Explanation of Osteoblastic Differentiation of Stem Cells by Photo Biomodulation Using the Resonant Recognition Model
Abstract
:Featured Application
Abstract
1. Introduction
2. Methods and Materials
2.1. Resonant Recognition Model (RRM)
2.2. Protein Sequences Analysed by RRM
- >sp|P01270|PTHY_HUMAN Parathyroid hormone OS = Homo sapiens GN = PTH PE = 1 SV = 1
- >sp|P04089|PTHY_RAT Parathyroid hormone OS = Rattus norvegicus GN = Pth PE = 2 SV = 1
- >sp|P01268|PTHY_BOVIN Parathyroid hormone OS = Bos taurus GN = PTH PE = 1 SV = 1
- >sp|P52212|PTHY_CANLF Parathyroid hormone OS = Canis lupus familiaris GN = PTH PE = 3 SV = 1
- >sp|Q27IM2|PTHY_HORSE Parathyroid hormone OS = Equus caballus GN = PTH PE = 3 SV = 1
- >sp|Q9GL67|PTHY_FELCA Parathyroid hormone OS = Felis catus GN = PTH PE = 3 SV = 1
- >sp|P15743|PTHY_CHICK Parathyroid hormone OS = Gallus gallus GN = PTH PE = 3 SV = 1
- >sp|P01269|PTHY_PIG Parathyroid hormone OS = Sus scrofa GN = PTH PE = 1 SV = 1
- >sp|Q9XT35|PTHY_MACFA Parathyroid hormone OS = Macaca fascicularis GN = PTH PE = 3 SV = 1
