Measuring Identification and Quantification Errors in Spectral CT Material Decomposition
Abstract
1. Introduction
2. Materials and Methods
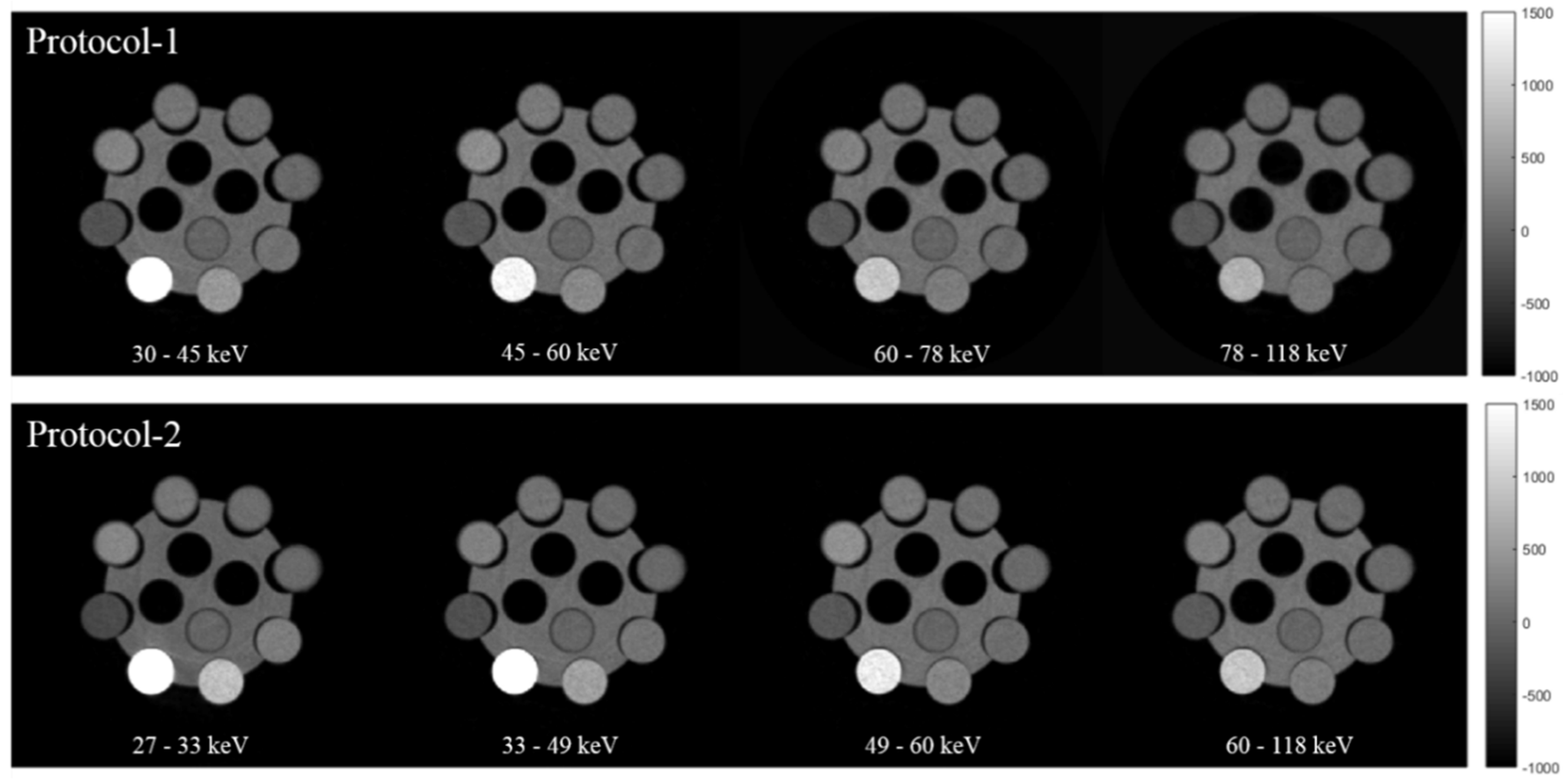
2.1. MARS Spectral Scanner
2.2. Experimental SETUP
2.3. Imaging Samples
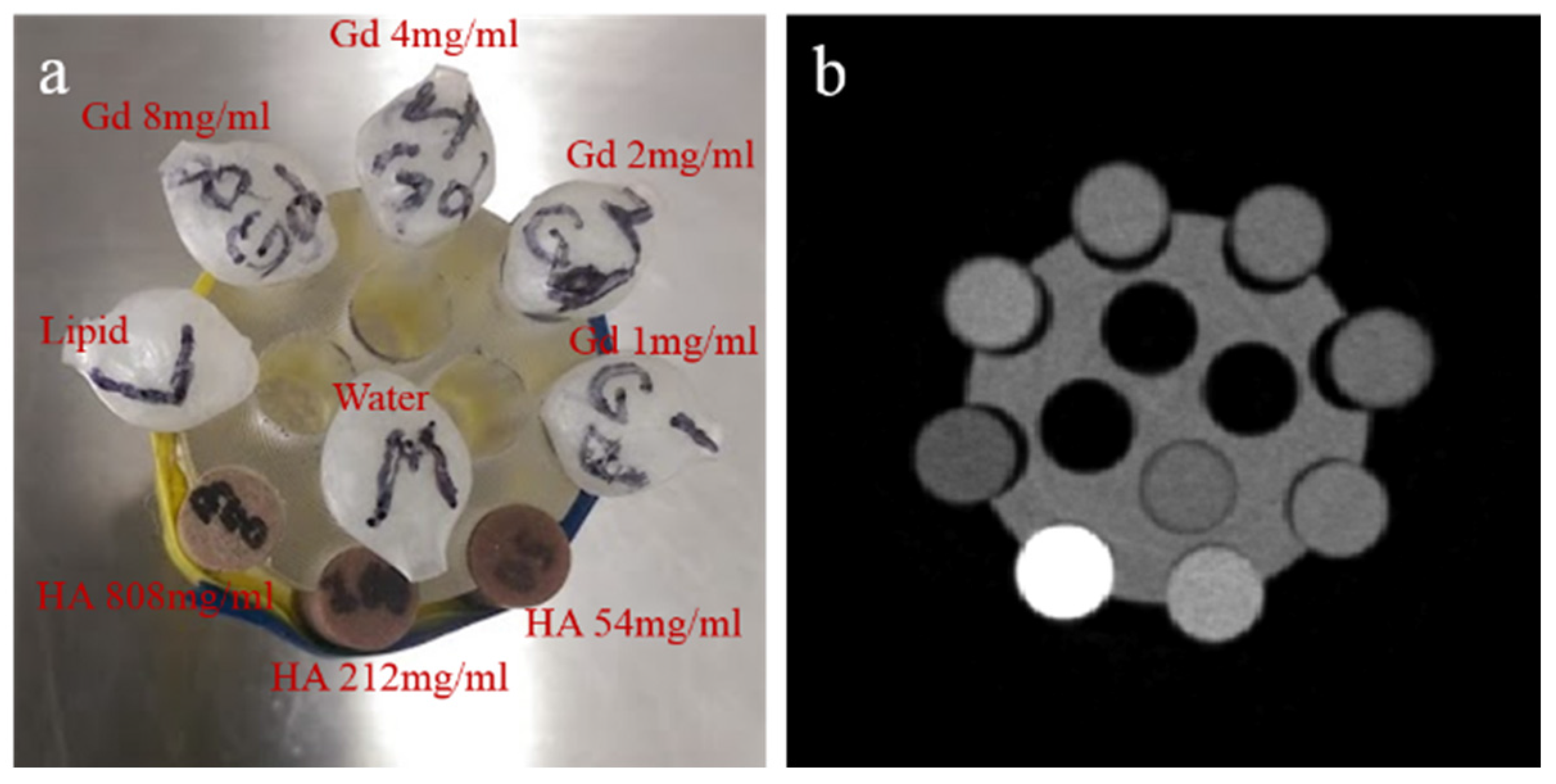
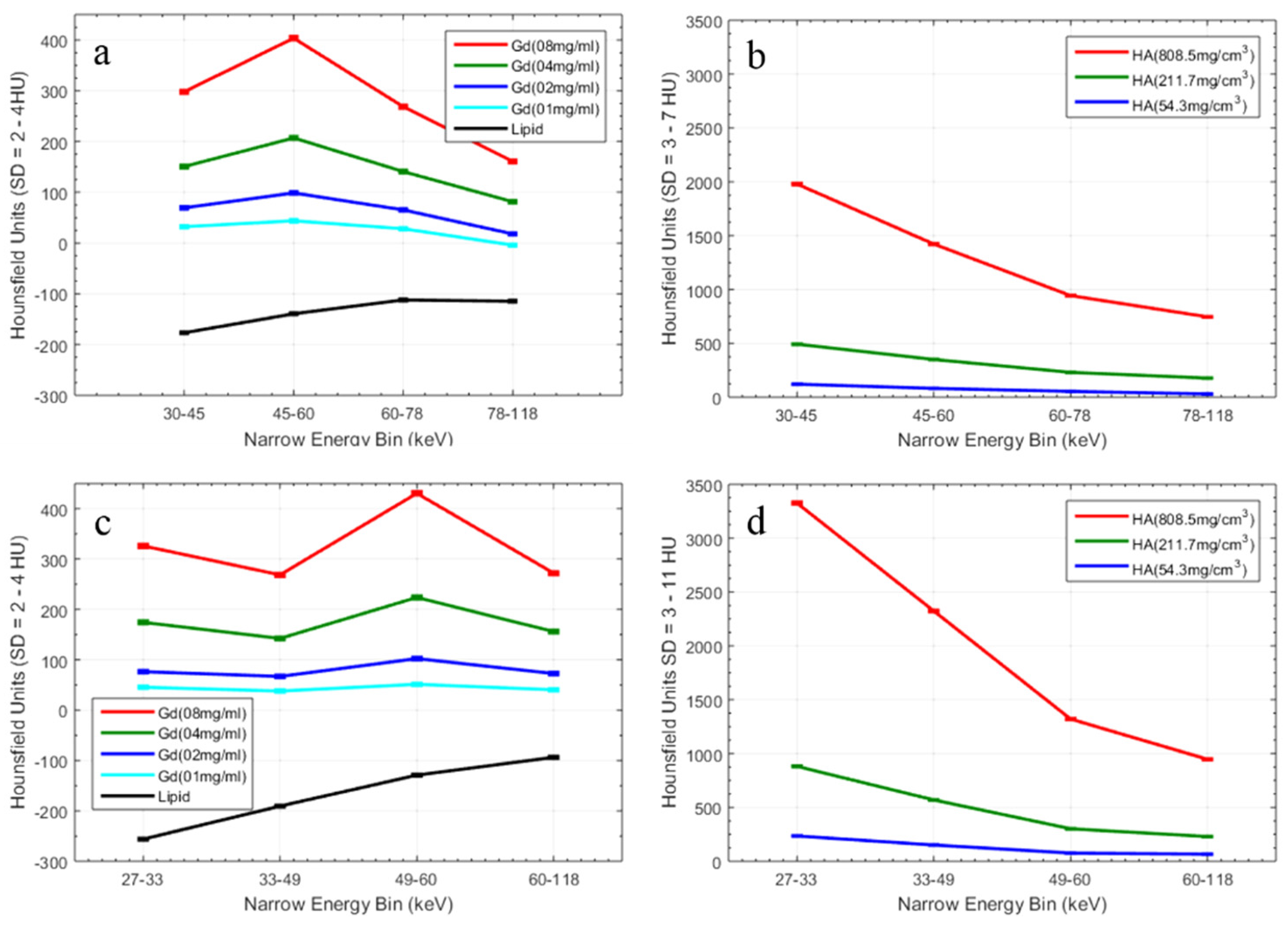
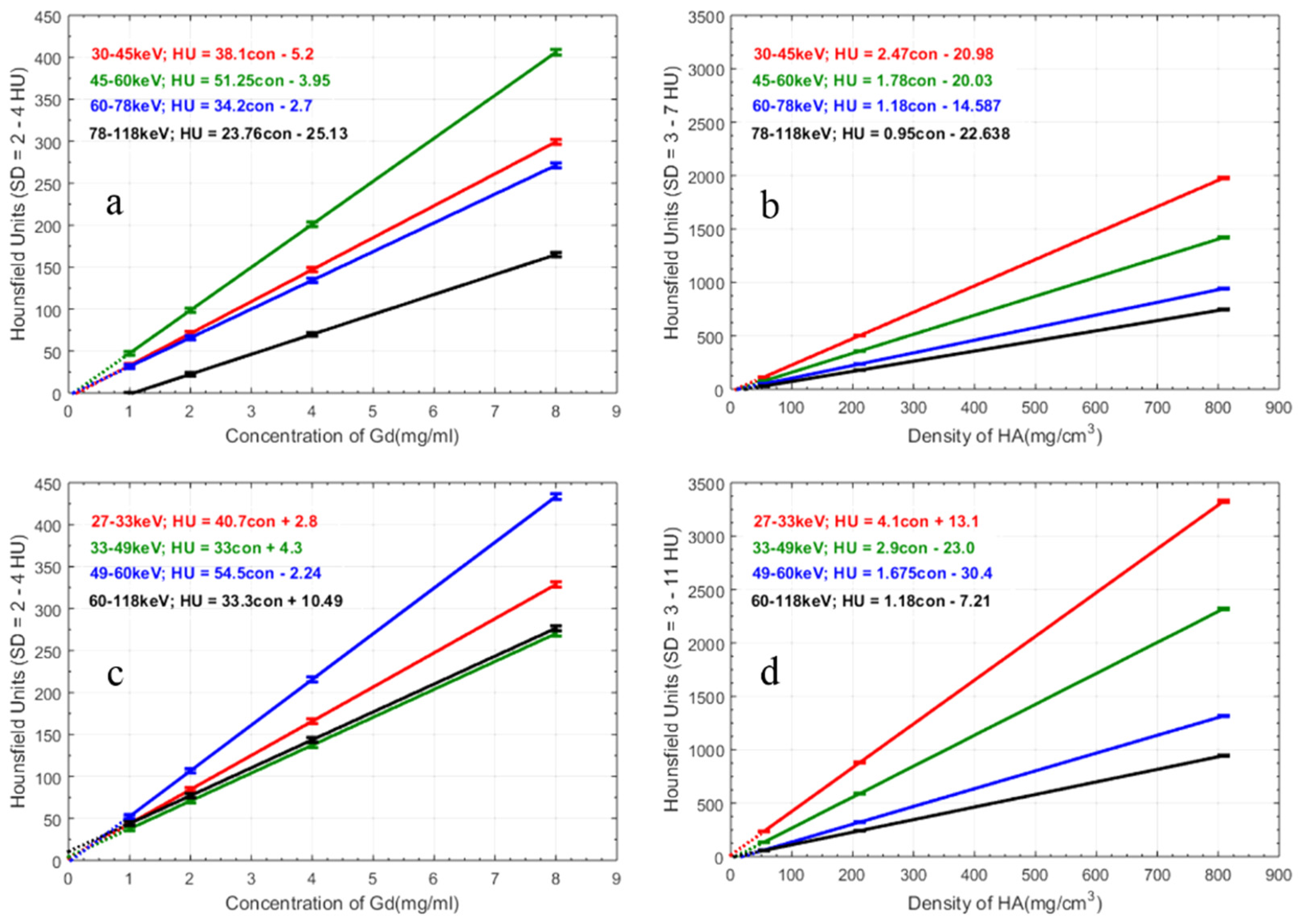
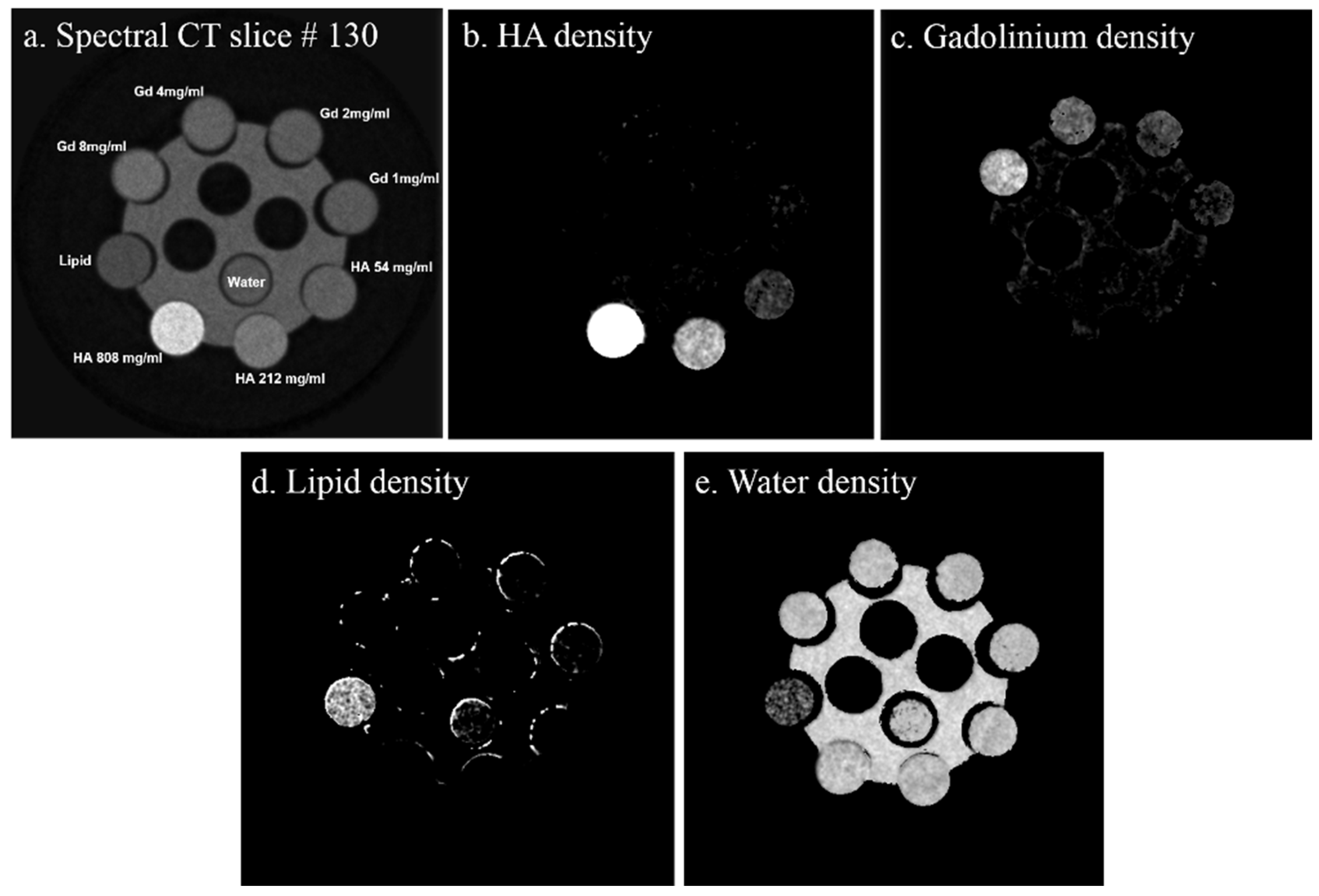
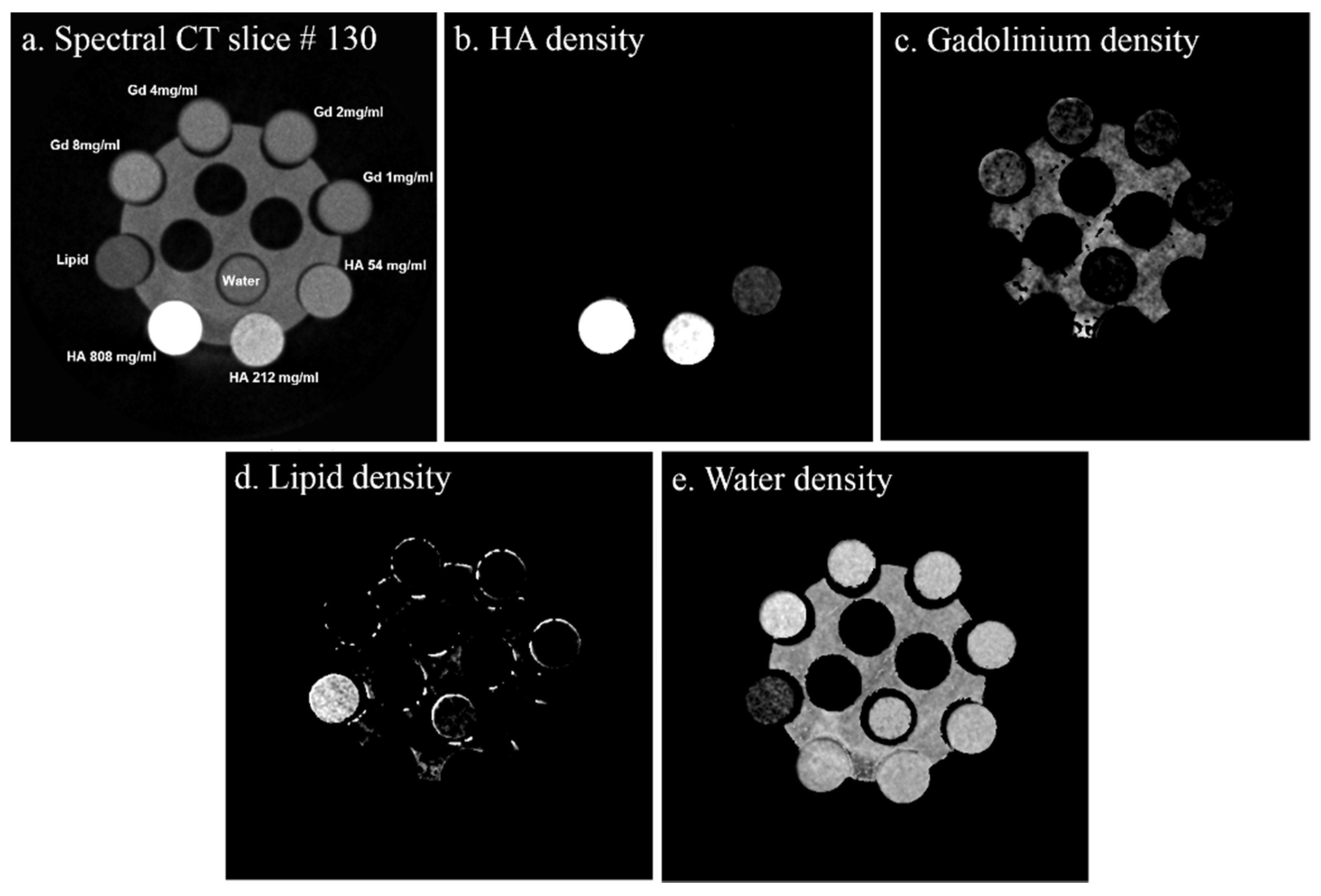
- Gd/HA phantom: A 31 mm-diameter polymethyl methacrylate (PMMA) phantom was custom-built, containing 6 mm diameter vials for a range of concentrations of gadolinium (1, 2, 4 and 8 mg/mL) and 6 mm diameter solid rods (20 mm long) of calcium hydroxyapatite (HA) (54.3, 211.7 and 808.5 mg/mL) (Quality Assurance in Radiology and Medicine (QRM) GmbH, Moehrendorf, Germany) along with water and a fat surrogate (vegetable oil) as shown in Figure 2a. Hydroxyapatite rods and water serve as bone-like and soft tissue-like material respectively. The gadolinium solutions (1, 2, 4, 8 mg/mL) were prepared by serially diluting Multihance (Gadobenate dimeglumine, Bracco Diagnostics Inc., Princeton, NJ, USA) in water.
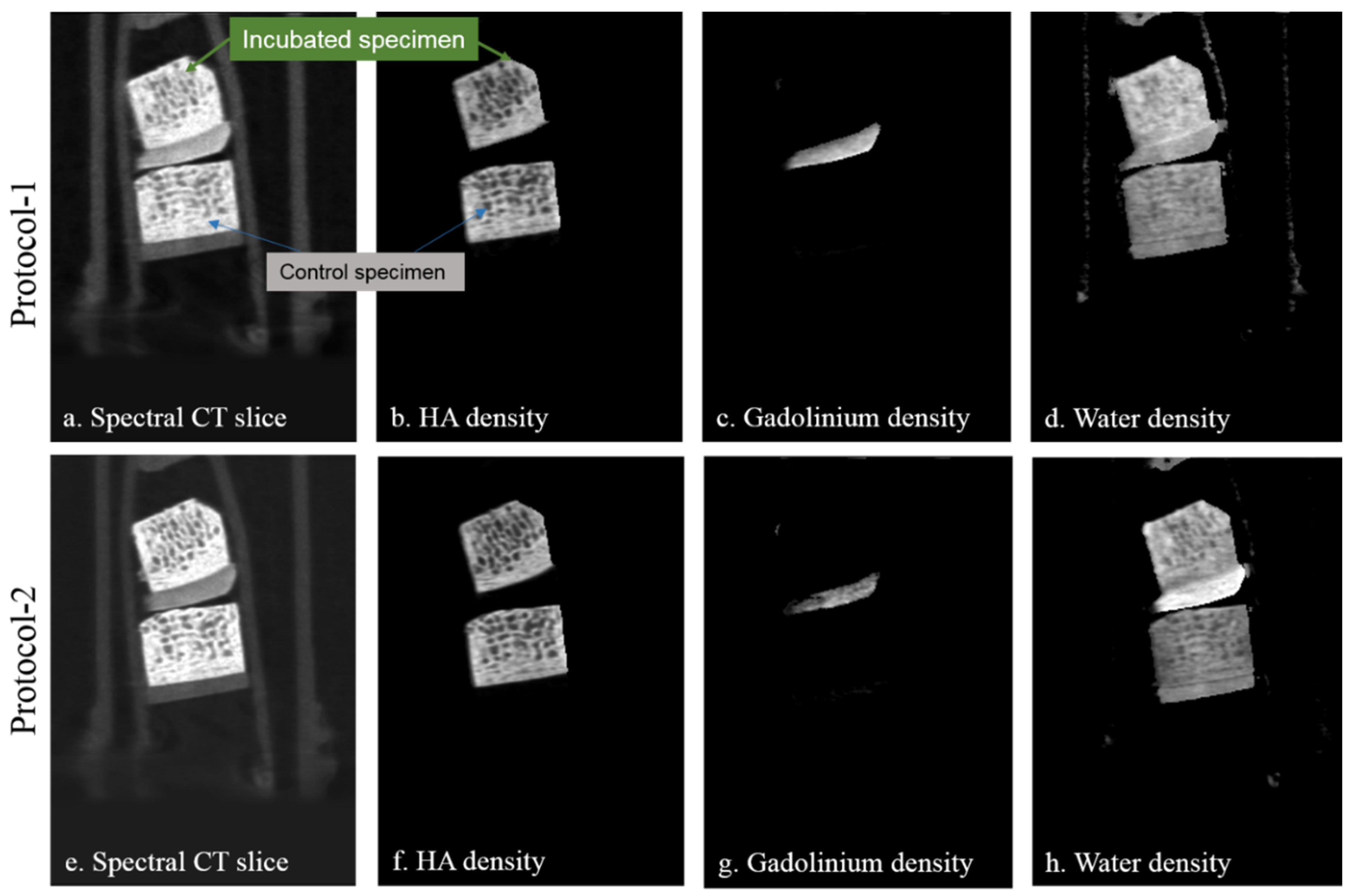
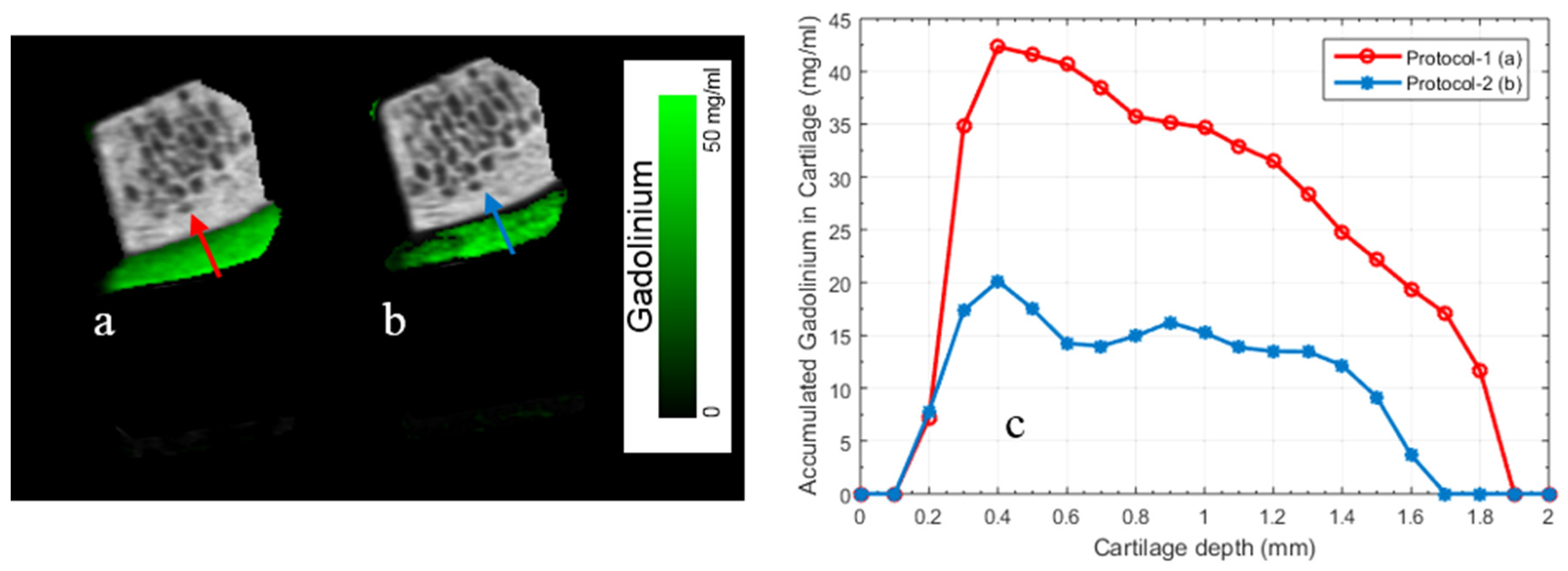
- Demonstration with biological specimens: Biological specimens were scanned to demonstrate the effectiveness of our proposed technique for correct material identification and quantification. Two cylindrical cartilage-bone plugs harvested from the bovine stifle joint, each with a diameter of 8 mm and length of 7 mm, were used to quantify gadolinium uptake in articular cartilage. The sample preparation setup was similar to that used in a previous study using iodinated contrast [9]. Samples were incubated for 24 h in Gd contrast agent solution (100% MultiHance) at 37 °C and then rinsed in PBS for 30 s. A non-Gd incubated sample was used as a control. Both incubated and control samples were placed side by side in a 15-mL Falcon tube, with PBS infused cotton wool at both ends, sealed to maintain a humid environment during scanning.
