A Critical Review of Spatial Predictive Modeling Process in Environmental Sciences with Reproducible Examples in R
Abstract
1. Introduction
2. Sampling Design, Sample Quality Control, and Spatial Reference Systems
2.1. Sampling Design
2.2. Sample Quality Control
2.3. Spatial Reference Systems
3. Selection of Spatial Predictive Methods
3.1. Spatial Predictive Methods
3.2. Selecting Spatial Predictive Methods
4. Pre-Selection of Predictive Variables
4.1. Principles for Pre-Selection of Predictive Variables and Limitations
4.2. Predictive Variables for Environmental Sciences
5. Exploratory Analysis for Variable Pre-Selection
5.1. Non-Machine Learning Methods
5.2. Machine Learning Methods and Hybrid Methods
5.3. Hybrid Methods
6. Parameter Selection
6.1. Parameter Selection for Non-Machine Learning Methods
6.2. Parameter Selection for Machine Learning Methods
6.3. Parameter Selection for Hybrid Methods
7. Variable Selection
- Important variable based on the predictive accuracy (IVPA).This refers to the variable for which exclusion during the variable selection process would reduce the accuracy of a predictive model based on cross-validation. It may be more appropriate to call it predictive accuracy boosting variable (PABV).
- Unimportant variable based on the predictive accuracy (UVPA).
8. Accuracy and Error Measures for Predictive Models
8.1. Relationship between Observed, Predicted, and True Values
8.2. Error and Accuracy Measures of Predictive Models
9. Model Validation
9.1. Model Validation Methods
- Hold-out validation;
- K-fold cross-validation;
- Leave-one-out cross-validation;
- Leave-q-out cross-validation;
- Bootstrapping cross-validation;
- Using any new samples that are not used for model training.
9.2. Randomness Associated with Cross-Validation Methods
10. Spatial Predictions, Prediction Uncertainty, and Their Visualization
10.1. Spatial Predictions
10.2. Prediction Uncertainty
10.3. Visualization
11. Reproducible Examples for Spatial Predictive Modeling
11.1. Accuracy of a Predictive Model for Seabed Gravel Content
- > library(spm)
- > data(petrel)
- > names(petrel)
- [1] “long” “lat” “mud” “sand” “gravel” “bathy” “dist” “relief” “slope”
- > set.seed(1234)
- > n <- 100
- > rfokvecv1 <- NULL
- > for (i in 1:n) {
- + rfokcv1 <- rfokcv(petrel[, c(1,2)], petrel[, c(1,2, 6:9)], petrel[, 5], predacc = “VEcv”)
- + rfokvecv1 [i] <- rfokcv1
- + }
- > mean(rfokvecv1)
- [1] 37.44799
11.2. Parameter Selection
- > library(spm)
- > data(petrel)
- > nmax <- c(5:12); vgm.args <- c(“Sph”, “Mat”, “Ste”, “Log”)
- > rfokopt3 <- array(0, dim = c(length(nmax), length(vgm.args)))
- > set.seed(1234)
- > for (i in 1:length(nmax)) {
- + for (j in 1:length(vgm.args)) {
- + rfokcv1.1 <- NULL
- + for (k in 1:100) {
- + rfokcv1.1[k] <- rfokcv(petrel[, c(1, 2)], petrel[, c(1, 2, 6:9)], petrel[, 5], nmax = nmax[i],
- + vgm.args = vgm.args[j], predacc = "VEcv") }
