Hyperspectral Reflectance Imaging Combined with Multivariate Analysis for Diagnosis of Sclerotinia Stem Rot on Arabidopsis Thaliana Leaves
Abstract
Featured Application
Abstract
1. Introduction
2. Materials and Methods
2.1. A. thaliana Cultivation and Pathogen Inoculation
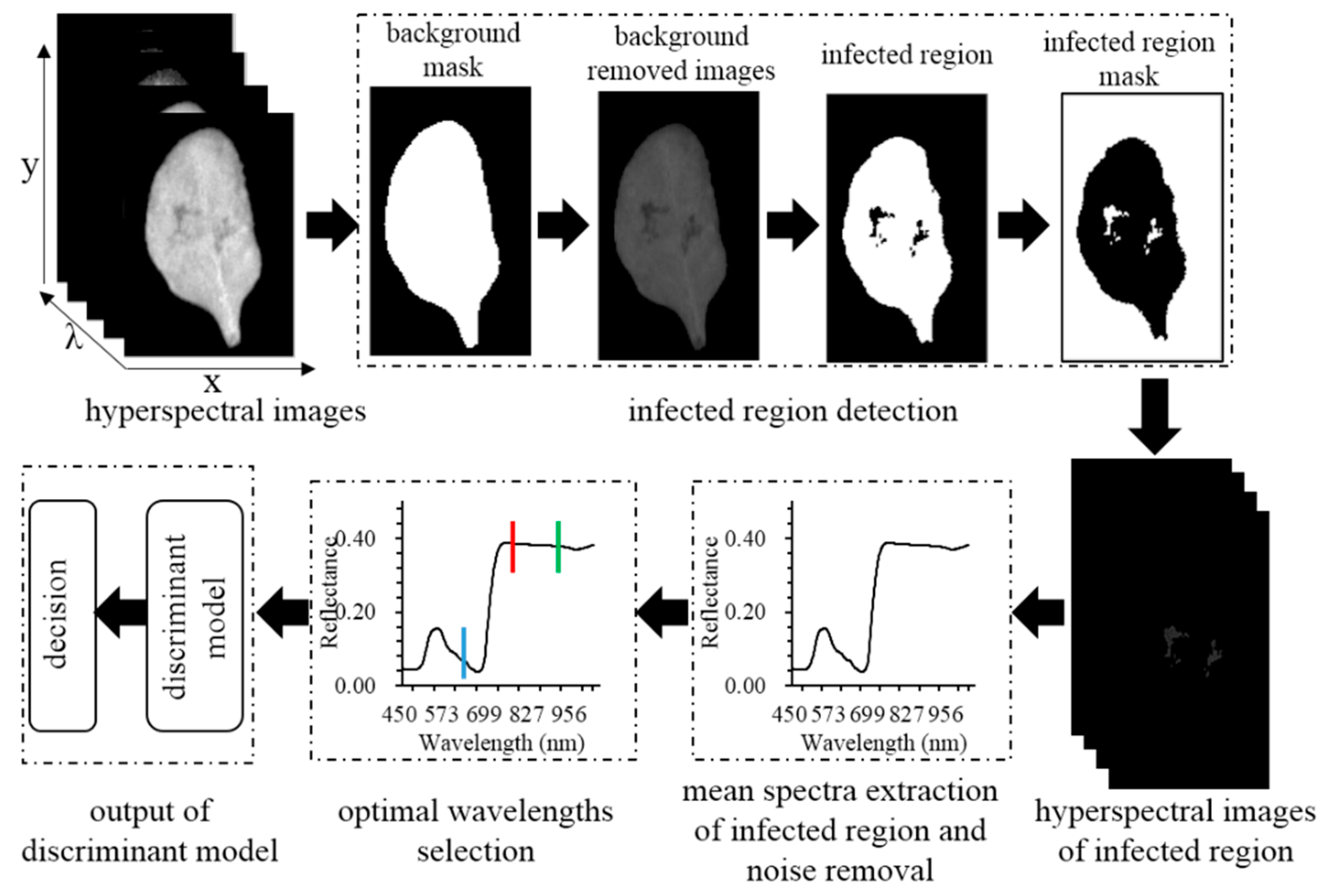
2.2. Hyperspectral Images Acquisition and Processing
2.3. Spectral and Textural Features Selection for Disease Detection
2.4. Discriminant Analysis Method for Disease Detection
3. Results and Discussion
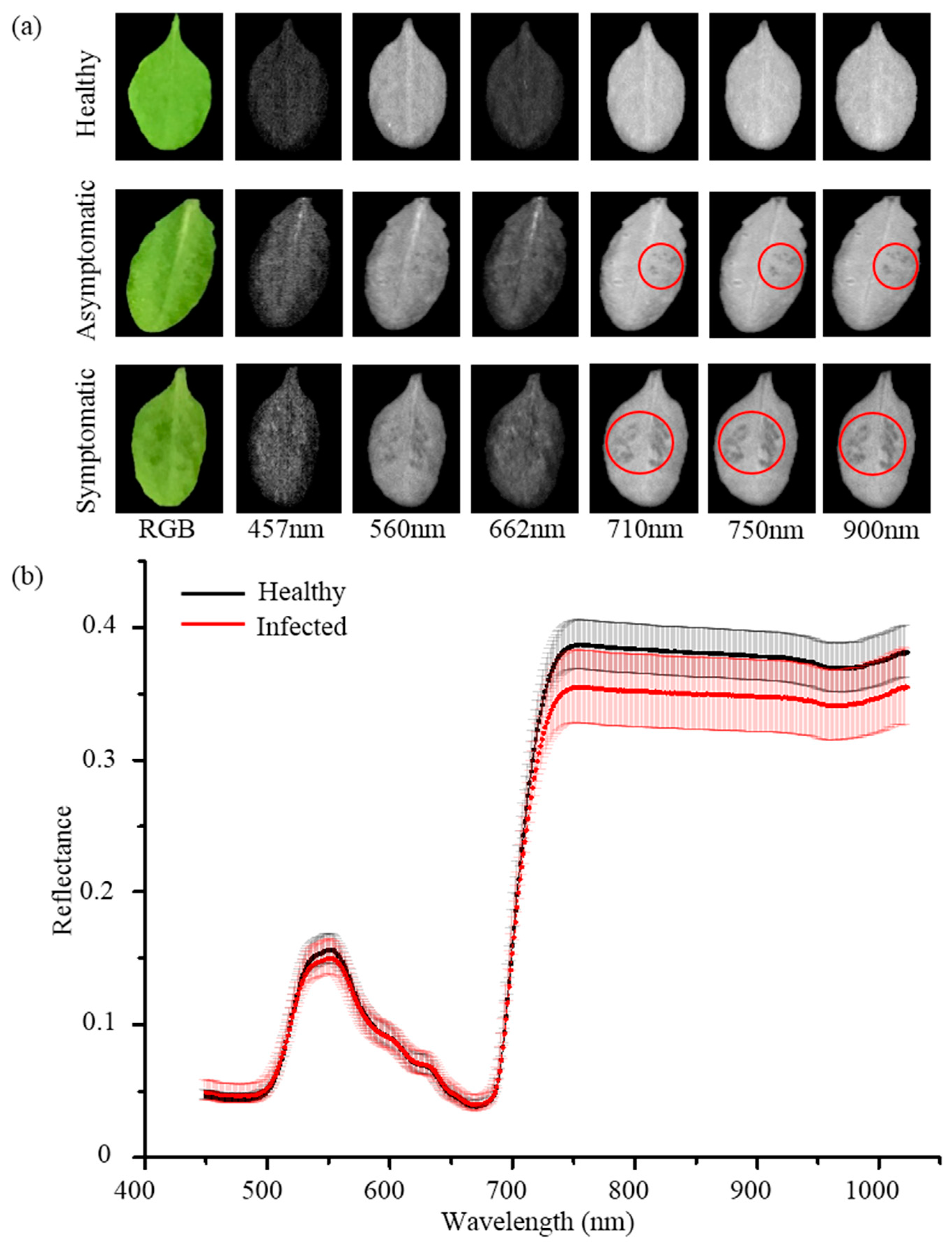
3.1. Analysis of Hyperspectral Reflectance
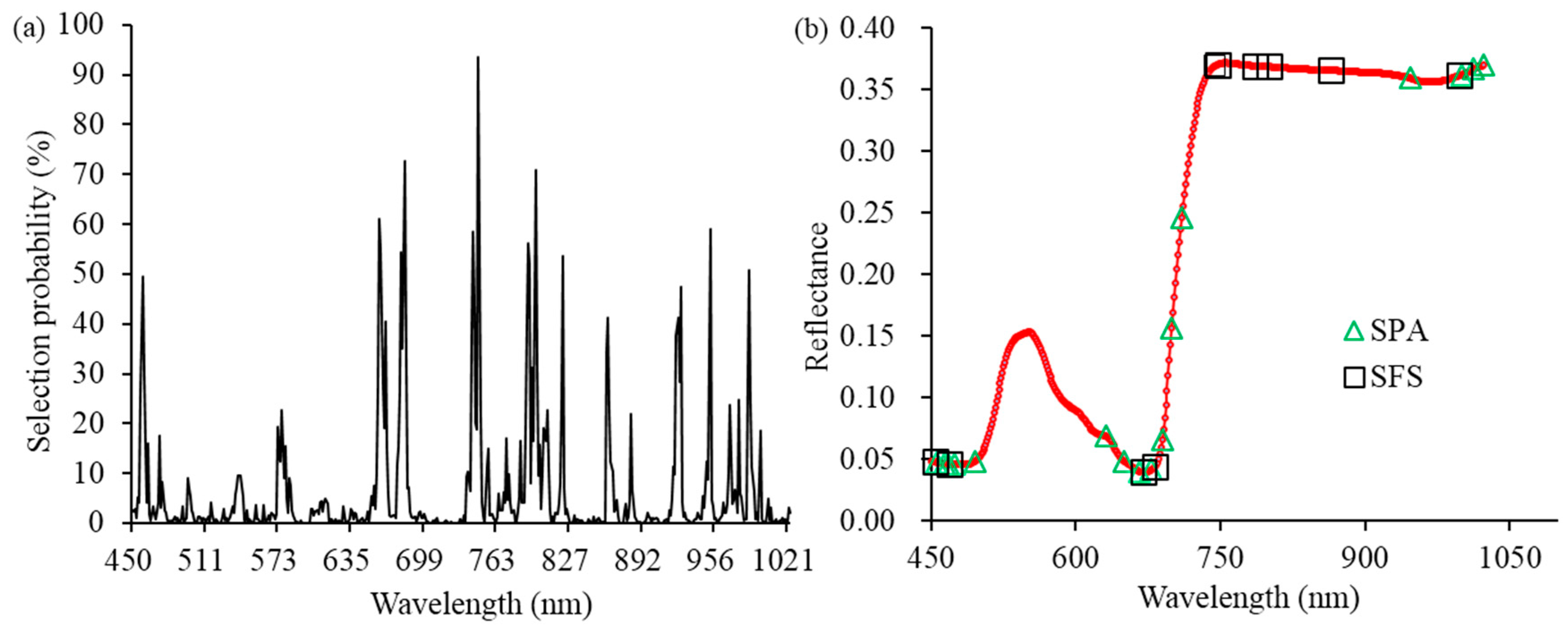
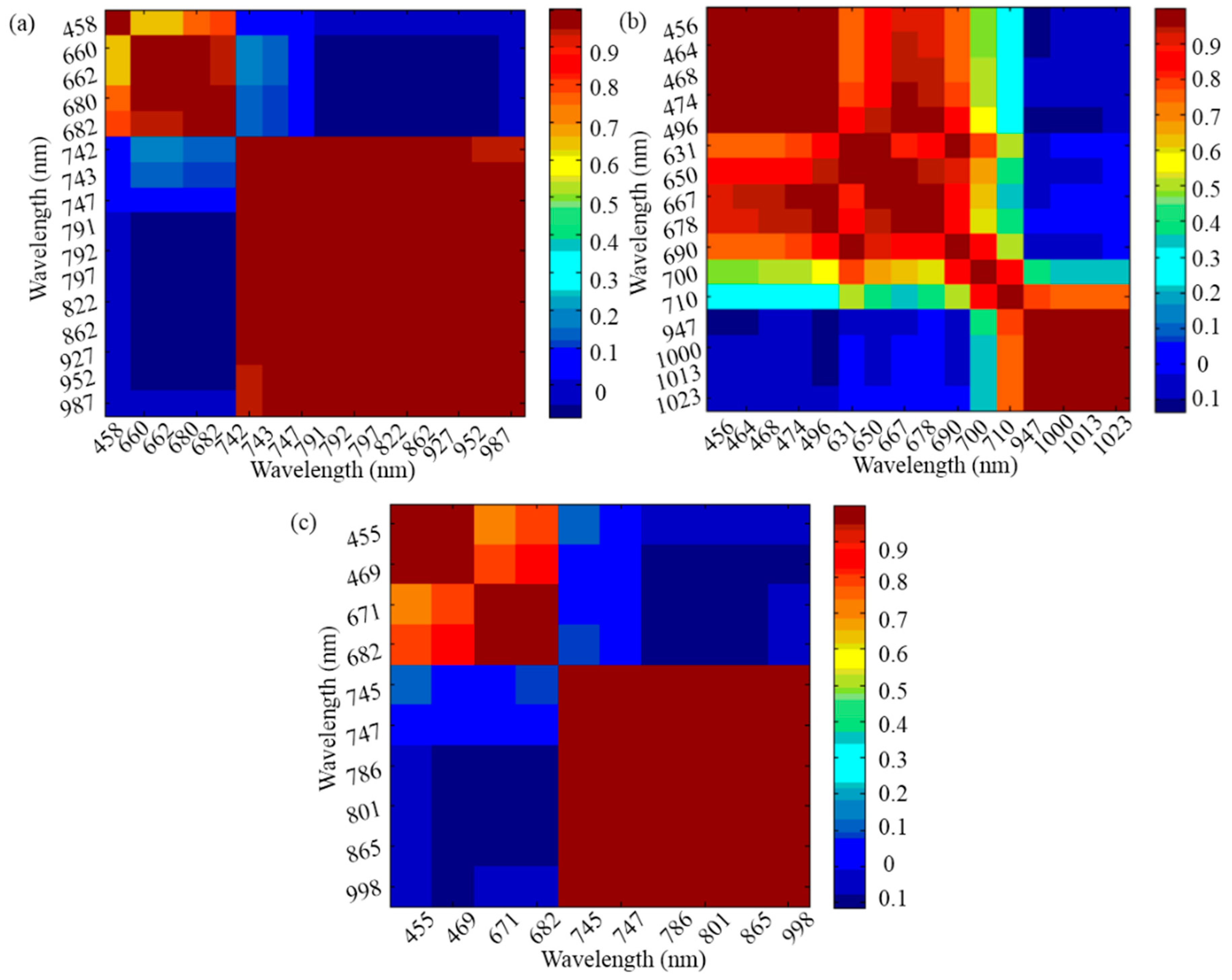
3.2. Optimal Wavelengths Selection for Disease Detection of A. thaliana Leaves
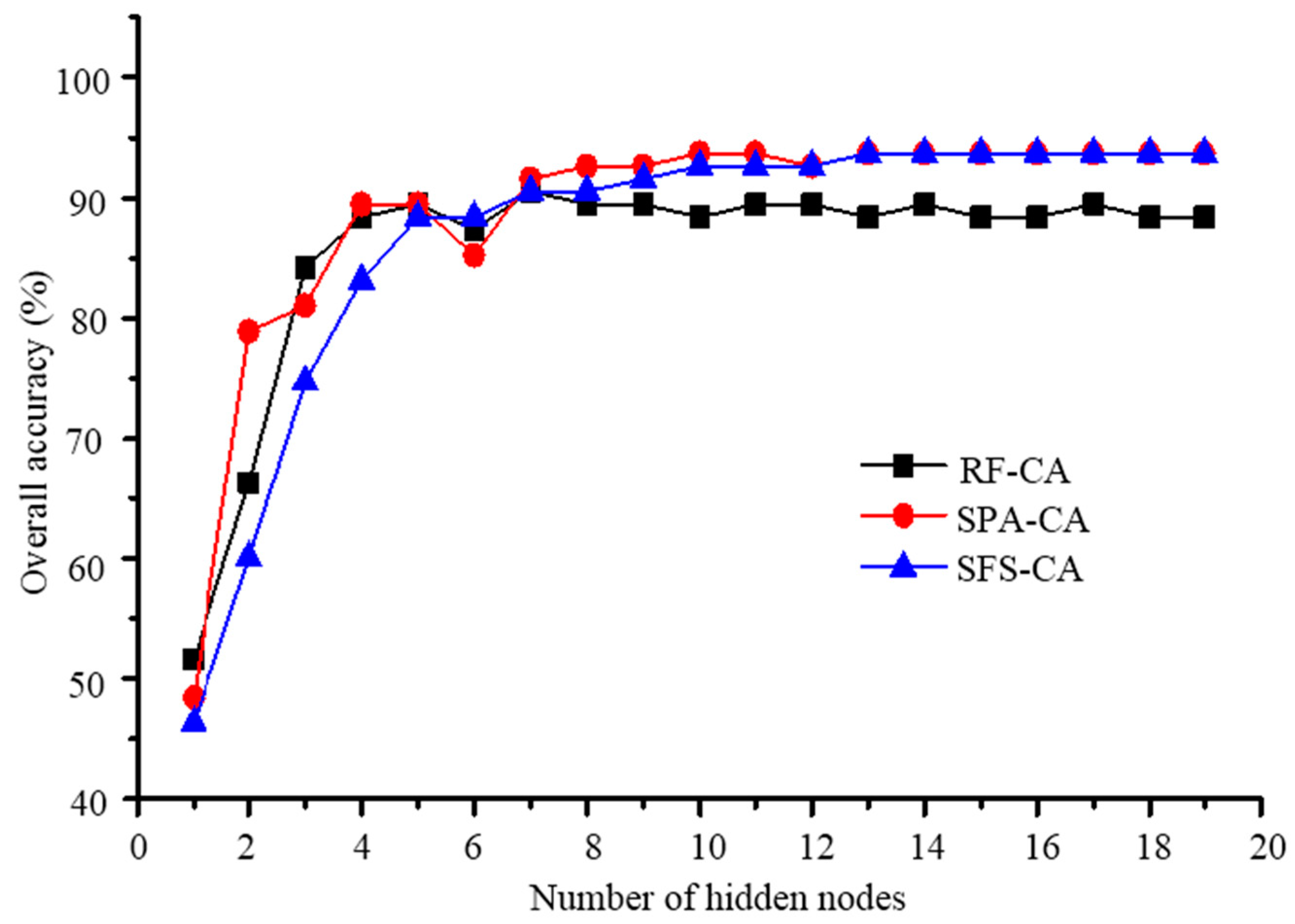
3.3. SSR Detection Based on Optimal Features
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Duan, Y.; Li, T.; Xiao, X.; Wu, J.; Li, S.; Wang, J.; Zhou, M. Pharmacological characteristics of the novel fungicide pyrisoxazole against Sclerotinia sclerotiorum. Pestic. Biochem. Physiol. 2018, 149, 61–66. [Google Scholar] [CrossRef] [PubMed]
- Melo, I.; Moretini, A.; Cassiolato, A.; Faull, J. Development of mutants of coniothyrium minitans with improved efficiency for control of Sclerotinia sclerotiorum. J. Plant Protect. Res. 2011, 51, 179–183. [Google Scholar] [CrossRef]
- Bardin, S.D.; Huang, H.C. Research on biology and control of Sclerotinia diseases in Canada. Can. J. Plant Pathol. 2001, 23, 88–98. [Google Scholar] [CrossRef]