2.4. Image Processing and Reconstruction
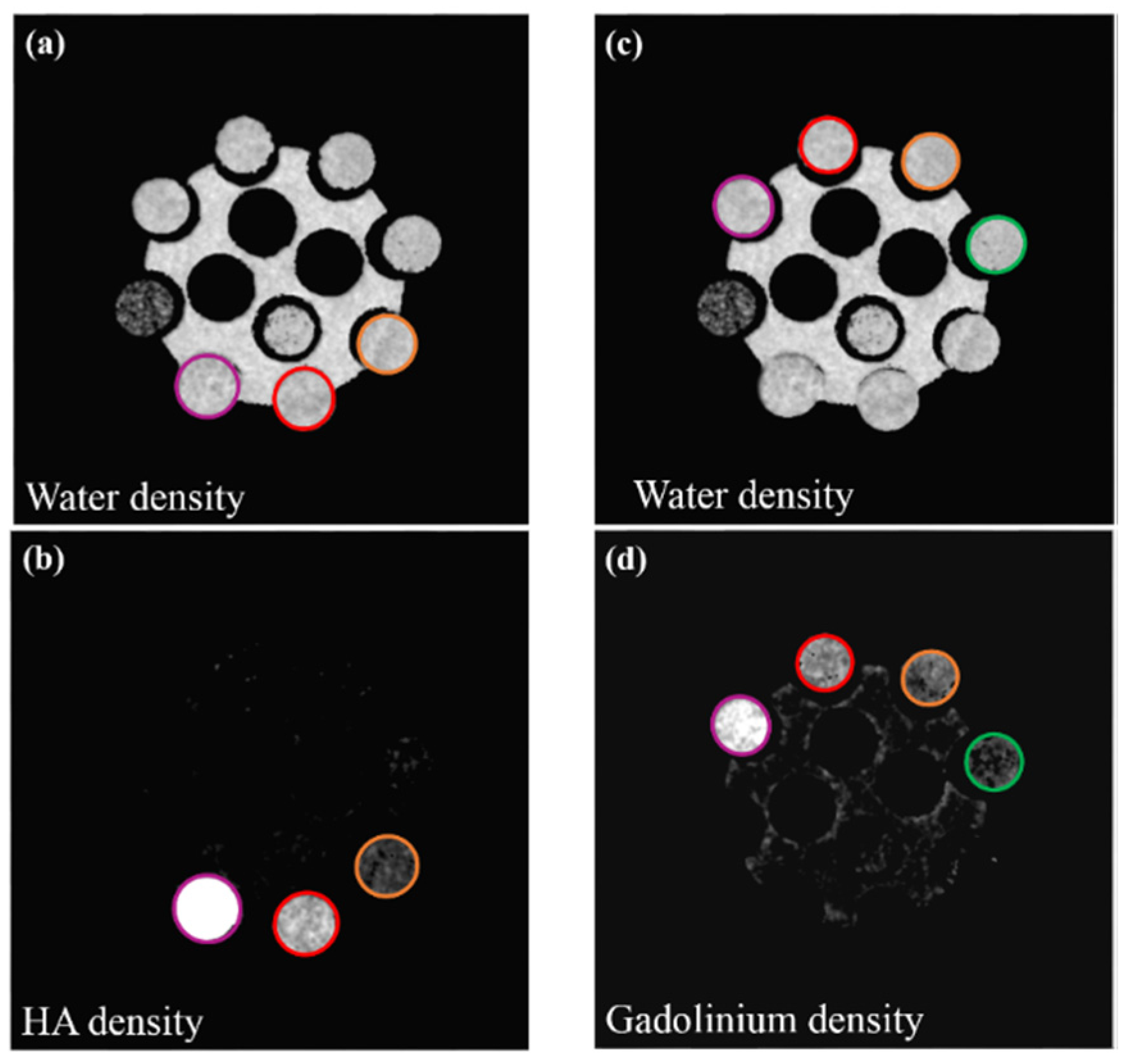
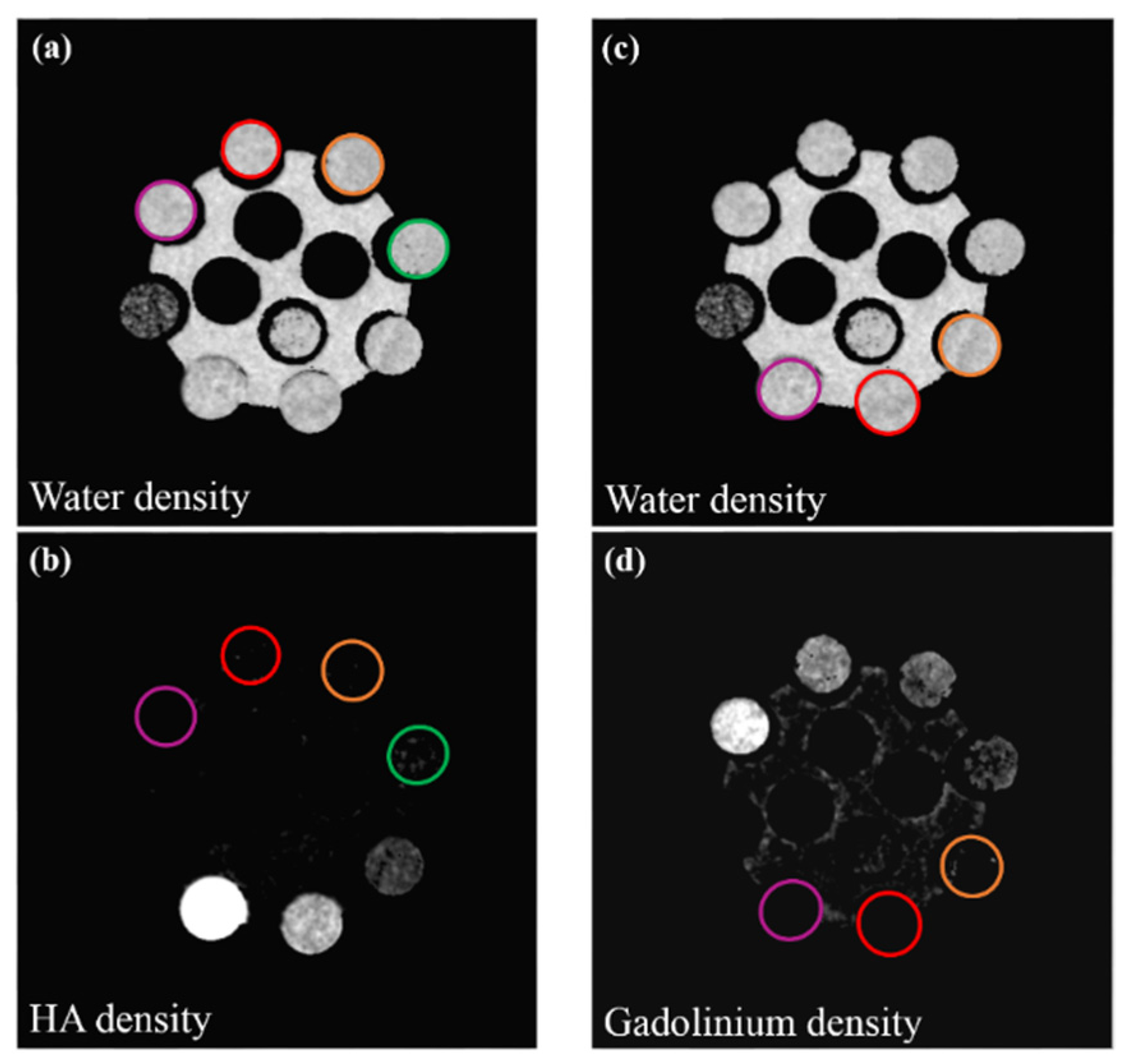
2.5. Material Decomposition
3. Results
4. Discussion
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Wang, A.S.; Pelc, N.J. Optimal energy thresholds and weights for separating materials using photon counting X-ray detectors with energy discriminating capabilities. Proc. SPIE 2009, 7258, 2101–2102. [Google Scholar]
- Baturin, P.; Alivov, Y.; Molloi, S. Spectral CT imaging of vulnerable plaque with two independent biomarkers. Phys. Med. Biol. 2012, 57, 4117–4138. [Google Scholar] [CrossRef] [PubMed]
- Zainon, R.; Ronaldson, J.; Janmale, T.; Scott, N.; Buckenham, T.; Butler, A.; Butler, P.; Doesburg, R.; Gieseg, S.; Roake, J.; et al. Spectral CT of carotid atherosclerotic plaque: Comparison with histology. Eur. Radiol. 2012, 22, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Anderson, N.; Butler, A.; Scott, N.; Cook, N.; Butzer, J.; Schleich, N.; Firsching, M.; Grasset, R.; de Ruiter, N.; Campbell, M.; et al. Spectroscopic (multi-energy) CT distinguishes iodine and barium contrast material in mice. Eur. Radiol. 2010, 20, 2126–2134. [Google Scholar] [CrossRef] [PubMed]
- Butler, A.P.H.; Butzer, J.; Schleich, N.; Cook, N.J.; Anderson, N.G.; Scott, N.; de Ruiter, N.; Grasset, R.; Tlustos, L.; Butler, P.H. Processing of spectral X-ray data with principal components analysis. Nucl. Instrum. Methods Phys. Res. Sec. A Accel. Spectrom. Detect. Assoc. Equip. 2011, 633 (Suppl. 1), S140–S142. [Google Scholar] [CrossRef]
- Ronaldson, J.P.; Zainon, R.; Scott, N.J.A.; Gieseg, S.P.; Butler, A.P.; Butler, P.H.; Anderson, N.G. Toward quantifying the composition of soft tissues by spectral CT with Medipix3. Med. Phys. 2012, 39, 6847–6857. [Google Scholar] [CrossRef] [PubMed]