- + rfokopt3[i, j] <- mean(rfokcv1.1) } }
- > which (rfokopt3 == max(rfokopt3, na.rm = T), arr.ind = T)
- [1,] 6 4
- > vgm.args[4]; nmax[6]
- [1] “Log”
- [1] 10
- > library(spm)
- > data(petrel)
- > set.seed(1234)
- > n <- 100
- > rfokvecv1 <- NULL
- > for (i in 1:n) {
- + rfokcv1 <- rfokcv(petrel[, c(1, 2)], petrel[, c(1, 2, 6:9)], petrel[, 5], vgm.args = “Log”,
- + nmax = 10,
- + predacc = “VEcv”)
- + rfokvecv1 [i] <- rfokcv1
- + }
- > mean(rfokvecv1)
- [1] 38.30175
11.3. Predictive Variable Selection
- > library(spm)
- > set.seed(1234)
- > rfokvecv1.1 <- NULL
- > for (i in 1:n) {
- > rfokcv1 <- rfokcv(petrel[, c(1, 2)], petrel[, c(1, 6:9)], petrel[, 5], vgm.args = “Log”,
- + nmax = 10,
- + predacc = “Vecv”)
- + rfokvecv1.1 [i] <- rfokcv1
- + }
- > mean(rfokvecv1.1, na.rm=T)
- [1] 39.00298
11.4. Generation of Spatial Predictions
- > set.seed(1234)
- > library(spm)
- > data(petrel); data(petrel.grid)
- > rfokpred1 <- rfokpred(petrel[, c(1, 2)], petrel[, c(1, 6:9)], petrel[, 5], petrel.grid[, c(1, 2)], + petrel.grid, ntree = 500, nmax = 10, vgm.args = (“Log”))
- > names(rfokpred1)
- [1] “LON” “LAT” “Predictions” “Variances”
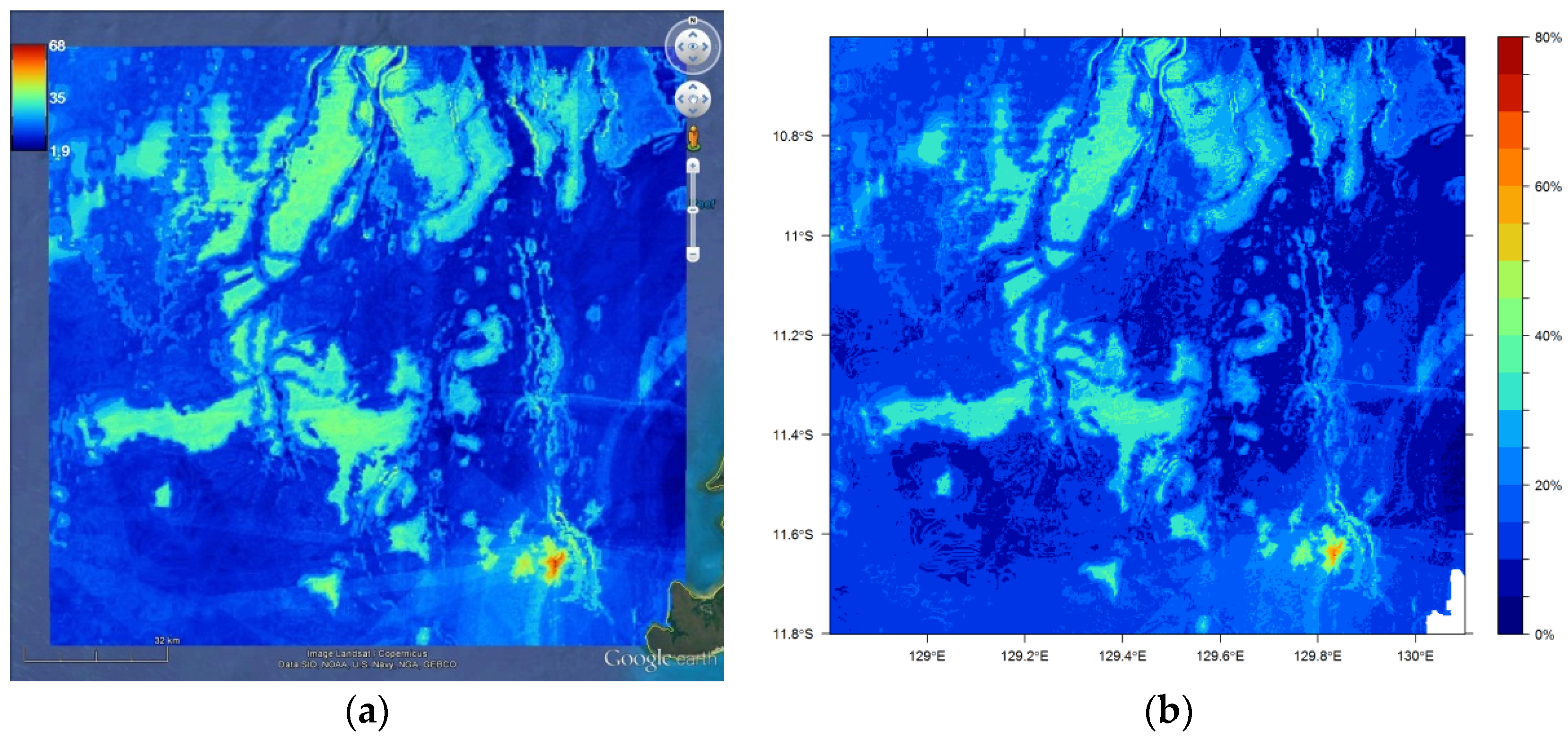
11.5. Visualisation of Spatial Predictions
- > library(sp); library(plotKML)
- > rfok1 <- rfokpred1
- > gridded(rfok1) <- ~ longitude + latitude
- > proj4string(rfok1) <- CRS(“+proj=longlat +datum=WGS84”)
- > plotKML(rfok1, colour_scale = SAGA_pal[[1]], grid2poly = TRUE)
- > par(font.axis=2, font.lab=2)
- > spplot(s1, c(“Predictions”), key.space=list(x=0.1,y=.95, corner=c(-1.2,2.8)),
- + col.regions = SAGA_pal[[1]], # this requires plotKML
- + scales=list(draw=T), colorkey = list(at = c(seq(0,80,5)), space=“right”,
- + labels = c("0%“,” “,“”,“”,“20%”,“”,“”,“”,“40%”,“”,“”,“”,“60%”,“”,“”,“”,“80%”)),
- + at=c(seq(0,80, 5)))
12. Summary
- Sampling design and data preparation;
- Selection of predictive methods;
- Pre-selection of predictive variables;
- Exploratory analysis;
- Parameter selection;
- Variable selection;
- Accuracy assessment;
- Model validation;
- Spatial predictions, prediction uncertainty, and their visualization.
Funding
Acknowledgments
Conflicts of Interest
References
- Marmion, M.; Luoto, M.; Heikkinen, R.K.; Thuiller, W. The performance of state-of-the-art modelling techniques depends on geographical distribution of species. Ecol. Model. 2009, 220, 3512–3520. [Google Scholar] [CrossRef]
- Maier, H.R.; Kapelan, Z.; Kasprzyk, J.; Kollat, J.; Matott, L.S.; Cunha, M.C.; Dandy, G.C.; Gibbs, M.S.; Keedwell, E.; Marchi, A.; et al. Evolutionary algorithms and other metaheuristics in water resources: Current status, research challenges and future directions. Environ. Model. Softw. 2014, 62, 271–299. [Google Scholar] [CrossRef]
- Li, J.; Heap, A. A Review of Spatial Interpolation Methods for Environmental Scientists; Record 2008/23; Geoscience Australia: Canberra, Australia, 2008; 137p.