- Kwon, J.H.; Choi, S.L.; Lee, H.S.; Shim, H.S. Stem rot of perilla caused by sclerotium rolfsii in Korea. Korean J. Mycol. 2012, 40, 177–178. [Google Scholar] [CrossRef]
- Lopez, M.M.; Bertolini, E.; Olmos, A.; Caruso, P.; Gorris, M.T.; Llop, P.; Penyalver, R.; Cambra, M. Innovative tools for detection of plant pathogenic viruses and bacteria. Int. Microbiol. 2003, 6, 233–243. [Google Scholar] [CrossRef]
- Sannakki, S.S.; Rajpurohit, V.S.; Nargund, V.B.; Arun, K.R. Leaf disease grading by machine vision and fuzzy logic. Int. J. Comput. Technol. Appl. 2011, 2, 1709–1716. [Google Scholar]
- Chaerle, L.; Hagenbeek, D.; Bruyne, E.D.; Straeten, D.V.D. Chlorophyll fluorescence imaging for disease-resistance screening of sugar beet. Plant Cell Tissue Organ Cult. 2007, 91, 97–106. [Google Scholar] [CrossRef]
- Xie, C.; Shao, Y.; Li, X.; He, Y. Detection of early blight and late blight diseases on tomato leaves using hyperspectral imaging. Sci. Rep. 2015, 5, 16564. [Google Scholar] [CrossRef]
- Oerke, E.C.; Herzog, K.; Toepfer, R. Hyperspectral phenotyping of the reaction of grapevine genotypes toPlasmopara viticola. J. Exp. Bot. 2016, 67, 5529–5543. [Google Scholar] [CrossRef]
- Weng, H.Y.; Lv, J.W.; Cen, H.Y.; He, M.B.; Zeng, Y.B.; Hua, S.J.; Li, H.Y.; Meng, Y.Q.; Fang, H.; He, Y. Hyperspectral reflectance imaging combined with carbohydrate metabolism analysis for diagnosis of citrus Huanglongbing in different seasons and cultivars. Sens. Actuators B 2018, 275, 50–60. [Google Scholar] [CrossRef]
