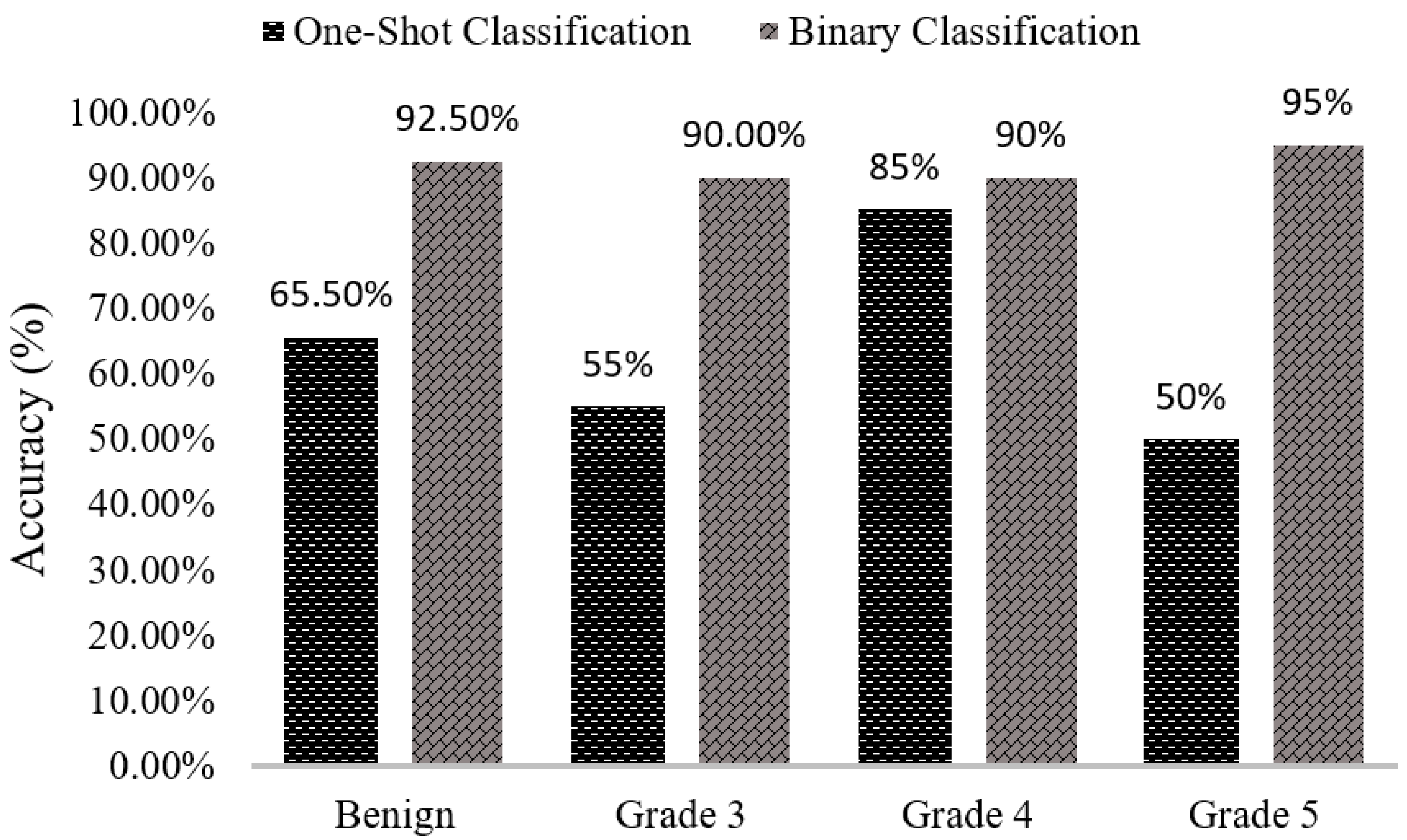
We can see that the results of the comparison between SVM classification accuracy in
Table 7 and
Figure 8 vary between one-shot and binary classifiers. When we classified our data using multi-class or one-shot classifiers, the classification accuracies for benign, Grade 3, Grade 4, and Grade 5 are 60%, 55%, 85%, and 50%, respectively. Using the proposed binary classification approach, the accuracies for the same groups are 92.5%, 90.0%, 90.0%, and 95.0%, respectively. Comparing both classifiers simultaneously, we can see that the results obtained using the binary classifier are better than those obtained using multi-class or one-shot classifier.
Table 8 shows the comparison results of MLP classifier between one-shot and binary classification. After comparing the results between SVM and MLP classification methods, we can say that the proposed method, SVM, achieved better results than MLP. In one-shot classification, the entire dataset is classified into four groups simultaneously. In this case, the errors in one class affect the performance of the others, negatively impacting the classification accuracy. Thus, the model cannot make correct predictions. Whereas, in binary classification, the entire dataset is separated into three groups and each group is classified separately and independently. In this case, the errors in one class do not affect the performance of the other class.
In
Table 9, we compare the accuracy of different standard classification methods with our proposed method. The classification accuracy achieved for the class low vs. high grade using the proposed method is higher than other methods described in the literature. On cancer diagnosis, when classified Malignant vs. Benign, our result is better than Nir et al. (2018) and Doyle et al. (2006), but not higher compared to Tabesh et al. (2017), because they used different types of features that are extracted from the tissue image, namely color channel histogram, fractal dimension, fractal code, wavelet, and MAGIC. The authors of Reference [
4] computed the features of epithelial nuclei objects in the tissue image, whereas, our method computed the features of all nuclei objects existing in the biopsy prostate tissue image.
5. Conclusions
In this study, we have developed a computerized grading system for digitized histopathology images using supervised learning methods. The segmentation process for biopsy tissue image was performed using the k-means algorithm and touching cells were separated using the watershed algorithm. Morphological features were selected for prostate cancer grading and diagnosis. Gaussian and linear kernels were used for the classification of prostate histopathological images. Using these kernels, we observed some improvements in the results, and gradually increased the performance of the model used for training and testing. The parameters of the kernel play a vital role in the classification process, and the best combination of and was selected for better classification accuracy. Satisfactory classification results were obtained using the extracted morphological features, and these features were extracted from the sub-images, viewable in 40× magnification. The quantitative analysis described here is remarkably flexible in terms of implementation. The SVM binary classification method presented in this paper is used to classify malignant vs. benign, Grade 3 vs. Grade 4+5, and Grade 4 vs. Grade 5. Our results are satisfactory and comparable with those reported in the literature and produced quantitative measures based on the features extracted from microscopic biopsy tissue images. In order to justify our proposed method, SVM, we also carried out features classification using MLP. One-shot and binary classification results were compared to show the differences in two classifications accuracies. In future studies, we will improve our classification accuracy using the combinations of multiple features. Deep learning and machine learning techniques will be used for comparative analysis, where, image classification will be performed using the convolutional neural network (CNN) and feature classification will be performed using support vector machine (SVM), respectively.