- Stephens, D.; Diesing, M. Towards quantitative spatial models of seabed sediment composition. PLoS ONE 2015, 10, e0142502. [Google Scholar] [CrossRef] [PubMed]
- Sanabria, L.A.; Cechet, R.P.; Li, J. Mapping of australian fire weather potential: Observational and modelling studies. In Proceedings of the 20th International Congress on Modelling and Simulation (MODSIM2013), Adelaide, Australia, 1–6 December 2013; pp. 242–248. [Google Scholar]
- Li, J.; Alvarez, B.; Siwabessy, J.; Tran, M.; Huang, Z.; Przeslawski, R.; Radke, L.; Howard, F.; Nichol, S. Application of random forest, generalised linear model and their hybrid methods with geostatistical techniques to count data: Predicting sponge species richness. Environ. Model. Softw. 2017, 97, 112–129. [Google Scholar] [CrossRef]
- Hastie, T.; Tibshirani, R.; Friedman, J. The Elements of Statistical Learning: Data Mining, Inference, and Prediction, 2nd ed.; Springer: New York, NY, USA, 2009; p. 763. [Google Scholar]
- Crawley, M.J. The R Book; John Wiley & Sons, Ltd.: Chichester, UK, 2007; p. 942. [Google Scholar]
- Kuhn, M.; Johnson, K. Applied Predictive Modeling; Springer: New York, NY, USA, 2013. [Google Scholar]
- Li, J.; Heap, A.D. Spatial interpolation methods applied in the environmental sciences: A review. Environ. Model. Softw. 2014, 53, 173–189. [Google Scholar] [CrossRef]
- Li, J.; Heap, A. A review of comparative studies of spatial interpolation methods in environmental sciences: Performance and impact factors. Ecol. Inform. 2011, 6, 228–241. [Google Scholar] [CrossRef]
- Li, J.; Potter, A.; Huang, Z.; Daniell, J.J.; Heap, A. Predicting Seabed Mud Content across the Australian Margin: Comparison of Statistical and Mathematical Techniques Using a Simulation Experiment; Record 2010/11; Geoscience Australia: Canberra, Australia, 2010; 146p.
- Sanabria, L.A.; Qin, X.; Li, J.; Cechet, R.P.; Lucas, C. Spatial interpolation of mcarthur’s forest fire danger index across australia: Observational study. Environ. Model. Softw. 2013, 50, 37–50. [Google Scholar] [CrossRef]
- Tadić, J.M.; Ilić, V.; Biraud, S. Examination of geostatistical and machine-learning techniques as interpolaters in anisotropic atmospheric environments. Atmos. Environ. 2015, 111, 28–38. [Google Scholar] [CrossRef]
- Wolpert, D.; Macready, W. No free lunch theorems for optimization. IEEE Trans. Evol. Comput. 1997, 1, 67–82. [Google Scholar] [CrossRef]
- Burrough, P.A.; McDonnell, R.A. Principles of Geographical Information Systems; Oxford University Press: Oxford, UK, 1998; p. 333. [Google Scholar]
- Jakeman, A.J.; Letcher, R.A.; Norton, J.P. Ten iterative steps in development and evaluation of environmental models. Environ. Model. Softw. 2006, 21, 602–614. [Google Scholar] [CrossRef]
- Li, J. Assessing spatial predictive models in the environmental sciences: Accuracy measures, data variation and variance explained. Environ. Model. Softw. 2016, 80, 1–8. [Google Scholar] [CrossRef]
- Leek, J.T.; Peng, R.D. What is the question? Science 2015, 347, 1314–1315. [Google Scholar] [CrossRef]
- Li, J. spm: Spatial Predictive Modelling. R Package Version 1.1.0. Available online: https://CRAN.R-project.org/package=spm: 2018 (accessed on 17 May 2019).
- Foster, S.D.; Hosack, G.R.; Lawrence, E.; Przeslawski, R.; Hedge, P.; Caley, M.J.; Barrett, N.S.; Williams, A.; Li, J.; Lynch, T.; et al. Spatially balanced designs that incorporate legacy sites. Methods Ecol. Evol. 2017, 8, 1433–1442. [Google Scholar] [CrossRef]
- Benedetti, R.; Piersimoni, F.; Postigione, P. Spatially balanced sampling: A review and a reappraisal. Int. Stat. Rev. 2017, 85, 439–454. [Google Scholar] [CrossRef]
- Stevens, D.L.; Olsen, A.R. Spatially balanced sampling of natural resources. J. Am. Stat. Assoc. 2004, 99, 262–278. [Google Scholar] [CrossRef]
- Benedetti, R.; Piersimoni, F. A spatially balanced design with probability function proportional to the within sample distance. Biom. J. 2017, 59, 1067–1084. [Google Scholar] [CrossRef]
- Wang, J.-F.; Stein, A.; Gao, B.-B.; Ge, Y. A review of spatial sampling. Spat. Stat. 2012, 2, 1–14. [Google Scholar] [CrossRef]
- Diggle, P.J.; Ribeiro, P.J., Jr. Model-Based Geostatistics; Springer: New York, NY, USA, 2010; p. 228. [Google Scholar]
- Przeslawski, R.; Daniell, J.; Anderson, T.; Vaughn Barrie, J.; Heap, A.; Hughes, M.; Li, J.; Potter, A.; Radke, L.; Siwabessy, J.; et al. Seabed Habitats and Hazards of the Joseph Bonaparte Gulf and Timor Sea, Northern Australia; Record 2008/23; Geoscience Australia: Canberra, Australia, 2011; 69p.
- Radke, L.C.; Li, J.; Douglas, G.; Przeslawski, R.; Nichol, S.; Siwabessy, J.; Huang, Z.; Trafford, J.; Watson, T.; Whiteway, T. Characterising sediments for a tropical sediment-starved shelf using cluster analysis of physical and geochemical variables. Environ. Chem. 2015, 12, 204–226. [Google Scholar] [CrossRef]
- Radke, L.; Nicholas, T.; Thompson, P.; Li, J.; Raes, E.; Carey, M.; Atkinson, I.; Huang, Z.; Trafford, J.; Nichol, S. Baseline biogeochemical data from australia’s continental margin links seabed sediments to water column characteristics. Mar. Freshw. Res. 2017. [Google Scholar] [CrossRef]
- Kincaid, T. GRTS Survey Designs for an Area Resource. 2019. Available online: https://cran.r-project.org/web/packages/spsurvey/vignettes/Area_Design.pdf (accessed on 17 May 2019).