- Zhou, R.Q.; Jin, J.J.; Li, Q.M.; Su, Z.Z.; Yu, X.J.; Tang, Y.; Luo, S.M.; He, Y.; Li, X.L. Early detection of Magnaporthe oryzae-infected Barley leaves and lesion visualization based on hyperspectral imaging. Front. Plant Sci. 2019, 9, 13. [Google Scholar] [CrossRef]
- Kong, W.W.; Yu, J.J.; Liu, F.; He, Y.; Bao, Y.D. Early diagnosis of gray mold on tomato stalks based on hyperspectral data. Spectrosc. Spectr. Anal. 2013, 33, 733–736. [Google Scholar]
- Xie, C.Q.; He, Y.; Li, X.L.; Liu, F.; Du, P.P.; Feng, L. Study of detection of SPAD value in tomato leaves stressed by grey mold based on hyperspectral technique. Spectrosc. Spectr. Anal. 2012, 32, 3324–3328. [Google Scholar]
- Zhang, C.; Feng, X.; Wang, J.; Liu, F.; He, Y.; Zhou, W. Mid-infrared spectroscopy combined with chemometrics to detect Sclerotinia stem rot on oilseed rape (Brassica napus L.) leaves. Plant Methods 2017, 13, 9. [Google Scholar] [CrossRef]
- Kong, W.; Zhang, C.; Huang, W.; Liu, F.; He, Y. Application of hyperspectral imaging to detect Sclerotinia sclerotiorum on oilseed rape stems. Sensors 2018, 18, 123. [Google Scholar] [CrossRef]
- Vala, H.J.; Baxi, A. A review on Otsu image segmentation algorithm. Int. J. Adv. Res. Comput. 2017, 2, 387–389. [Google Scholar]
- Li, H.D.; Xu, Q.S.; Liang, Y.Z. Random frog: An efficient reversible jump Markov Chain Monte Carlo-like approach for variable selection with applications to gene selection and disease classification. Anal. Chim. Acta 2012, 740, 20–26. [Google Scholar] [CrossRef] [PubMed]