- Aamir, R.; Chernoglazov, A.; Bateman, C.J.; Butler, A.P.H.; Butler, P.H.; Anderson, N.G.; Bell, S.T.; Panta, R.K.; Healy, J.L.; Mohr, J.L.; et al. MARS spectral molecular imaging of lamb tissue: Data collection and image analysis. J. Instrum. 2014, 9, P02005. [Google Scholar] [CrossRef]
- Rajendran, K.; Walsh, M.F.; Ruiter, N.J.A.D.; Chernoglazov, A.I.; Panta, R.K.; Butler, A.P.H.; Butler, P.H.; Bell, S.T.; Anderson, N.G.; Woodfield, T.B.F.; et al. Reducing beam hardening effects and metal artefacts in spectral CT using Medipix3RX. J. Instrum. 2014, 9, P03015. [Google Scholar] [CrossRef]
- Rajendran, K.; Löbker, C.; Schon, B.S.; Bateman, C.J.; Younis, R.A.; de Ruiter, N.J.A.; Chernoglazov, A.I.; Ramyar, M.; Hooper, G.J.; Butler, A.P.H.; et al. Quantitative imaging of excised osteoarthritic cartilage using spectral CT. Eur. Radiol. 2017, 27, 384–392. [Google Scholar] [CrossRef] [PubMed]
- Feuerlein, S.; Roessl, E.; Proksa, R.; Martens, G.; Klass, O.; Jeltsch, M.; Rasche, V.; Brambs, H.-J.; Hoffmann, M.H.K.; Schlomka, J.-P. Multienergy Photon-counting K-edge Imaging: Potential for Improved Luminal Depiction in Vascular Imaging. Radiology 2008, 249, 1010–1016. [Google Scholar] [CrossRef] [PubMed]
- He, P.; Wei, B.; Cong, W.; Wang, G. Optimization of K-edge imaging with spectral CT. Med. Phys. 2012, 39, 6572–6579. [Google Scholar] [CrossRef] [PubMed]
- Rink, K.; Oelfke, U.; Fiederle, M.; Zuber, M.; Koenig, T. Investigating the feasibility of photon-counting K-edge imaging at high X-ray fluxes using nonlinearity corrections. Med. Phys. 2013, 40, 101908. [Google Scholar] [CrossRef] [PubMed]
- Roessl, E.; Proksa, R. K-edge imaging in X-ray computed tomography using multi-bin photon counting detectors. Phys. Med. Biol. 2007, 52, 4679–4696. [Google Scholar] [CrossRef] [PubMed]