- Kincaid, T.M.; Olsen, A.R. spsurvey: Spatial Survey Design and Analysis. R Package Version 3.3. 2016. Available online: https://cran.r-project.org/web/packages/spsurvey/index.html (accessed on 17 May 2019).
- Hengl, T. GSIF: Global Soil Information Facilities. R Package Version 0.4-1. 2014. Available online: https://cran.r-project.org/web/packages/GSIF/index.html (accessed on 17 May 2019).
- Walvoort, D.J.J. Spatial Coverage Sampling and Random Sampling from Compact Geographical Strata. R Package Version 0.3-6. Available online: https://cran.r-project.org/web/packages/spcosa/index.html (accessed on 17 May 2019).
- Roudier, P. CLHS: A R Package for Conditioned Latin Hypercube Sampling. 2011. Available online: https://cran.r-project.org/web/packages/clhs/index.html (accessed on 17 May 2019).
- Grafströn, A.; Lisic, J. Balancedsampling: Balanced and Saptially Balanced Sampling. R Package Version 1.5.4. 2018. Available online: https://cran.r-project.org/web/packages/BalancedSampling/index.html (accessed on 17 May 2019).
- Radke, L.; Smit, N.; Li, J.; Nicholas, T.; Picard, K. Outer Darwin Harbour Shallow Water Sediment Survey 2016: Ga0356—Post-Survey Report; Record 2017/06; Geoscience Australia: Canberra, Australia, 2017. [CrossRef]
- Siwabessy, P.J.W.; Smit, N.; Atkinson, I.; Dando, N.; Harries, S.; Howard, F.J.F.; Li, J.; Nicholas, W.A.; Picard, K.; Radke, L.C.; et al. Bynoe Harbour Marine Survey 2016: Ga4452/sol6432—Post-Survey Report; Record 2017/04; Geoscience Australia: Canberra, Australia, 2017.
- Foster, S.D. MBHdesign: Spatial Designs for Ecological and Environmental Surveys. R Package Version 1.0.76. 2017. Available online: https://cran.r-project.org/web/packages/MBHdesign/index.html (accessed on 17 May 2019).
- Cai, L.; Zhu, Y. The challenges of data quality and data quality assessment in the big data era. Data Sci. J. 2015, 14, 1–10. [Google Scholar]
- Pipino, L.L.; Lee, Y.W.; Wang, R.Y. Data quality assessment. Commun. ACM 2002, 45, 211–218. [Google Scholar] [CrossRef]
- Li, J.; Potter, A.; Huang, Z.; Heap, A. Predicting Seabed sand Content across the Australian Margin Using Machine Learning and Geostatistical Methods; Record 2012/48; Geoscience Australia: Canberra, Australia, 2012; 115p.
- Li, J.; Hilbert, D.W.; Parker, T.; Williams, S. How do species respond to climate change along an elevation gradient? A case study of the grey-headed robin (Heteromyias albispecularis). Glob. Chang. Biol. 2009, 15, 255–267. [Google Scholar] [CrossRef]
- Jiang, W.; Li, J. The Effects of Spatial Reference Systems on the Predictive Accuracy of Spatial Interpolation Methods; Record 2014/01; Geoscience Australia: Canberra, Australia, 2014; p. 33.
- Jiang, W.; Li, J. Are Spatial Modelling Methods Sensitive to Spatial Reference Systems for Predicting Marine Environmental Variables. In Proceedings of the 20th International Congress on Modelling and Simulation, Adelaide, Australia, 1–6 December 2013; pp. 387–393. [Google Scholar]
- Turner, A.J.; Li, J.; Jiang, W. Effects of spatial reference systems on the accuracy of spatial predictive modelling along a latitudinal gradient. In Proceedings of the 22nd International Congress on Modelling and Simulation, Hobart, Australia, 3–8 December 2017; pp. 106–112. [Google Scholar]
- Purss, M. Topic 21: Discrete Global Grid Systems Abstract Specification, Open Geospatial Consortium [OGC 15-104r5]. 2017. Available online: https://www.google.com.au/url?sa=t&rct=j&q=&esrc=s&source=web&cd=4&cad=rja&uact=8&ved=2ahUKEwiHmPmnrqHiAhWFfisKHfTlB18QFjADegQIABAC&url=https%3A%2F%2Fportal.opengeospatial.org%2Ffiles%2F15-104r5&usg=AOvVaw3Ww2TasQntx17y99VlHwig (accessed on 17 May 2019).