- >sp|Q03431|PTH1R_HUMAN Parathyroid hormone/parathyroid hormone-related peptide receptor OS = Homo sapiens GN = PTH1R PE = 1 SV = 1
- >sp|P25961|PTH1R_RAT Parathyroid hormone/parathyroid hormone-related peptide receptor OS = Rattus norvegicus GN = Pth1r PE = 1 SV = 1
- >sp|P41593|PTH1R_MOUSE Parathyroid hormone/parathyroid hormone-related peptide receptor OS = Mus musculus GN = Pth1r PE = 1 SV = 2
- >sp|Q9TU31|PTH1R_CANLF Parathyroid hormone/parathyroid hormone-related peptide receptor OS = Canis lupus familiaris GN = PTH1R PE = 1 SV = 1
- >sp|P50133|PTH1R_PIG Parathyroid hormone/parathyroid hormone-related peptide receptor OS = Sus scrofa GN = PTH1R PE = 1 SV = 1
- >sp|Q1LZF7|PTH1R_BOVIN Parathyroid hormone/parathyroid hormone-related peptide receptor OS = Bos taurus GN = PTH1R PE = 2 SV = 1
- >sp|Q5RAQ1|PTH1R_PONAB Parathyroid hormone/parathyroid hormone-related peptide receptor OS = Pongo abelii GN = PTH1R PE = 2 SV = 1
- >sp|P48061|SDF1_HUMAN Stromal cell-derived factor 1 OS = Homo sapiens GN = CXCL12 PE = 1 SV = 1
- >sp|P40224|SDF1_MOUSE Stromal cell-derived factor 1 OS = Mus musculus GN = Cxcl12 PE = 2 SV = 2