- Yun, Y.H.; Li, H.D.; Wood, L.R.E.; Fan, W.; Wang, J.J.; Cao, D.S.; Xu, Q.S.; Liang, Y.Z. An efficient method of wavelength interval selection based on random frog for multivariate spectral calibration. Spectrochim. Acta Part A 2013, 111, 31–36. [Google Scholar] [CrossRef]
- Liu, D.; Sun, D.W.; Zeng, X.A. Recent advances in wavelength selection techniques for hyperspectral image processing in the food industry. Food Bioprocess Technol. 2013, 7, 307–323. [Google Scholar] [CrossRef]
- Sun, K.; Geng, X.R.; Ji, L.Y. An efficient unsupervised band selection method based on an autocorrelation matrix for a hyperspectral image. Int. J. Remote Sens. 2014, 35, 7458–7476. [Google Scholar] [CrossRef]
- Kong, W.W.; Zhang, C.; Cao, F.; Liu, F.; Luo, S.M.; Tang, Y.; He, Y. Detection of Sclerotinia Stem Rot on Oilseed Rape (Brassica napus L.) Leaves Using Hyperspectral Imaging. Sensors 2018, 18, 1764. [Google Scholar] [CrossRef]
- Ravikanth, L.; Chelladurai, V.; Jayas, D.S.; White, N.D.G. Detection of Broken Kernels Content in Bulk Wheat Samples Using Near-Infrared Hyperspectral Imaging. Agric. Res. 2016, 5, 285–292. [Google Scholar] [CrossRef]
- Cen, H.; Lu, R.; Zhu, Q.; Mendoza, F. Nondestructive detection of chilling injury in cucumber fruit using hyperspectral imaging with feature selection and supervised classification. Postharvest Biol. Technol. 2016, 111, 352–361. [Google Scholar] [CrossRef]
- Zheng, Z.X.; Qi, L.; Ma, X.; Zhu, X.Y.; Wang, W.J. Grading method of rice leaf blast using hyperspectral imaging technology. Trans. Chin. Soc. Agric. Eng. 2013, 29, 138–144. [Google Scholar]