- Li, J. Predictive modelling using random forest and its hybrid methods with geostatistical techniques in marine environmental geosciences. In Proceedings of the Eleventh Australasian Data Mining Conference (AusDM 2013), Canberra, Australia, 13–15 November 2013; Volume 146. [Google Scholar]
- Stephens, D.; Diesing, M. A comparison of supervised classification methods for the prediction of substrate type using multibeam acoustic and legacy grain-size data. PLoS ONE 2014, 9, e93950. [Google Scholar] [CrossRef] [PubMed]
- Hengl, T.; Heuvelink, G.B.M.; Kempen, B.; Leenaars, J.G.B.; Walsh, M.G.; Shepherd, K.D.; Sila, A.; MacMillan, R.A.; de Jesus, J.M.; Tamene, L.; et al. Mapping soil properties of africa at 250 m resolution: Random forests significantly improve current predictions. PLoS ONE 2015, 10, e0125814. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Liu, G.; Wang, H.; Li, X. Application of a hybrid interpolation method based on support vector machine in the precipitation spatial interpolation of basins. Water 2017, 9, 760. [Google Scholar] [CrossRef]
- Seo, Y.; Kim, S.; Singh, V.P. Estimating spatial precipitation using regression kriging and artificial neural network residual kriging (rknnrk) hybrid approach. Water Resour. Manag. 2015, 29, 2189–2204. [Google Scholar] [CrossRef]
- Demyanov, V.; Kanevsky, M.; Chernov, S.; Savelieva, E.; Timonin, V. Neural network residual kriging application for climatic data. J. Geogr. Inf. Decis. Anal. 1998, 2, 215–232. [Google Scholar]
- Appelhans, T.; Mwangomo, E.; Hardy, D.R.; Hemp, A.; Nauss, T. Evaluating machine learning approaches for the interpolation of monthly air temperature at mt. Kilimanjaro, tanzania. Spat. Stat. 2015, 14, 91–113. [Google Scholar] [CrossRef]
- Leathwick, J.R.; Elith, J.; Francis, M.P.; Hastie, T.; Taylor, P. Variation in demersal fish species richness in the oceans surrounding new zealand: An analysis using boosted regression trees. Mar. Ecol. Prog. Ser. 2006, 321, 267–281. [Google Scholar] [CrossRef]
- Leathwick, J.R.; Elith, J.; Hastie, T. Comparative performance of generalised additive models and multivariate adaptive regression splines for statistical modelling of species distributions. Ecol. Model. 2006, 199, 188–196. [Google Scholar] [CrossRef]
- Isaaks, E.H.; Srivastava, R.M. Applied Geostatistics; Oxford University Press: New York, NY, USA, 1989; p. 561. [Google Scholar]
- Hengl, T. A Practical Guide to Geostatistical Mapping of Environmental Variables; Office for Official Publication of the European Communities: Luxembourg, 2007; p. 143. [Google Scholar]
- Pebesma, E.J. Multivariable geostatistics in s: The gstat package. Comput. Geosci. 2004, 30, 683–691. [Google Scholar] [CrossRef]
- Bivand, R.S.; Pebesma, E.J.; Gómez-Rubio, V. Applied Spatial Data Analysis with R; Springer: New York, NY, USA, 2008; p. 374. [Google Scholar]
- Lark, R.M.; Ferguson, R.B. Mapping risk of soil nutrient deficiency or excess by disjunctive and indicator kriging. Geoderma 2004, 118, 39–53. [Google Scholar] [CrossRef]
- Huang, H.; Chen, C. Optimal geostatistical model selection. J. Am. Stat. Assoc. 2007, 102, 1009–1024. [Google Scholar] [CrossRef]
- Hernandez-Stefanoni, J.L.; Ponce-Hernandez, R. Mapping the spatial variability of plant diversity in a tropical forest: Comparison of spatial interpolation methods. Environ. Monit. Assess. 2006, 117, 307–334. [Google Scholar] [CrossRef] [PubMed]
- Stein, A.; Hoogerwerf, M.; Bouma, J. Use of soil map delineations to improve (co-)kriging of point data on moisture deficits. Geoderma 1988, 43, 163–177. [Google Scholar] [CrossRef]
- Voltz, M.; Webster, R. A comparison of kriging, cubic splines and classification for predicting soil properties from sample information. J. Soil Sci. 1990, 41, 473–490. [Google Scholar] [CrossRef]
- Bennett, N.D.; Croke, B.F.W.; Guariso, G.; Guillaume, J.H.A.; Hamilton, S.H.; Jakeman, A.J.; Marsili-Libelli, S.; Newham, L.T.H.; Norton, J.P.; Perrin, C.; et al. Characterising performance of environmental models. Environ. Model. Softw. 2013, 40, 1–20. [Google Scholar] [CrossRef]
- Gneiting, T.; Balabdaoui, F.; Raftery, A.E. Probabilistic forecasts, calibration and sharpness. J. R. Stat. Soc. Ser. B 2007, 69, 243–268. [Google Scholar] [CrossRef]
- Austin, M. Species distribution models and ecological theory: A critical assessment and some possible new approaches. Ecol. Model. 2007, 200, 1–19. [Google Scholar] [CrossRef]
- Elith, J.; Leathwick, J. Species distribution models: Ecological explanation and prediction across space and time. Annu. Rev. Ecol. Evol. Syst. 2009, 40, 677–697. [Google Scholar] [CrossRef]
- McArthur, M.A.; Brooke, B.P.; Przeslawski, R.; Ryan, D.A.; Lucieer, V.L.; Nichol, S.; McCallum, A.W.; Mellin, C.; Cresswell, I.D.; Radke, L.C. On the use of abiotic surrogates to describe marine benthic biodiversity. Estuar. Coast. Shelf Sci. 2010, 88, 21–32. [Google Scholar] [CrossRef]
- Huston, M.A. Hidden treatments in ecological experiments: Re-evaluating the ecosystem function of biodiversity. Oecologia 1997, 110, 449–460. [Google Scholar] [CrossRef]
- Arthur, A.D.; Li, J.; Henry, S.; Cunningham, S.A. Influence of woody vegetation on pollinator densities in oilseed brassica fields in an australian temperate landscape. Basic Appl. Ecol. 2010, 11, 406–414. [Google Scholar] [CrossRef]
- Elith, J.; Graham, C.H.; Anderson, R.P.; Dulik, M.; Ferrier, S.; Guisan, A.; Hijmans, R.J.; Huettmann, F.; Leathwick, J.R.; Lehmann, A.; et al. Novel methods improve prediction of species’ distributions from occurrence data. Ecography 2006, 29, 129–151. [Google Scholar] [CrossRef]
- Miller, K.; Puotinen, M.; Przeslawski, R.; Huang, Z.; Bouchet, P.; Radford, B.; Li, J.; Kool, J.; Picard, K.; Thums, M.; et al. Ecosystem Understanding to Support Sustainable Use, Management and Monitoring of Marine Assets in the North and North-West Regions: Final Report for NESP d1 2016e; Report to the National Environmental Science Program, Marine Biodiversity Hub; Australian Institute of Marine Science: Townsville, Australia, 2016; 146p. Available online: https://www.nespmarine.edu.au/system/files/Miller%20et%20al%20Project%20D1%20Report%20summarising%20outputs%20from%20synthesis%20of%20datasets%20and%20predictive%20models%20for%20N%20and%20NW_Milestone%204_RPv3.pdf (accessed on 17 May 2019).