- >sp|Q8UUJ9|SDF1_XENLA Stromal cell-derived factor 1 OS = Xenopus laevis GN = cxcl12 PE = 2 SV = 1
- >sp|O62657|SDF1_FELCA Stromal cell-derived factor 1 OS = Felis catus GN = CXCL12 PE = 3 SV = 1
- >sp|Q5EBF6|SDF1_XENTR Stromal cell-derived factor 1 OS = Xenopus tropicalis GN = cxcl12 PE = 3 SV = 1
3. Results
4. Discussion and Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Yu, B.; Zhao, X.; Crane, J.; Xian, L.; Lu, W.; Wan, M.; Cao, X. Parathyroid Hormone Induces Differentiation of Mesenchymal Stromal/Stem Cells by Enhancing Bone Morphogenetic Protein Signalling. J. Bone Min. Res. 2012, 27, 2001–2014. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Du, L.; Ge, S. PTH/SDF-1α Cotherapy Induces CD90+CD34- Stromal Cells Migration and Promotes Tissue Regeneration in a Rat Periodontal Defect Model. Sci. Rep. 2016, 2, 30403. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Huang, Y.Y.; Wang, Y.; Lyu, P.; Hamblin, M.R. Photobiomodulation (Blue and Green Light) Encourages Osteoblastic-Differentiation of Human Adipose-Derived Stem Cells: Role of Intracellular Calcium and Light-Gated Ion Channels. Sci. Rep. 2016, 6, 33719. [Google Scholar] [CrossRef] [PubMed]