- Aamir, R. Using MARS Spectral CT for Identifying Biomedical Nanoparticles. Ph.D. Thesis, University of Canterbury, Christchurch, New Zealand, 2013. [Google Scholar]
- Roessl, E.; Cormode, D.; Brendel, B.; Jürgen Engel, K.; Martens, G.; Thran, A.; Fayad, Z.; Proksa, R. Preclinical spectral computed tomography of gold nano-particles. Nucl. Instrum. Methods Phys. Res. Sec. A Accel. Spectrom. Detect. Assoc. Equip. 2011, 648 (Suppl. 1), S259–S264. [Google Scholar] [CrossRef]
- Badea, C.T.; Johnston, S.M.; Qi, Y.; Ghaghada, K.; Johnson, G.A. Dual-energy micro-CT imaging for differentiation of iodine- and gold-based nanoparticles. Med. Imaging 2011 Phys. Med. Imaging 2011, 7961, 79611X. [Google Scholar] [CrossRef]
- Ashton, J.R.; West, J.L.; Badea, C.T. In vivo small animal micro-CT using nanoparticle contrast agents. Front. Pharmacol. 2015, 6, 256. [Google Scholar] [CrossRef] [PubMed]
- Cormode, D.P.; Roessl, E.; Thran, A.; Skajaa, T.; Gordon, R.E.; Schlomka, J.-P.; Fuster, V.; Fisher, E.A.; Mulder, W.J.M.; Proksa, R.; et al. Atherosclerotic Plaque Composition: Analysis with Multicolor CT and Targeted Gold Nanoparticles. Radiology 2010, 256, 774–782. [Google Scholar] [CrossRef] [PubMed]
- Fornaro, J.; Leschka, S.; Hibbeln, D.; Butler, A.; Anderson, N.; Pache, G.; Scheffel, H.; Wildermuth, S.; Alkadhi, H.; Stolzmann, P. Dual- and multi-energy CT: Approach to functional imaging. Insights Imaging 2011, 2, 149–159. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, R.E.; Macovski, A. Energy-selective reconstructions in X-ray computerised tomography. Phys. Med. Biol. 1976, 21, 733. [Google Scholar] [CrossRef] [PubMed]
- Brooks, R.A. A Quantitative Theory of the Hounsfield Unit and Its Application to Dual Energy Scanning. J. Comput. Assist. Tomogr. 1977, 1, 487–493. [Google Scholar] [CrossRef] [PubMed]
- Riederer, S.J.; Mistretta, C.A. Selective iodine imaging using K-edge energies in computerized X-ray tomography. Med. Phys. 1977, 4, 474–481. [Google Scholar] [CrossRef] [PubMed]