- Li, J. Predicting the spatial distribution of seabed gravel content using random forest, spatial interpolation methods and their hybrid methods. In Proceedings of the International Congress on Modelling and Simulation (MODSIM) 2013, Adelaide, Austrialia, 1–6 December 2013; pp. 394–400. [Google Scholar]
- Verfaillie, E.; Van Lancker, V.; Van Meirvenne, M. Multivariate geostatistics for the predictive modelling of the surficial sand distribution in shelf seas. Cont. Shelf Res. 2006, 26, 2454–2468. [Google Scholar] [CrossRef]
- Verfaillie, E.; Du Four, I.; Van Meirvenne, M.; Van Lancker, V. Geostatistical modeling of sedimentological parameters using multi-scale terrain variables: Application along the belgian part of the north sea. Int. J. Geogr. Inf. Sci. 2008. [Google Scholar] [CrossRef]
- Huang, Z.; Nichol, S.; Siwabessy, P.J.W.; Daniell, J.; Brooke, B.P. Predictive modelling of seabed sediment parameters using multibeam acoustic data: A case study on the carnarvon shelf, western australia. Int. J. Geogr. Inf. Sci. 2012, 26, 283–307. [Google Scholar] [CrossRef]
- Li, J.; Siwabessy, J.; Tran, M.; Huang, Z.; Heap, A. Predicting seabed hardness using random forest in R. In Data Mining Applications with R; Zhao, Y., Cen, Y., Eds.; Elsevier: Amsterdam, The Netherlands, 2014; pp. 299–329. [Google Scholar]
- Li, J.; Tran, M.; Siwabessy, J. Selecting optimal random forest predictive models: A case study on predicting the spatial distribution of seabed hardness. PLoS ONE 2016, 11, e0149089. [Google Scholar] [CrossRef]
- Siwabessy, P.J.W.; Daniell, J.; Li, J.; Huang, Z.; Heap, A.D.; Nichol, S.; Anderson, T.J.; Tran, M. Methodologies for Seabed Substrate Characterisation Using Multibeam Bathymetry, Backscatter and Video Data: A Case Study from the Carbonate Banks of the Timor Sea, Northern Australia; Record 2013/11; Geoscience Australia: Canberra, Australia, 2013; 82p.
- Huang, Z.; Brooke, B.; Li, J. Performance of predictive models in marine benthic environments based on predictions of sponge distribution on the australian continental shelf. Ecol. Inform. 2011, 6, 205–216. [Google Scholar] [CrossRef]
- Lark, R.M.; Marchant, B.P.; Dove, D.; Green, S.L.; Stewart, H.; Diesing, M. Combining observations with acoustic swath bathymetry and backscatter to map seabed sediment texture classes: The empirical best linear unbiased predi. Sediment. Geol. 2015, 328, 17–32. [Google Scholar] [CrossRef]
- Diesing, M.; Mitchell, P.; Stephens, D. Image-based seabed classification: What can we learn from terrestrial remote sensing? ICES J. Mar. Sci. 2016, fsw 118. [Google Scholar] [CrossRef]
- Fisher, P.; Wood, J.; Cheng, T. Where is helvellyn? Fuzziness of multi-scale landscape morphometry. Trans. Inst. Br. Geogr. 2004, 29, 106–128. [Google Scholar] [CrossRef]
- Zuur, A.; Leno, E.N.; Elphick, C.S. A protocol for data exploration to avoid common statistical problems. Methods Ecol. Evol. 2010, 1, 3–14. [Google Scholar] [CrossRef]
- O’Brien, R.M. A caution regarding rules of thumb for variance inflation factors. Qual. Quant. 2007, 41, 673–690. [Google Scholar] [CrossRef]
- Harrell, F.E., Jr. Regression modelling strategies: with applications to linear models, logistic regression, and survival analysis; Springer: New York, NY, USA, 1997. [Google Scholar]
- Li, J.; Heap, A.D.; Potter, A.; Daniell, J. Application of machine learning methods to spatial interpolation of environmental variables. Environ. Model. Softw. 2011, 26, 1647–1659. [Google Scholar] [CrossRef]
- Cutler, D.R.; Edwards, T.C.J.; Beard, K.H.; Cutler, A.; Hess, K.T.; Gibson, J.; Lawler, J.J. Random forests for classification in ecology. Ecography 2007, 88, 2783–2792. [Google Scholar] [CrossRef]
- Collins, F.C.; Bolstad, P.V. A comparison of spatial interpolation techniques in temperature estimation. In Proceedings of the Third International Conference/Workshop on Integrating GIS and Environmental Modeling, Santa Fe, NM, USA, 21–25 January 1996. [Google Scholar]
- Ripley, B.D. Spatial Statistics; John Wiley & Sons: New York, NY, USA, 1981; p. 252. [Google Scholar]
- Wu, J.; Norvell, W.A.; Welch, R.M. Kriging on highly skewed data for dtpa-extractable soil zn with auxiliary information for ph and organic carbon. Geoderma 2006, 134, 187–199. [Google Scholar] [CrossRef]
- Meul, M.; Van Meirvenne, M. Kriging soil texture under different types of nonstationarity. Geoderma 2003, 112, 217–233. [Google Scholar] [CrossRef]
- Liaw, A.; Wiener, M. Classification and regression by randomforest. R News 2002, 2, 18–22. [Google Scholar]
- Ridgeway, G. gbm: Generalized Boosted Regression Models. R Package Version 2.1.3. 2017. Available online: https://cran.r-project.org/web/packages/gbm/index.html (accessed on 17 May 2019).