- Zhao, Y.R.; Yu, K.Q.; Li, X.L.; He, Y. Detection of fungus infection on petals of rapeseed (Brassica napus L.) using NIR hyperspectral imaging. Sci. Rep. 2016, 6, 38879. [Google Scholar] [CrossRef]
- López-Fandiño, J.; Quesada-Barriuso, P.; Heras, D.B.; Argüello, F. Efficient ELM-based techniques for the classification of hyperspectral remote sensing images on commodity GPUs. IEEE J. Sel. Top Appl. 2017, 8, 2884–2893. [Google Scholar] [CrossRef]
- Jiang, J.L.; Cen, H.Y.; Zhang, C.; Lyu, X.H.; Weng, H.Y.; Xu, H.X.; He, Y. Nondestructive quality assessment of chili peppers using near-infrared hyperspectral imaging combined with multivariate analysis. Postharvest Biol. Technol. 2018, 146, 147–154. [Google Scholar] [CrossRef]
- Bo, C.; Lu, H.; Wang, D. Spectral-spatial K-Nearest Neighbor approach for hyperspectral image classification. Multimed. Tools Appl. 2017, 77, 10419–10436. [Google Scholar] [CrossRef]
- Bandos, T.V.; Bruzzone, L.; Camps-Valls, G. Classification of hyperspectral images with regularized linear discriminant analysis. IEEE Trans. Geosci. Remote Sens. 2009, 47, 862–873. [Google Scholar] [CrossRef]
- Wei, X.; Liu, F.; Qiu, Z.; Shao, Y.; He, Y. Ripeness classification of astringent persimmon using hyperspectral imaging technique. Food Bioprocess Technol. 2014, 7, 1371–1380. [Google Scholar] [CrossRef]
- Pant, B.; Pant, K.; Pardasani, K.R. Naïve Bayes classifier for classification of plantand animal miRNA. Int. J. Comput. Theor. 2010, 2, 1793–8201. [Google Scholar] [CrossRef]
