Biopurification of Oligosaccharides by Immobilized Kluyveromyces Lactis
Abstract
1. Introduction
2. Materials and Methods
2.1. Culture Conditions for the Growth of Kluyveromyces lactis
2.2. Immobilization of K. lactis with Sodium Alginate
2.3. The Extraction of Semi-Purified Human Milk Oligosaccharides (HMOs) from Human Milk
2.4. Analysis of Mono-, Di-, and Oligosaccharides
3. Results
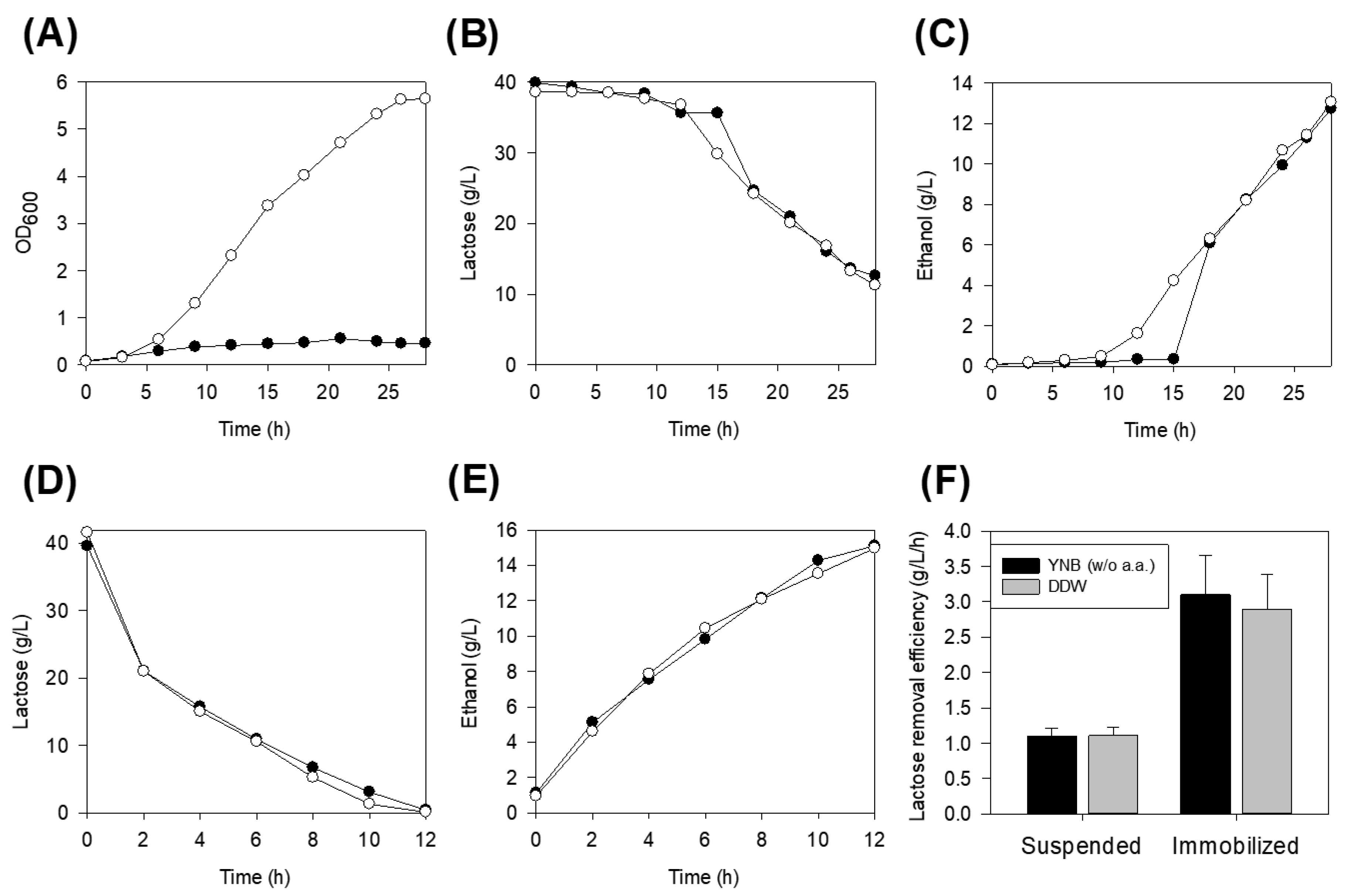
3.1. Sugar Utilization in the Nutrient-Rich and Defined Salt Media
3.2. Lactose Removal on Defined Salt Media and Distilled Water
3.3. Effect of Immobilization
3.4. Impact of Pre-Activation of Immobilized Cells
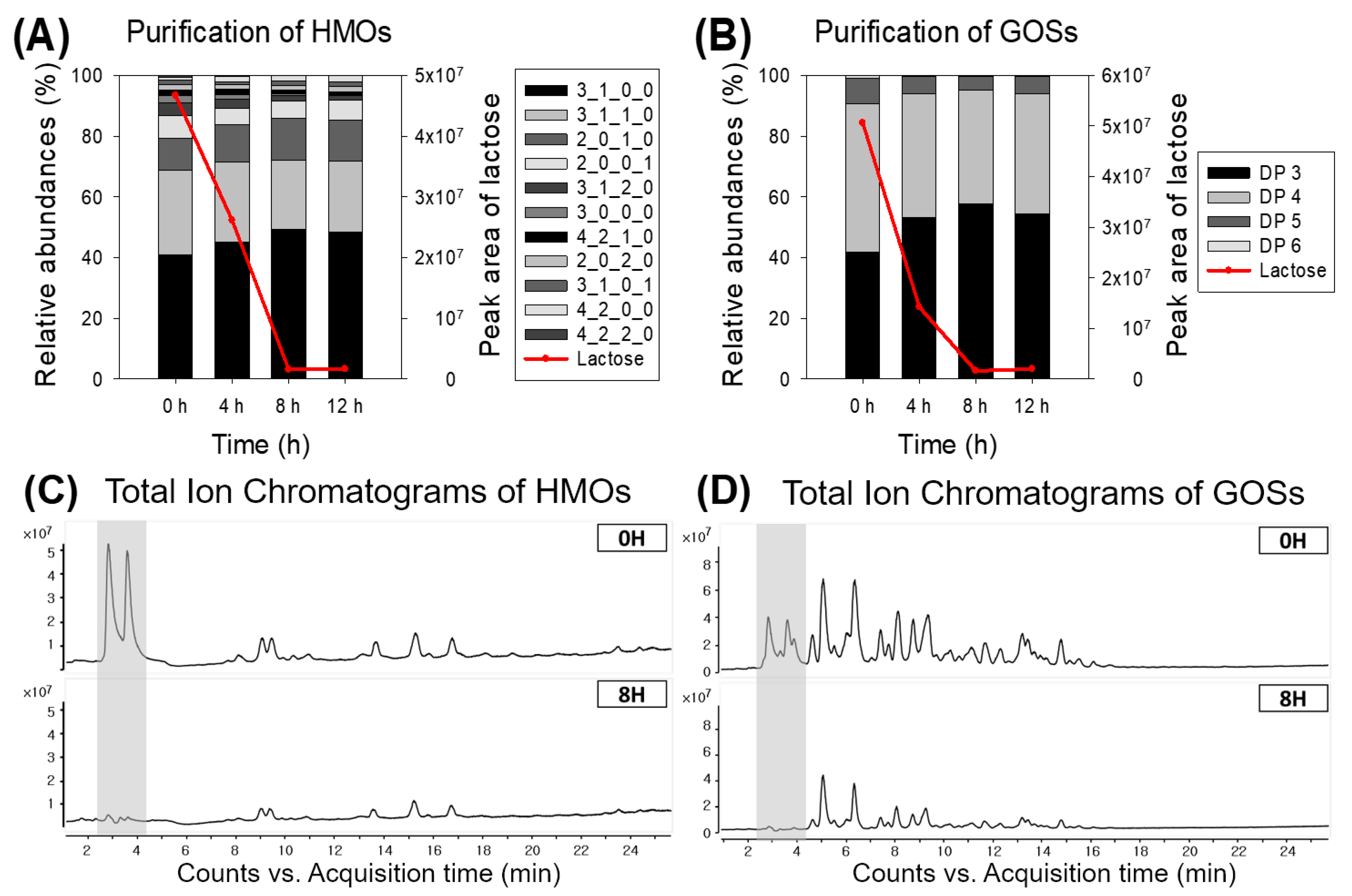
3.5. Lactose Removal of Human Milk Oligosaccharides (HMOs) and Galactooligosaccharides (GOSs)
4. Discussion
Author Contributions
Funding
Conflicts of Interest
References
- Smilowitz, J.T.; Lebrilla, C.B.; Mills, D.A.; German, J.B.; Freeman, S.L. Breast milk oligosaccharides: Structure-function relationships in the neonate. Annu. Rev. Nutr. 2014, 34, 143–169. [Google Scholar] [CrossRef] [PubMed]
- Fong, B.; Ma, K.; McJarrow, P. Quantification of bovine milk oligosaccharides using liquid chromatography-selected reaction monitoring-mass spectrometry. J. Agric. Food Chem. 2011, 59, 9788–9795. [Google Scholar] [CrossRef] [PubMed]
- Engfer, M.B.; Stahl, B.; Finke, B.; Sawatzki, G.; Daniel, H. Human milk oligosaccharides are resistant to enzymatic hydrolysis in the upper gastrointestinal tract. Am. J. Clin. Nutr. 2000, 71, 1589–1596. [Google Scholar] [CrossRef] [PubMed]
- Gnoth, M.J.; Kunz, C.; Kinne-Saffran, E.; Rudloff, S. Human milk oligosaccharides are minimally digested in vitro. J. Nutr. 2000, 130, 3014–3020. [Google Scholar] [CrossRef] [PubMed]