- Elith, J.; Leathwick, J.R.; Hastie, T. A working guide to boosted regression trees. J. Anim. Ecol. 2008, 77, 802–813. [Google Scholar] [CrossRef]
- Breiman, L.; Friedman, J.H.; Olshen, R.A.; Stone, C.J. Classification and Regression Trees; Belmont: Wadsworth, OH, USA, 1984. [Google Scholar]
- Li, J.; Hilbert, D.W. Lives: A new habitat modelling technique for predicting the distributions of species‘ occurrence using presence-only data based on limiting factor theory. Biodivers. Conserv. 2008, 17, 3079–3095. [Google Scholar] [CrossRef]
- Johnson, J.B.; Omland, K.S. Model selection in ecology and evolution. Trends Ecol. Evol. 2004, 19, 101–108. [Google Scholar] [CrossRef]
- Venables, W.N.; Ripley, B.D. Modern Applied Statistics with S-Plus, 4th ed.; Springer: New York, NY, USA, 2002; p. 495. [Google Scholar]
- Chambers, J.M.; Hastie, T.J. Statistical Models in S; Wadsworth and Brooks/Cole Advanced Books and Software: Pacific Grove, CA, USA, 1992; p. 608. [Google Scholar]
- Lumley, T.; Miller, A. leaps: Regression Subset Selection. R Package Version 3.0. 2009. Available online: https://cran.r-project.org/web/packages/leaps/index.html (accessed on 17 May 2019).
- McLeod, A.I.; Xu, C. bestglm: Best Subset GLM. R Package Version 0.36. 2017. Available online: https://cran.r-project.org/web/packages/bestglm/index.html (accessed on 17 May 2019).
- Li, J.; Alvarez, B.; Siwabessy, J.; Tran, M.; Huang, Z.; Przeslawski, R.; Radke, L.; Howard, F.; Nichol, S. Selecting predictors to form the most accurate predictive model for count data. In Proceedings of the International Congress on Modelling and Simulation (MODSIM) 2017, Hobart, Australia, 3–8 December 2017. [Google Scholar]
- Kursa, M.B.; Rudnicki, W.R. Feature selection with the boruta package. J. Stat. Softw. 2010, 36, 1–13. [Google Scholar] [CrossRef]
- Kuhn, M. caret: Classification and Regression Training. R Package Version 6.0-81. 2018. Available online: https://cran.r-project.org/web/packages/caret/index.html (accessed on 17 May 2019).
- Genuer, R.; Poggi, J.M.; Tuleau-Malot, C. VSURF: Variable Selection Using Random Forests. R Package Version 1.0.2. 2015. Available online: https://cran.r-project.org/web/packages/VSURF/index.html (accessed on 17 May 2019).
- Li, J.; Siwabessy, J.; Huang, Z.; Nichol, S. Developing an optimal spatial predictive model for seabed sand content using machine learning, geostatistics and their hybrid methods. Geosciences 2019, 9, 180. [Google Scholar] [CrossRef]
- Han, J.; Kamber, M. Data Mining: Concept and Techniques, 2nd ed.; Elsevier: Amsterdam, The Netherlands, 2006; p. 770. [Google Scholar]
- Moriasi, D.N.; Arnold, J.G.; Van Liew, M.W.; Bingner, R.L.; Harmel, R.D.; Veith, T.L. Model evaluation guidelines for systematic quantification of accuracy in watershed simulations. Am. Soc. Agric. Biol. Eng. 2007, 50, 885–900. [Google Scholar]
- Li, J. Assessing the accuracy of predictive models for numerical data: Not r nor r2, why not? Then what? PLoS ONE 2017, 12, e0183250. [Google Scholar] [CrossRef] [PubMed]
- Allouche, O.; Tsoar, A.; Kadmon, R. Assessing the accuracy of species distribution models: Prevalence, kappa and true skill statistic (tss). J. Appl. Ecol. 2006, 43, 1223–1232. [Google Scholar] [CrossRef]
- Fielding, A.H.; Bell, J.F. A review of methods for the assessment of prediction errors in conservation presence/absence models. Environ. Conserv. 1997, 24, 38–49. [Google Scholar] [CrossRef]
- Thibaud, E.; Petitpierre, B.; Broennimann, O.; Davison, A.C.; Guisan, A. Measuring the relative effect of factors affecting species distribution model predictions. Methods Ecol. Evol. 2014, 5, 947–955. [Google Scholar] [CrossRef]
- Lobo, J.M.; Jiménez-Valverde, A.; Real, R. Auc: A misleading measure of the performance of predictive distribution models. Glob. Ecol. Biogeogr. 2008, 7, 145–151. [Google Scholar] [CrossRef]
- Kohavi, R. A study of cross-validation and bootstrap for accuracy estimation and model selection. In Proceedings of the International Joint Conference on Artificial Intelligence (IJCAI), Montreal, QC, Canada, 20–25 August 1995; pp. 1137–1143. [Google Scholar]
- Refsgaard, J.C.; van der Sluijs, J.P.; Højberg, A.L.; Vanrolleghem, P.A. Uncertainty in the environmental modelling process - a framework and guidance. Environ. Model. Softw. 2007, 22, 1543–1556. [Google Scholar] [CrossRef]
- Hayes, K.R. Uncertainty and Uncertainty Analysis Methods; CSIRO: Canberra, Australia, 2011; p. 131. Available online: https://publications.csiro.au/rpr/download?pid=csiro:EP102467&dsid=DS3 (accessed on 17 May 2019).