- Kumar, D.; Singh, R.; Kumar, A.; Sharma, N. An adaptive method of PCA for minimization of classification error using Naïve Bayes classifier. Procedia Comput. Sci. 2015, 70, 9–15. [Google Scholar] [CrossRef][Green Version]
- Sankaran, S.; Ehsani, R. Detection of huanglongbing disease in citrus using fluorescence spectroscopy. Trans. ASABE 2012, 55, 313–320. [Google Scholar] [CrossRef]
- Cen, H.; Weng, H.; Yao, J.; He, M.; Lv, J.; Hua, S.; Li, H.; He, Y. Chlorophyll fluorescence imaging uncovers photosynthetic fingerprint of citrus Huanglongbing. Front. Plant Sci. 2017, 8, 1509. [Google Scholar] [CrossRef]
- Macho, S.; Rius, A.; Callao, M.P.; Larrechi, M.S. Monitoring ethylene content in heterophasic copolymers by near-infrared spectroscopy: Standardisation of the calibration model. Anal. Chim. Acta 2001, 445, 213–220. [Google Scholar] [CrossRef]
- Feng, Y.Z.; Sun, D.W. Near-infrared hyperspectral imaging in tandem with partial least squares regression and genetic algorithm for non-destructive determination and visualization of Pseudomonas loads in chicken fillets. Talanta 2013, 109, 74–83. [Google Scholar] [CrossRef]
| Feature Selection | Number | Wavelength (nm) |
|---|---|---|
| RF | 16 | 458, 660, 662, 680, 682, 742, 743, 747, 791, 792, 797, 822, 862, 927, 952, 987 |
| SPA | 16 | 456, 464, 468, 474, 496, 631, 650, 667, 678, 690, 700, 710, 947, 1000, 1013, 1023 |
| SFS | 10 | 455, 469, 671, 682, 745, 747, 786, 801, 865, 998 |
| RF-CA | 3 | 458, 660, 742 |
| SPA-CA | 4 | 456, 678, 710, 947 |