- Bode, L. Human milk oligosaccharides: Every baby needs a sugar mama. Glycobiology 2012, 22, 1147–1162. [Google Scholar] [CrossRef] [PubMed]
- Zivkovic, A.M.; Barile, D. Bovine milk as a source of functional oligosaccharides for improving human health. Adv. Nutr. 2011, 2, 284–289. [Google Scholar] [CrossRef] [PubMed]
- Mehra, R.; Barile, D.; Marotta, M.; Lebrilla, C.B.; Chu, C.; German, J.B. Novel high-molecular weight fucosylated milk oligosaccharides identified in dairy streams. PLoS ONE 2014, 9, e96040. [Google Scholar] [CrossRef] [PubMed]
- Smilowitz, J.T.; Lemay, D.G.; Kalanetra, K.M.; Chin, E.L.; Zivkovic, A.M.; Breck, M.A.; German, J.B.; Mills, D.A.; Slupsky, C.; Barile, D. Tolerability and safety of the intake of bovine milk oligosaccharides extracted from cheese whey in healthy human adults. J. Nutr. Sci. 2017, 6, e6. [Google Scholar] [CrossRef]
- Barile, D.; Meyrand, M.; Lebrilla, C.B.; German, J.B. Examining bioactive components of milk Sources of complex oligosaccharides (Part 2). Agro. Food Ind. Hi Tech. 2011, 22, 37–39. [Google Scholar]
- Morr, C.V.; Brandon, S.C. Membrane fractionation processes for removing 90% to 95% of the lactose and sodium from skim milk and for preparing lactose and sodium-reduced skim milk. J. Food Sci. 2008, 73, C639–C647. [Google Scholar] [CrossRef]
- Geisser, A.; Hendrich, T.; Boehm, G.; Stahl, B. Separation of lactose from human milk oligosaccharides with simulated moving bed chromatography. J. Chromatogr. A 2005, 1092, 17–23. [Google Scholar] [CrossRef] [PubMed]
- Vente, J.A.; Bosch, H.; de Haan, A.B.; Bussmann, P.J.T. Comparison of sorption isotherms of mono- and disaccharides relevant to oligosaccharide separations for Na, K, and Ca loaded cation exchange resins. Chem. Eng. Commun. 2005, 192, 23–33. [Google Scholar] [CrossRef]