- Barry, S.; Elith, J. Error and uncertainty in habitat models. J. Appl. Ecol. 2006, 43, 413–423. [Google Scholar] [CrossRef]
- Oxley, T.; ApSimon, H. A conceptual framework for mapping uncertainty in integrated assessment. In Proceedings of the 19th International Congress on Modelling and Simulation, Perth, Australia, 12–16 December 2011. [Google Scholar]
- Walker, W.E.; Harremoes, P.; Rotmans, J.; Van der Sluijs, J.P.; van Asselt, M.B.A.; Janssen, P.; Krayer von Krauss, M.P. Defining uncertainty: A conceptual basis for uncertainty management in model-based decision support. Integr. Assess. 2003, 4, 5–17. [Google Scholar] [CrossRef]
- Goovaerts, P. Geostatistics for Natural Resources Evaluation; Oxford University Press: New York, NY, USA, 1997; p. 483. [Google Scholar]
- Mentch, L.; Hooker, G. Quantifying uncertainty in random forests via confidence intervals and hypothesis tests. J. Mach. Learn. Res. 2016, 17, 1–41. [Google Scholar]
- Slaets, J.I.F.; Piepho, H.-P.; Schmitter, P.; Hilger, T.; Cadisch, G. Quantifying uncertainty on sediment loads using bootstrap confidence intervals. Hydrol. Earth Syst. Sci. 2017, 21, 571–588. [Google Scholar] [CrossRef]
- Wager, S.; Hastie, T.; Efron, B. Confidence intervals for random forests: The jackknife and the infinitesimal jackknife. J. Mach. Learn. Res. 2014, 15, 1625–1651. [Google Scholar] [PubMed]
- Wright, M.N.; Ziegler, A. Ranger: A fast implementation of random forests for high dimensional data in c++ and r. J. Stat. Softw. 2017, 77, 1–17. [Google Scholar] [CrossRef]
- Coulston, J.W.; Blinn, C.E.; Thomas, V.A.; Wynne, R.H. Approximating prediction uncertainty for random forest regression models. Photogramm. Eng. Remote Sens. 2016, 82, 189–197. [Google Scholar] [CrossRef]
- Chen, J.; Li, M.-C.; Wang, W. Statistical uncertainty estimation using random forests and its application to drought forecast. Math. Probl. Eng. 2012, 2012, 915053. [Google Scholar] [CrossRef]
- Bishop, T.F.A.; Minasny, B.; McBratney, A.B. Uncertainty analysis for soil-terrain models. Int. J. Geogr. Inf. Sci. 2006, 20, 117–134. [Google Scholar] [CrossRef]
- Hijmans, R.J. raster: Geographic Data Analysis and Modeling. Available online: http://CRAN.R-project.org/package=raster (accessed on 17 May 2019).
| Non-Machine Learning Method and Hybrid Methods | Machine Learning Method and Hybrid Methods | ||
|---|---|---|---|
| Non-machine learning method | Hybrid methods | Machine learning method | Hybrid methods |
| Generalized additive models | Cubist | Cubist and OK (cubistOK) | |
| Generalized least squares trend estimation (GLS) | GLS and OK | Generalized boosted regression modeling (GBM) | GBM and IDS (GBMIDS) |
| Generalized linear models (GLM) | GLM and IDW (GLMIDW) | GBM and OK (GBMOK) | |
| GLM and OK (GLMOK) | General regression neural network (GRNN) | GRNN and IDS (GRNNIDS) | |
| GLM with lasso or elastic net regularization | GRNN and OK (GRNNOK) | ||
| Linear models and OK | Multivariate adaptive regression splines | ||
| RT and IDS (RTIDS) | Naïve Bayes | ||
| RT and OK (RTOK) | Random forest (RF) | RF and IDS (RKIDS) | |
| RF and OK (RKOK) | |||
| Support vector machine (SVM) | SVM and OK (SVMOK) | ||
| SVM and OK (SVMIDS) | |||
| No | Predictive Variables | Seabed Sediment/Grain Size | Seabed Hardness | Sponge Species Richness | Window/Kernel Size(s) |
|---|---|---|---|---|---|
| 1 | Longitude (long) | yes | yes | yes | |
| 2 | Latitude (lat) | yes | yes | yes | |
| 3 | Distance to coast (dist) | yes | yes | ||
| 4 | Bathymetry (bathy) | yes | yes | yes | |
| 5 | Local Moran’s I from bathymetry | yes | yes | yes | yes |
| 6 | Mean curvature | yes | yes | ||
| 7 | Planar curvature | yes | yes | yes | yes |
| 8 | Profile curvature | yes | yes | yes | yes |
| 9 | Relief | yes | yes | yes | yes |
| 10 | Rugosity (or surface, surface complexity) | yes | yes | yes | yes |
| 11 | Slope | yes | yes | yes | yes |
| 12 | Topographic or bathymetric position index (tpi or bpi) | yes | yes | yes | yes |
| 13 | Fuzzy morphometric features | yes | yes | ||
| 14 | Backscatter (bs) 10–36 | yes | yes | yes | |
| 15 | Entropy from bs | yes | yes | ||
| 16 | Homogeneity from bs | yes | yes | yes | |
| 17 | Local Moran’s I from bs | yes | yes | yes | |
| 18 | Prock from bs | yes | |||
| 19 | Variance from bs | yes | yes | yes | |
| 20 | Suspended particulate matter | yes | |||
| 21 | Mean tidal current velocity | yes | |||
| 22 | Peak orbital velocity of waves at seabed | yes | |||
| 23 | Roughness from bathy * | yes | |||
| 24 | Roughness from bs * | yes | |||
| 25 | Sobel filter from bathy # | yes |
© 2019 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, J. A Critical Review of Spatial Predictive Modeling Process in Environmental Sciences with Reproducible Examples in R. Appl. Sci. 2019, 9, 2048. https://doi.org/10.3390/app9102048
Li J. A Critical Review of Spatial Predictive Modeling Process in Environmental Sciences with Reproducible Examples in R. Applied Sciences. 2019; 9(10):2048. https://doi.org/10.3390/app9102048
Chicago/Turabian StyleLi, Jin. 2019. "A Critical Review of Spatial Predictive Modeling Process in Environmental Sciences with Reproducible Examples in R" Applied Sciences 9, no. 10: 2048. https://doi.org/10.3390/app9102048
APA StyleLi, J. (2019). A Critical Review of Spatial Predictive Modeling Process in Environmental Sciences with Reproducible Examples in R. Applied Sciences, 9(10), 2048. https://doi.org/10.3390/app9102048