| SFS-CA | 3 | 455, 671, 747 |
| Feature Selection | Classifiers | ||||
|---|---|---|---|---|---|
| NB | KNN | LDA | ELM | SVM | |
| RF-CA | 86.3 | 89.5 | 88.4 | 90.5 | 89.5 |
| SPA-CA | 86.3 | 86.3 | 89.5 | 93.7 | 94.7 |
| SFS-CA | 88.4 | 85.3 | 89.5 | 93.7 | 94.7 |
| Prediction | KNN | |||||
| RF-CA | SPA-CA | SFS-CA | ||||
| Infected | Healthy | Infected | Healthy | Infected | Healthy | |
| Infected | 39 (79.6%) | 10 (20.4%) | 32 (69.6%) | 14 (30.4%) | 32 (69.6%) | 14 (30.4%) |
| Healthy | 0 | 46 (100%) | 0 | 49 (100%) | 0 | 49 (100%) |
| Kappa value | 0.791 | 0.702 | 0.702 | |||
| Prediction | LDA | |||||
| RF-CA | SPA-CA | SFS-CA | ||||
| Infected | Healthy | Infected | Healthy | Infected | Healthy | |
| Infected | 44 (89.8%) | 5 (10.2%) | 39 (88.6%) | 5 (11.4%) | 41 (89.1%) | 5 (10.9%) |
| Healthy | 6 (13%) | 40 (87%) | 5 (9.8%) | 46 (90.2%) | 5 (10.2%) | 44 (89.8%) |
| Kappa value | 0.768 | 0.788 | 0.789 | |||
| Prediction | NB | |||||
| RF-CA | SPA-CA | SFS-CA | ||||
| Infected | Healthy | Infected | Healthy | Infected | Healthy | |
| Infected | 38 (77.6%) | 11 (22.4%) | 32 (72.7%) | 12 (27.3%) | 35 (76.1%) | 11 (23.9%) |
| Healthy | 2 (4.3%) | 44 (95.7%) | 1 (2%) | 50 (98%) | 0 | 49 (100%) |
| Kappa value | 0.728 | 0.720 | 0.766 | |||
| Prediction | ELM | |||||
| RF-CA | SPA-CA | SFS-CA | ||||
| Infected | Healthy | Infected | Healthy | Infected | Healthy | |
| Infected | 44 (91.7%) | 4 (8.3%) | 39 (97.5%) | 1 (2.5%) | 41 (97.6%) | 1 (2.4%) |
| Healthy | 5 (10.6%) | 42 (89.4%) | 5 (9.1%) | 50 (90.9%) | 5 (9.4%) | 48 (90.6%) |