- Strum, J.S.; Aldredge, D.; Barile, D.; Lebrilla, C.B. Coupling flash liquid chromatography with mass spectrometry for enrichment and isolation of milk oligosaccharides for functional studies. Anal. Biochem. 2012, 424, 87–96. [Google Scholar] [CrossRef]
- Robinson, R.C.; Colet, E.; Tian, T.; Poulsen, N.A.; Barile, D. An improved method for the purification of milk oligosaccharides by graphitised carbon-solid phase extraction. Int. Dairy J. 2018, 80, 62–68. [Google Scholar] [CrossRef] [PubMed]
- Santibanez, L.; Fernandez-Arrojo, L.; Guerrero, C.; Plou, F.J.; Illanes, A. Removal of lactose in crude galacto-oligosaccharides by beta-galactosidase from Kluyveromyces lactis. J. Mol. Catal. B-Enzym. 2016, 133, 85–91. [Google Scholar] [CrossRef]
- Yu, S.; Liu, J.J.; Yun, E.J.; Kwak, S.; Kim, K.H.; Jin, Y.S. Production of a human milk oligosaccharide 2’-fucosyllactose by metabolically engineered Saccharomyces cerevisiae. Microb. Cell. Fact. 2018, 17, 101. [Google Scholar] [CrossRef]
- Hollands, K.; Baron, C.M.; Gibson, K.J.; Kelly, K.J.; Krasley, E.A.; Laffend, L.A.; Lauchli, R.M.; Maggio-Hall, L.A.; Nelson, M.J.; Prasad, J.C.; et al. Engineering two species of yeast as cell factories for 2’-fucosyllactose. Metab. Eng. 2019, 52, 232–242. [Google Scholar] [CrossRef]
- Chin, Y.W.; Seo, N.; Kim, J.H.; Seo, J.H. Metabolic engineering of Escherichia coli to produce 2’-fucosyllactose via salvage pathway of guanosine 5’-diphosphate (GDP)-l-fucose. Biotechnol. Bioeng. 2016, 113, 2443–2452. [Google Scholar] [CrossRef]
- Chin, Y.W.; Kim, J.Y.; Kim, J.H.; Jung, S.M.; Seo, J.H. Improved production of 2’-fucosyllactose in engineered Escherichia coli by expressing putative alpha-1,2-fucosyltransferase, WcfB from Bacteroides fragilis. J. Biotechnol. 2017, 257, 192–198. [Google Scholar] [CrossRef]