- Cosic, I. Macromolecular Bioactivity: Is it Resonant Interaction between Macromolecules?—Theory and Applications. IEEE Trans. Biomed. Eng. 1994, 41, 1101–1114. [Google Scholar] [CrossRef] [PubMed]
- Cosic, I. The Resonant Recognition Model of Macromolecular Bioactivity: Theory and Applications; Birkhauser Verlag: Basel, Switzerland, 1997. [Google Scholar]
- Cosic, I.; Cosic, D.; Lazar, K. Analysis of Tumor Necrosis Factor Function Using the Resonant Recognition Model. Cell Biochem. Biophys. 2016, 74, 175–180. [Google Scholar] [CrossRef] [PubMed]
- Pirogova, E.; Simon, G.P.; Cosic, I. Investigation of the applicability of Dielectric Relaxation properties of amino acid solutions within the Resonant Recognition Model. IEEE Trans. NanoBiosci. 2003, 2, 63–69. [Google Scholar] [CrossRef]
- Pirogova, E.; Cosic, I. The Use of Ionisation Constants of Amino Acids for Protein Signal Analysis within the RRM—Application to Proteases. Mol. Simul. 2002, 28, 845–851. [Google Scholar] [CrossRef]
- Cosic, I.; Lazar, K.; Cosic, D. Prediction of Tubulin resonant frequencies using the Resonant Recognition Model (RRM). IEEE Trans. Nanobiosci. 2015, 12, 491–496. [Google Scholar] [CrossRef] [PubMed]
- Cosic, I.; Cosic, D.; Lazar, K. Is it possible to predict electromagnetic resonances in proteins, DNA and RNA? Nonlinear Biomed. Phys. 2015, 3, 5. [Google Scholar] [CrossRef]
- Cosic, I.; Cosic, D.; Lazar, K. Environmental Light and Its Relationship with Electromagnetic Resonances of Biomolecular Interactions, as Predicted by the Resonant Recognition Model. Int. J. Environ. Res. Public Health 2016, 13, 647. [Google Scholar] [CrossRef] [PubMed]
- Cosic, I.; Cosic, D. The Treatment of Crigler-Najjar Syndrome by Blue Light as Explained by Resonant Recognition Model. EPJ Nonlinear Biomed. Phys. 2016, 4, 9. [Google Scholar] [CrossRef]
- Cosic, I.; Cosic, D.; Lazar, K. Cancer Related BRCA-1 and BRCA-2 Mutations as Analysed by the Resonant Recognition Model. J. Adv. Mol. Biol. 2017, 1. [Google Scholar] [CrossRef]
- Cosic, I.; Cosic, D. Influence of Tuning Element Relief Patches on Pain as Analyzed by the Resonant Recognition Model. IEEE Trans. Nanobiosci. 2017, 16, 822–827. [Google Scholar] [CrossRef] [PubMed]