| Kappa value | 0.810 | 0.872 | 0.873 | |||
| Prediction | SVM | |||||
| RF-CA | SPA-CA | SFS-CA | ||||
| Infected | Healthy | Infected | Healthy | Infected | Healthy | |
| Infected | 48 (88.9%) | 6 (11.1%) | 49 (96.1%) | 2 (3.9%) | 53 (94.6%) | 3 (5.4%) |
| Healthy | 4 (9.8%) | 37 (90.2%) | 3 (6.8%) | 41 (93.2%) | 2 (5.4%) | 37 (94.6%) |
| Kappa value | 0.787 | 0.894 | 0.892 | |||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liang, J.; Li, X.; Zhu, P.; Xu, N.; He, Y. Hyperspectral Reflectance Imaging Combined with Multivariate Analysis for Diagnosis of Sclerotinia Stem Rot on Arabidopsis Thaliana Leaves. Appl. Sci. 2019, 9, 2092. https://doi.org/10.3390/app9102092
Liang J, Li X, Zhu P, Xu N, He Y. Hyperspectral Reflectance Imaging Combined with Multivariate Analysis for Diagnosis of Sclerotinia Stem Rot on Arabidopsis Thaliana Leaves. Applied Sciences. 2019; 9(10):2092. https://doi.org/10.3390/app9102092
Chicago/Turabian StyleLiang, Jing, Xiaoli Li, Panpan Zhu, Ning Xu, and Yong He. 2019. "Hyperspectral Reflectance Imaging Combined with Multivariate Analysis for Diagnosis of Sclerotinia Stem Rot on Arabidopsis Thaliana Leaves" Applied Sciences 9, no. 10: 2092. https://doi.org/10.3390/app9102092
APA StyleLiang, J., Li, X., Zhu, P., Xu, N., & He, Y. (2019). Hyperspectral Reflectance Imaging Combined with Multivariate Analysis for Diagnosis of Sclerotinia Stem Rot on Arabidopsis Thaliana Leaves. Applied Sciences, 9(10), 2092. https://doi.org/10.3390/app9102092