- Rycroft, C.E.; Jones, M.R.; Gibson, G.R.; Rastall, R.A. A comparative in vitro evaluation of the fermentation properties of prebiotic oligosaccharides. J. Appl. Microbiol. 2001, 91, 878–887. [Google Scholar] [CrossRef]
- Lamsal, B.P. Production, health aspects and potential food uses of dairy prebiotic galactooligosaccharides. J. Sci. Food Agric. 2012, 92, 2020–2028. [Google Scholar] [CrossRef] [PubMed]
- Depeint, F.; Tzortzis, G.; Vulevic, J.; I’Anson, K.; Gibson, G.R. Prebiotic evaluation of a novel galactooligosaccharide mixture produced by the enzymatic activity of Bifidobacterium bifidum NCIMB 41171, in healthy humans: A randomized, double-blind, crossover, placebo-controlled intervention study. Am. J. Clin. Nutr. 2008, 87, 785–791. [Google Scholar] [CrossRef] [PubMed]
- Moreno, F.J.; Montilla, A.; Villamiel, M.; Corzo, N.; Olano, A. Analysis, structural characterization, and bioactivity of oligosaccharides derived from lactose. Electrophoresis 2014, 35, 1519–1534. [Google Scholar] [CrossRef]
- Urrutia, P.; Rodriguez-Colinas, B.; Fernandez-Arrojo, L.; Ballesteros, A.O.; Wilson, L.; Illanes, A.; Plou, F.J. Detailed analysis of galactooligosaccharides synthesis with beta-galactosidase from Aspergillus oryzae. J. Agric. Food Chem. 2013, 61, 1081–1087. [Google Scholar] [CrossRef] [PubMed]
- Crittenden, R.G.; Playne, M.J. Purification of food-grade oligosaccharides using immobilised cells of Zymomonas mobilis. Appl. Microbiol. Biotechnol. 2002, 58, 297–302. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.C.; Yu, M.C.; Cheng, T.C.; Sheu, D.C.; Duan, K.J.; Tai, W.L. Production of high-content galacto-oligosaccharide by enzyme catalysis and fermentation with Kluyveromyces marxianus. Biotechnol. Lett. 2006, 28, 793–797. [Google Scholar] [CrossRef] [PubMed]