- Cosic, I.; Cosic, D.; Uzelac, B.; Brasovan, S. Possible Mechanism of Titanium Salt Infused Patches Remediation of Autism and Attention Deficit Disorders. Md-Med. Data 2018, 10, 97–102. [Google Scholar]
- Cosic, I.; Paspaliaris, V.; Cosic, D.; Kolios, G. Analysis of Interleukin-12 and Interleukin-23 Pathways to Distinguish between Immune Activation and Inflammation Functions. MD-Med. Data 2019, 11, 7–14. [Google Scholar]
- Vojisavljevic, V.; Pirogova, E.; Cosic, I. The Effect of Electromagnetic Radiation (550–850 nm) on I-Lactate Dehydrogenase Kinetics. Int. J. Radiat. Biol. 2007, 83, 221–230. [Google Scholar] [CrossRef] [PubMed]
- Dotta, B.T.; Murugan, N.J.; Karbowski, L.M.; Lafrenie, R.M.; Persinger, M.A. Shifting wavelength of ultraweak photon emissions from dying melanoma cells: Their chemical enhancement and blocking are predicted by Cosic’s theory of resonant recognition model for macromolecules. Naturwissenschaften 2014, 101, 87–94. [Google Scholar] [CrossRef] [PubMed]
- Murugan, N.J.; Karbowski, L.M.; Persinger, M.A. Cosic’s Resonance Recognition Model for Protein Sequences and Photon Emission Differentiates Lethal and Non-Lethal Ebola Strains: Implications for Treatment. Open J. Biophys. 2014, 5, 35. [Google Scholar] [CrossRef]
- Karbowski, L.M.; Murugan, N.J.; Persinger, M.A. Novel Cosic resonance (standing wave) solutions for components of the JAK-STAT cellular signalling pathway: A convergence of spectral density profiles. FEBS Open Biol. 2015, 5, 245–250. [Google Scholar] [CrossRef] [PubMed]
- Jin, L.; Briggs, S.L.; Chandrasekhar, S.; Chirgadze, N.Y.; Clawson, D.K.; Schevitz, R.W.; Smiley, D.L.; Tashjian, A.H.; Zheng, F. Crystal Structure of Human Parathyroid Hormone 1-34 at 0.9Å Resolution. JBC 2000, 275, 27238–27244. [Google Scholar]
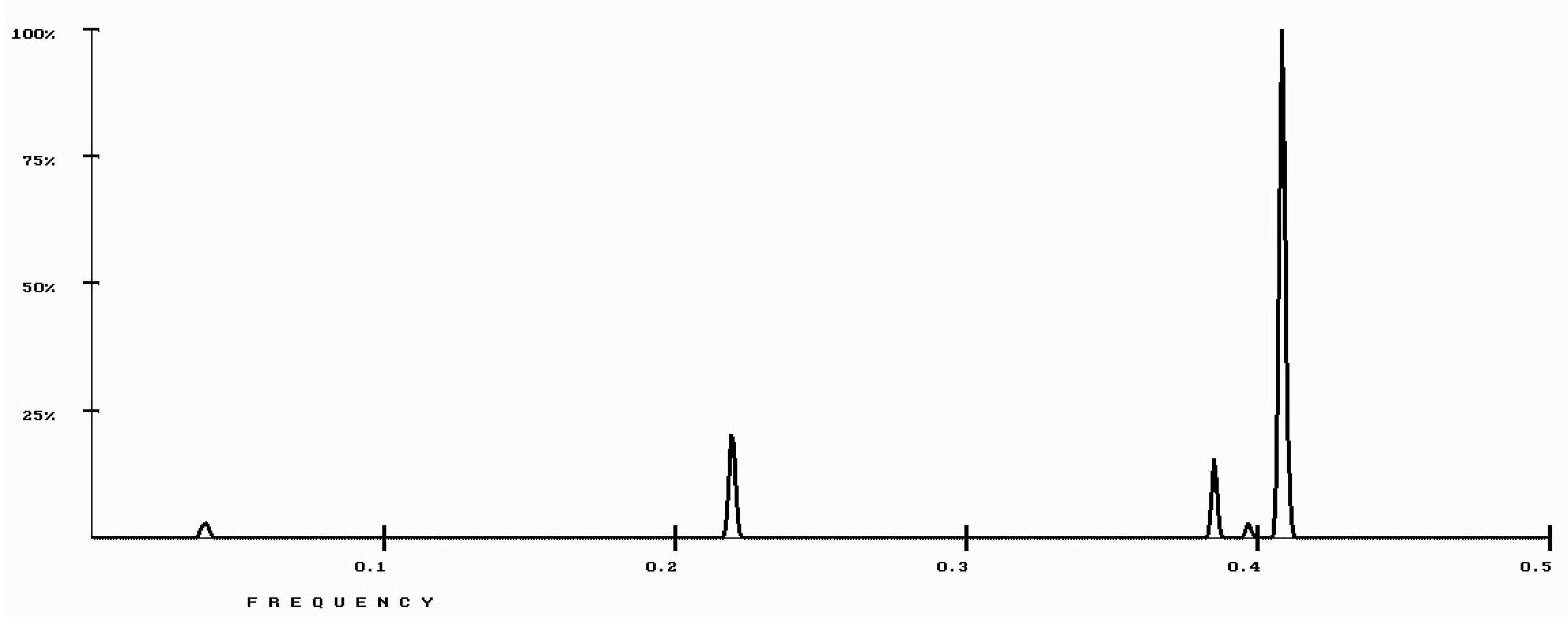
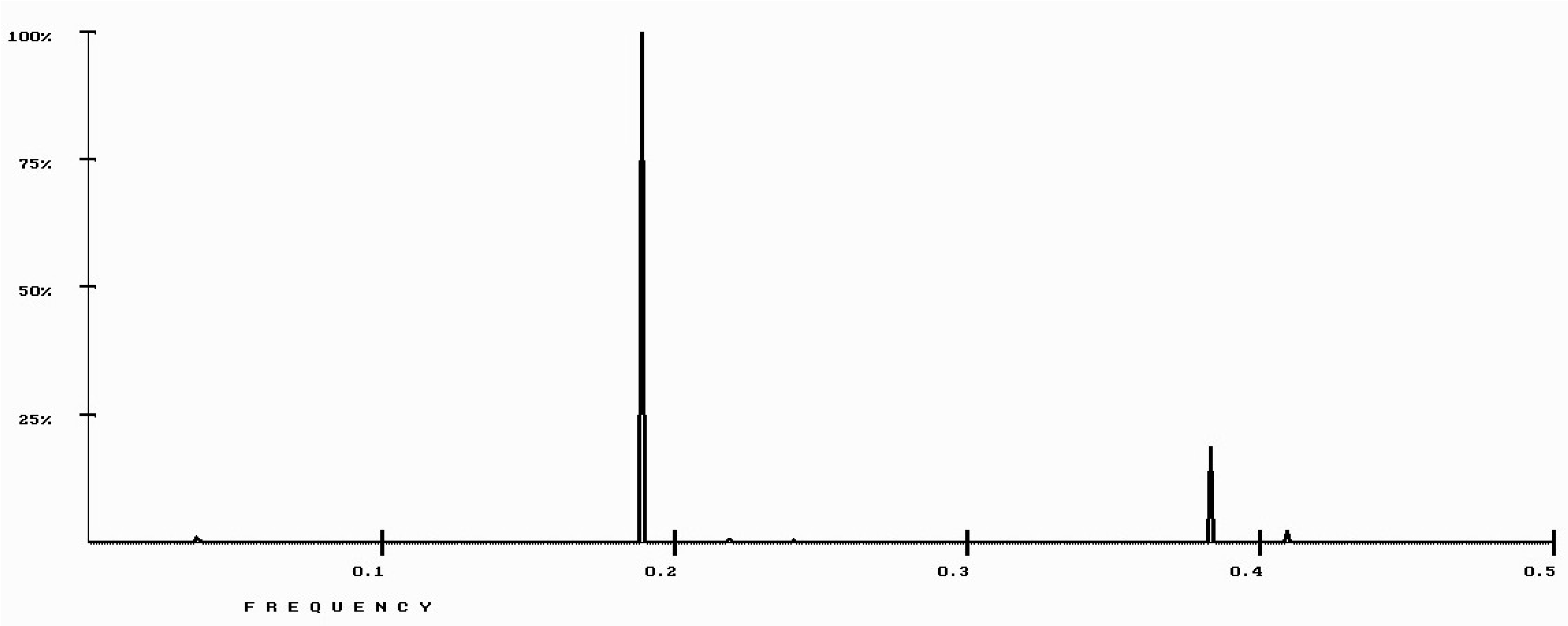
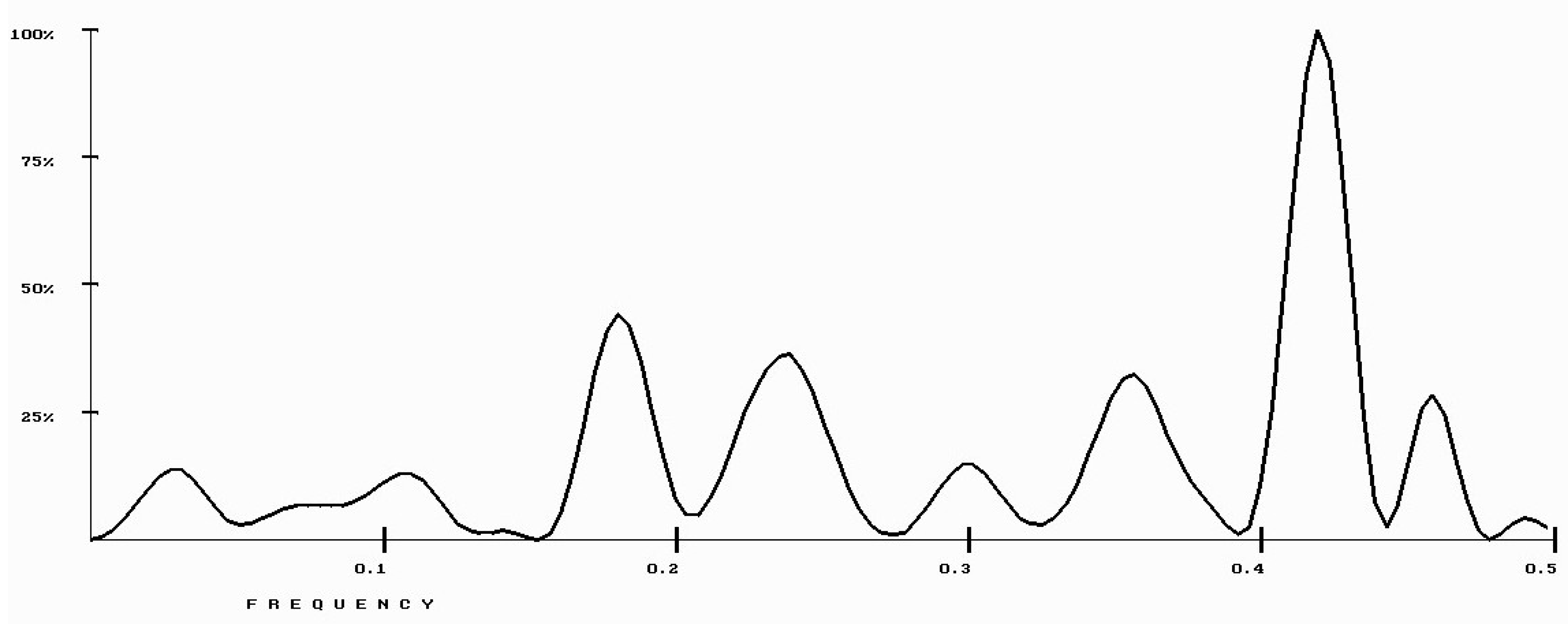
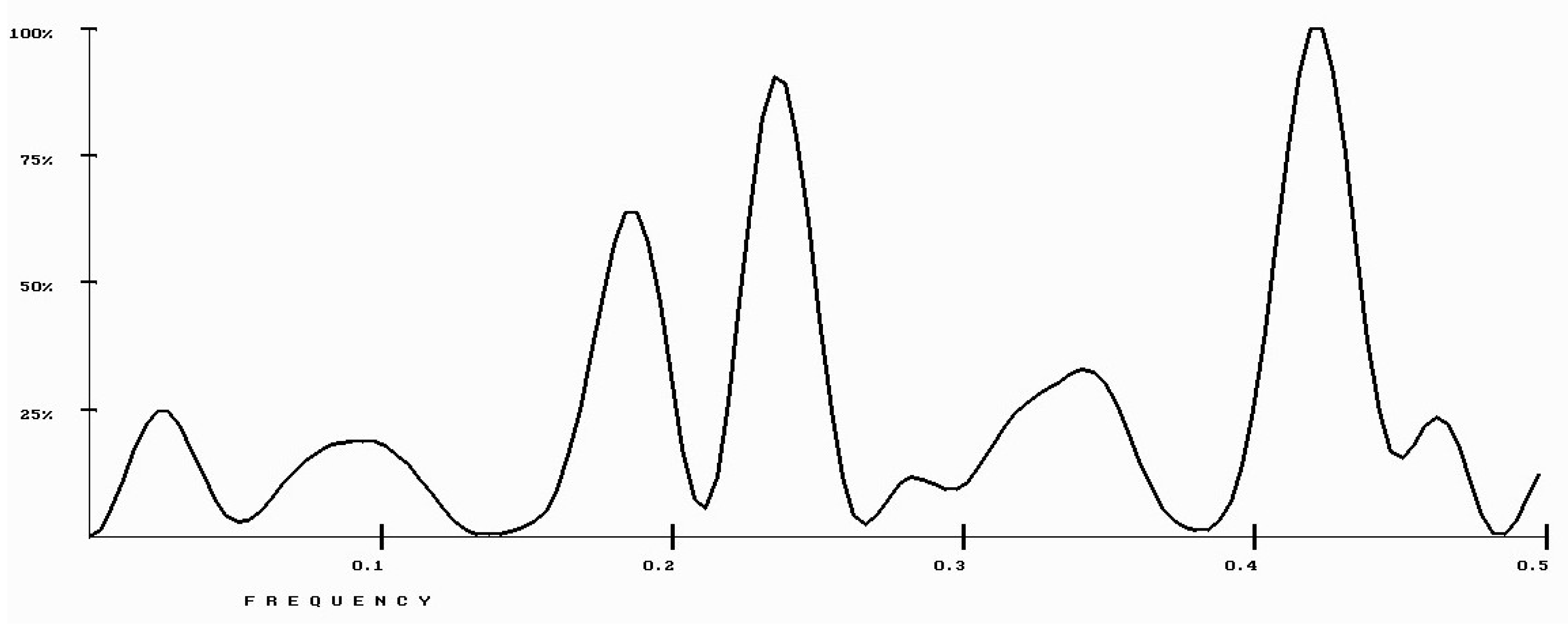
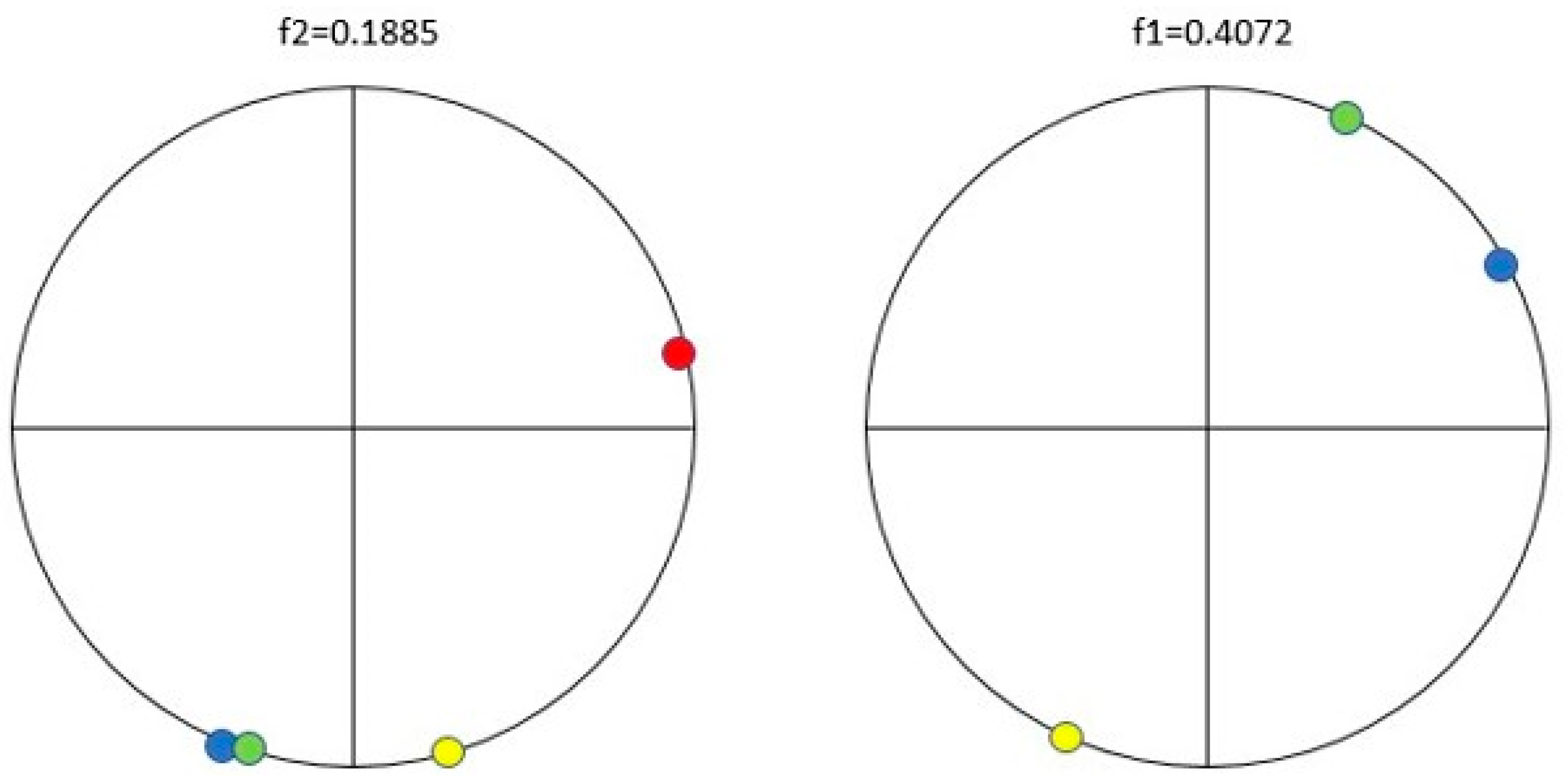
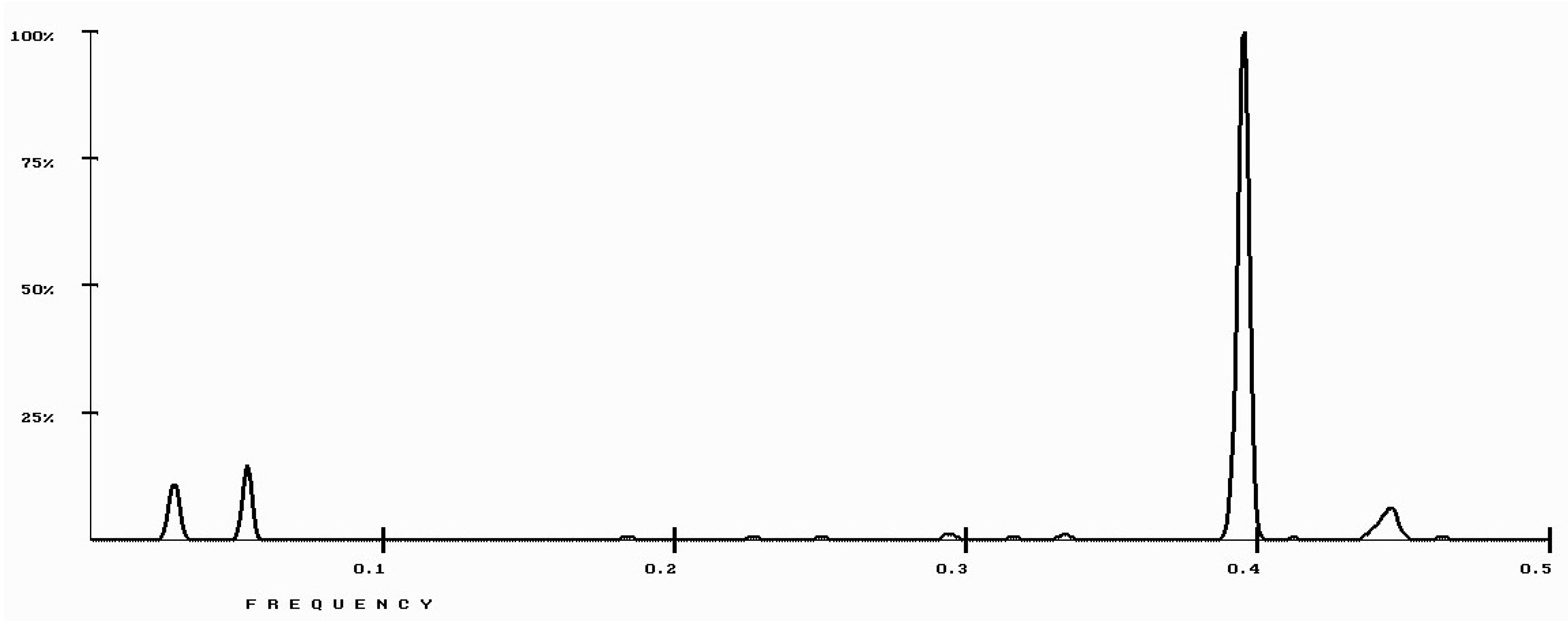
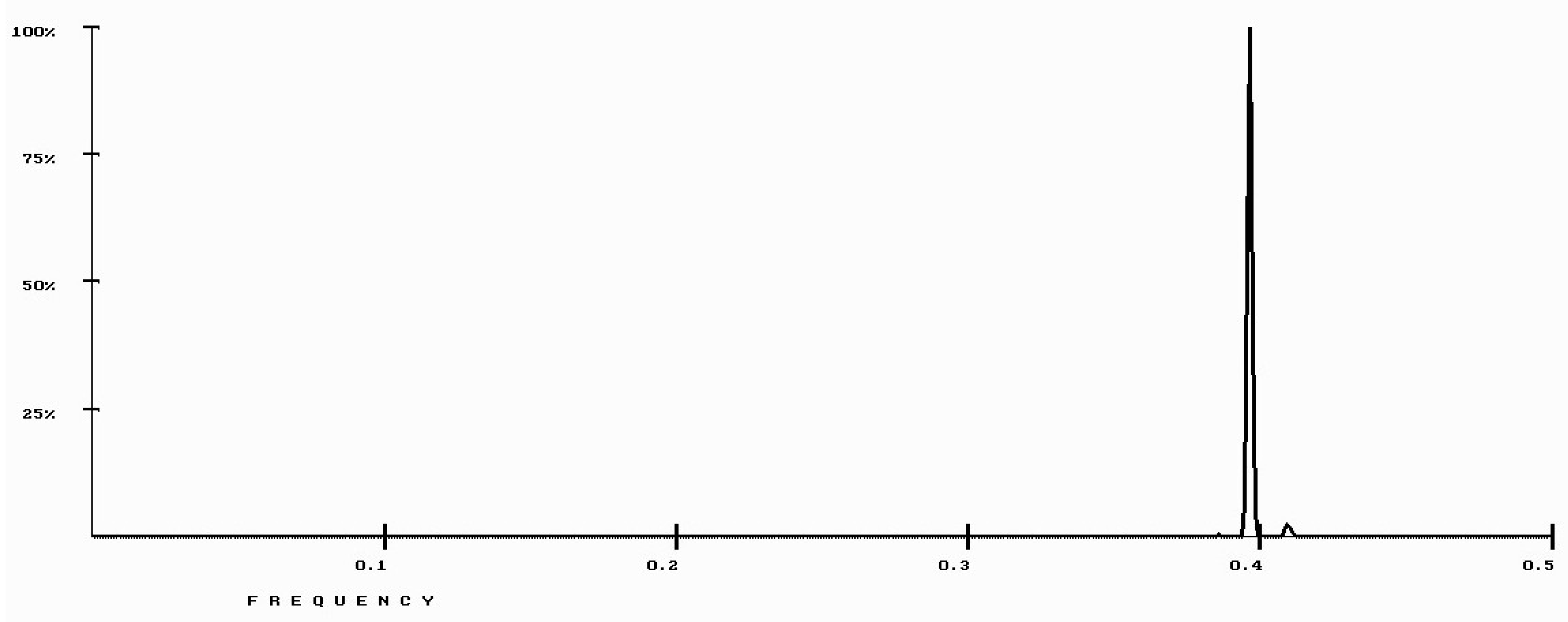
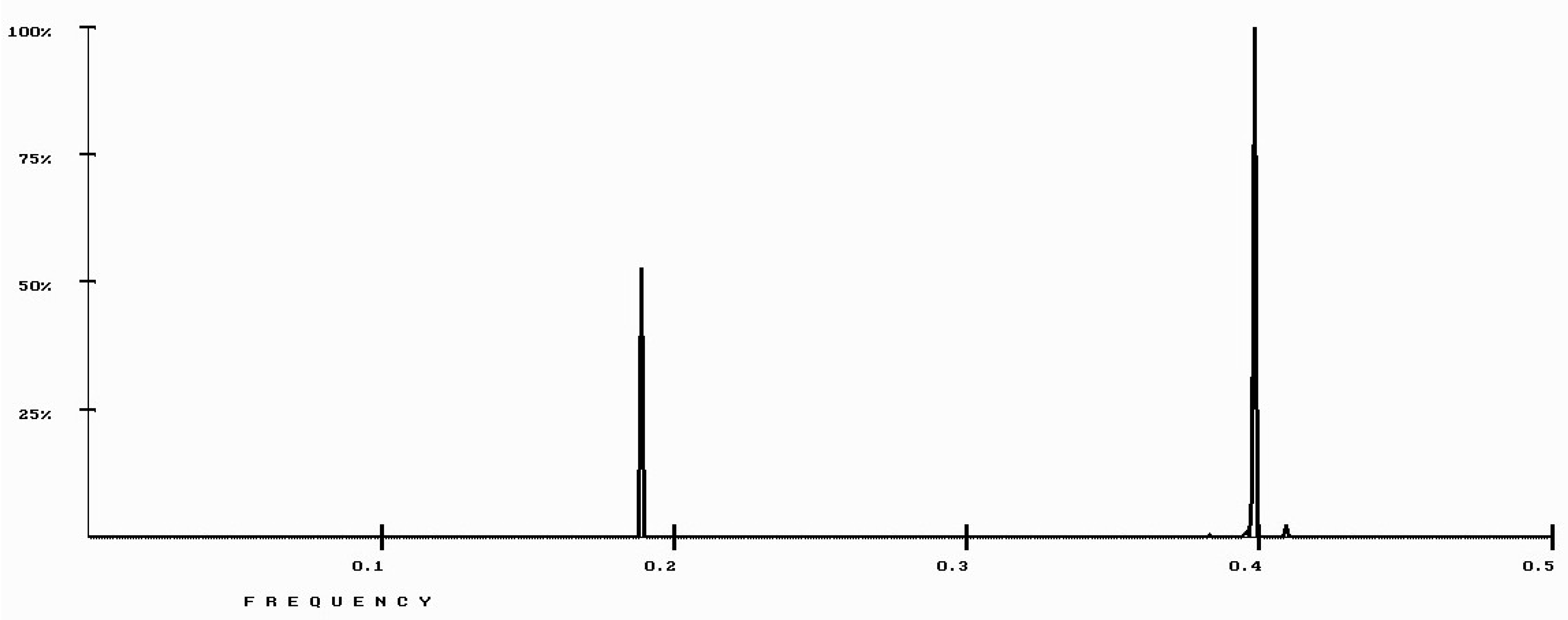
| Protein/Frequency | f2 = 0.1885 | f1 = 0.4072 |
|---|---|---|