- Schlomka, J.P.; Roessl, E.; Dorscheid, R.; Dill, S.; Martens, G.; Istel, T.; Bäumer, C.; Herrmann, C.; Steadman, R.; Zeitler, G.; et al. Experimental feasibility of multi-energy photon-counting K-edge imaging in pre-clinical computed tomography. Phys. Med. Biol. 2008, 53, 4031–4047. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, T.G.; Pektas, F. Region-of-interest material decomposition from truncated energy-resolved CT. Med. Phys. 2011, 38, 5657–5666. [Google Scholar] [CrossRef] [PubMed]
- Huy, Q.; Le, S.M. Segmentation and quantification of materials with energy discriminating computed tomography: A phantom study. Med. Phys. 2011, 38, 228–237. [Google Scholar] [CrossRef]
- Nik, S.J.; Meyer, J.; Watts, R. Optimal material discrimination using spectral X-ray imaging. Phys. Med. Biol. 2011, 56, 5969–5983. [Google Scholar] [CrossRef] [PubMed]
- Jakubek, J. Energy-sensitive X-ray radiography and charge sharing effect in pixelated detector. Nucl. Instrum. Methods Phys. Res. Sec. A Accel. Spectrom. Detect. Assoc. Equip. 2009, 607, 192–195. [Google Scholar] [CrossRef]
- Fink, J.; Kraft, E.; Kruger, H.; Wermes, N.; Engel, K.J.; Herrmann, C. Comparison of Pixelated CdZnTe, CdTe and Si Sensors With the Simultaneously Counting and Integrating CIX Chip. IEEE Trans. Nucl. Sci. 2009, 56, 3819–3827. [Google Scholar] [CrossRef]
- Funaki, M.; Ozaki, T.; Satoh, K.; Ohno, R. Growth and characterization of CdTe single crystals for radiation detectors. Nucl. Instrum. Methods Phys. Res. Sec. A Accel. Spectrom. Detect. Assoc. Equip. 1999, 436, 120–126. [Google Scholar] [CrossRef]
- Sellin, P.J.; Davies, A.W.; Lohstroh, A.; Ozsan, M.E.; Parkin, J. Drift mobility and mobility-lifetime products in CdTe:Cl grown by the travelling heater method. IEEE Trans. Nucl. Sci. 2005, 52, 3074–3078. [Google Scholar] [CrossRef]