- Spohner, S.C.; Schaum, V.; Quitmann, H.; Czermak, P. Kluyveromyces lactis: An emerging tool in biotechnology. J. Biotechnol. 2016, 222, 104–116. [Google Scholar] [CrossRef]
- Gnoth, M.J.; Rudloff, S.; Kunz, C.; Kinne, R.K. Investigations of the in vitro transport of human milk oligosaccharides by a Caco-2 monolayer using a novel high performance liquid chromatography-mass spectrometry technique. J. Biol. Chem. 2001, 276, 34363–34370. [Google Scholar] [CrossRef]

| Media | Substrate | Maximum OD600 (AU) | μmax (h−1) | Substrate Consumption 1 (g/L) | Ethanol Production 1 (g/L) |
|---|---|---|---|---|---|
| YPD | Glucose | 7.9 | 0.33 | 20.0 g/L | 10.0 g/L |
| Galactose | 5.7 | 0.20 | 11.0 g/L | 4.2 g/L | |
| Lactose | 6.9 | 0.31 | 20.0 g/L | 12.0 g/L | |
| YNB (with amino acids) | Glucose | 5.4 | 0.31 | 17.0 g/L | 7.3 g/L |
| Galactose | 3.5 | 0.29 | 6.8 g/L | 3.5 g/L | |
| Lactose | 5.1 | 0.31 | 19.0 g/L | 9.3 g/L | |
| YNB (without amino acids) | Lactose | 5.0 | 0.27 | 14.0 g/L | 9.5 g/L |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yeo, I.-S.; Yoon, Y.-J.; Seo, N.; An, H.J.; Kim, J.-H. Biopurification of Oligosaccharides by Immobilized Kluyveromyces Lactis. Appl. Sci. 2019, 9, 2845. https://doi.org/10.3390/app9142845
Yeo I-S, Yoon Y-J, Seo N, An HJ, Kim J-H. Biopurification of Oligosaccharides by Immobilized Kluyveromyces Lactis. Applied Sciences. 2019; 9(14):2845. https://doi.org/10.3390/app9142845
Chicago/Turabian StyleYeo, In-Seok, Yeo-Jin Yoon, Nari Seo, Hyun Joo An, and Jae-Han Kim. 2019. "Biopurification of Oligosaccharides by Immobilized Kluyveromyces Lactis" Applied Sciences 9, no. 14: 2845. https://doi.org/10.3390/app9142845
APA StyleYeo, I.-S., Yoon, Y.-J., Seo, N., An, H. J., & Kim, J.-H. (2019). Biopurification of Oligosaccharides by Immobilized Kluyveromyces Lactis. Applied Sciences, 9(14), 2845. https://doi.org/10.3390/app9142845