| PTH | −1.98 | +0.58 |
| PTH 1-34 | −1.94 | +1.26 |
| PTH 7-34 | −1.38 | −1.88 |
| PTH receptor | +0.28 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cosic, I.; Paspaliaris, V.; Cosic, D. Explanation of Osteoblastic Differentiation of Stem Cells by Photo Biomodulation Using the Resonant Recognition Model. Appl. Sci. 2019, 9, 1979. https://doi.org/10.3390/app9101979
Cosic I, Paspaliaris V, Cosic D. Explanation of Osteoblastic Differentiation of Stem Cells by Photo Biomodulation Using the Resonant Recognition Model. Applied Sciences. 2019; 9(10):1979. https://doi.org/10.3390/app9101979
Chicago/Turabian StyleCosic, Irena, Vasilis Paspaliaris, and Drasko Cosic. 2019. "Explanation of Osteoblastic Differentiation of Stem Cells by Photo Biomodulation Using the Resonant Recognition Model" Applied Sciences 9, no. 10: 1979. https://doi.org/10.3390/app9101979
APA StyleCosic, I., Paspaliaris, V., & Cosic, D. (2019). Explanation of Osteoblastic Differentiation of Stem Cells by Photo Biomodulation Using the Resonant Recognition Model. Applied Sciences, 9(10), 1979. https://doi.org/10.3390/app9101979