- Panta, R.K.; Walsh, M.F.; Bell, S.T.; Anderson, N.G.; Butler, A.P.; Butler, P.H. Energy Calibration of the Pixels of Spectral X-ray Detectors. IEEE Trans. Med. Imaging 2015, 34, 697–706. [Google Scholar] [CrossRef] [PubMed]
- Aamir, R.; Lansley, S.P.; Zainon, R.; Fiederle, M.; Fauler, A.; Greiffenberg, D.; Butler, P.H.; Butler, A.P.H. Pixel sensitivity variations in a CdTe-Medipix2 detector using poly-energetic X-rays. J. Instrum. 2011, 6, C01059. [Google Scholar] [CrossRef]
- Frey, E.C.; Wang, X.; Du, Y.; Taguchi, K.; Xu, J.; Tsui, B.M.W. Investigation of the use of photon counting X-ray detectors with energy discrimination capability for material decomposition in micro-computed tomography. Proc SPIE 2007, 6510, 65100A. [Google Scholar] [CrossRef]
- Endrizzi, M.; Nesterets, Y.I.; Mayo, S.C.; Trinchi, A.; Gureyev, T.E. Multi-energy computed tomography using pre-reconstruction decomposition and iterative reconstruction algorithms. J. Phys. D Appl. Phys. 2012, 45, 475103. [Google Scholar] [CrossRef]
- Ehn, S.; Sellerer, T.; Mechlem, K.; Fehringer, A.; Epple, M.; Herzen, J.; Pfeiffer, F.; Noël, P.B. Basis material decomposition in spectral CT using a semi-empirical, polychromatic adaption of the Beer–Lambert model. Phys. Med. Biol. 2017, 62, N1–N17. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Ding, H.; Molloi, S.; Zhang, X.; Gao, H. TICMR: Total Image Constrained Material Reconstruction via Nonlocal Total Variation Regularization for Spectral CT. IEEE Trans. Med. Imaging 2016, 35, 2578–2586. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Xing, Z. A practical material decomposition method for X-ray dual spectral computed tomography. J. X-Ray Sci. Technol. 2016, 24, 207–425. [Google Scholar] [CrossRef] [PubMed]
- Butler, P.H.; Bell, A.J.; Butler, A.P.H.; Cook, N.J.; Reinisch, L.; Butzer, J.S.; Anderson, N. Applying CERN’s detector technology to health: MARS Biomedical 3D spectroscopic X-ray imaging. In Proceedings of the International Symposium on Peaceful Applications of Nuclear Technologies in the GCC Countries, Jeddah, Saudi Arabia, 3–5 November 2008. [Google Scholar]
- Ronaldson, J.P.; Butler, A.P.; Anderson, N.G.; Zainon, R.; Butler, P.H. The performance of MARS-CT using Medipix3 for spectral imaging of soft-tissue. In Proceedings of the 2011 IEEE Nuclear Science Symposium and Medical Imaging Conference (NSS/MIC), Valencia, Spain, 23–29 October 2011; pp. 4002–4008. [Google Scholar]
- Walsh, M.F.; Opie, A.M.T.; Ronaldson, J.P.; Doesburg, R.M.N.; Nik, S.J.; Mohr, J.L.; Ballabriga, R.; Butler, A.P.H.; Butler, P.H. First CT using Medipix3 and the MARS-CT-3 spectral scanner. J. Instrum. 2011, 6, C01095. [Google Scholar] [CrossRef]
- Ballabriga, R.; Alozy, J.; Blaj, G.; Campbell, M.; Fiederle, M.; Frojdh, E.; Heijne, E.H.M.; Llopart, X.; Pichotka, M.; Procz, S.; et al. The Medipix3RX: A high resolution, zero dead-time pixel detector readout chip allowing spectroscopic imaging. JINST 2013, 8. [Google Scholar] [CrossRef]
- Gimenez, E.N.; Ballabriga, R.; Campbell, M.; Horswell, I.; Llopart, X.; Marchal, J.; Sawhney, K.J.S.; Tartoni, N.; Turecek, D. Study of charge-sharing in MEDIPIX3 using a micro-focused synchrotron beam. J. Instrum. 2011, 6, C01031. [Google Scholar] [CrossRef]
- Ballabriga, R.; Campbell, M.; Heijne, E.; Llopart, X.; Tlustos, L.; Wong, W. Medipix3: A 64 k pixel detector readout chip working in single photon counting mode with improved spectrometric performance. Nucl. Instrum. Methods Phys. Res. Sec. A Accel. Spectrom. Detect. Assoc. Equip. 2011, 633 (Suppl. 1), S15–S18. [Google Scholar] [CrossRef]
- Ronaldson, J.P.; Walsh, M.; Nik, S.J.; Donaldson, J.; Doesburg, R.M.N.; van Leeuwen, D.; Ballabriga, R.; Clyne, M.N.; Butler, A.P.H.; Butler, P.H. Characterization of Medipix3 with the MARS readout and software. J. Instrum. 2011, 6, C01056. [Google Scholar] [CrossRef]
- Walsh, M.F. Spectral Computed Tomography Development; University of Otago: Christchurch, New Zealand, 2014. [Google Scholar]
- Jakubek, J. Semiconductor Pixel detectors and their applications in life sciences. J. Instrum. 2009, 4, P03013. [Google Scholar] [CrossRef]
- Jan, S.; Andrei, P. Reduction of ring artefacts in high resolution micro-CT reconstructions. Phys. Med. Biol. 2004, 49, N247–N253. [Google Scholar]
- Hudson, H.M.; Larkin, R.S. Accelerated image reconstruction using ordered subsets of projection data. IEEE Trans. Med. Imaging 1994, 13, 601–609. [Google Scholar] [CrossRef] [PubMed]
- Gordon, R.; Bender, R.; Herman, G.T. Algebraic Reconstruction Techniques (ART) for three-dimensional electron microscopy and X-ray photography. J. Theor. Biol. 1970, 29, 471–481. [Google Scholar] [CrossRef]
- De Ruiter, N.J.A.; Butler, P.H.; Butler, A.P.H.; Bell, S.T.; Chernoglazov, A.I.; Walsh, M.F. MARS imaging and reconstruction challenges. In Proceedings of the 14th International Meeting on Fully Three-Dimensional Image Reconstruction in Radiology and Nuclear Medicine, Xi’an, China, 18–23 June 2017; pp. 852–857. [Google Scholar]
- Hurrell, M.; Butler, A.; Cook, N.; Butler, P.; Ronaldson, J.P.; Zainon, R. Spectral Hounsfield units: A new radiological concept. Eur. Radiol. 2012, 22, 1008–1013. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Yu, L.; Primak, A.N.; McCollough, C.H. Quantitative imaging of element composition and mass fraction using dual-energy CT: Three-material decomposition. Med. Phys. 2009, 36, 1602–1609. [Google Scholar] [CrossRef] [PubMed]
- White, D.R. An analysis of the Z-dependence of photon and electron interactions. Phys. Med. Biol. 1977, 22, 219–228. [Google Scholar] [CrossRef] [PubMed]
- Heismann, B.J.; Leppert, J.; Stierstorfer, K. Density and atomic number measurements with spectral X-ray attenuation method. J. Appl. Phys. 2003, 94, 2073–2079. [Google Scholar] [CrossRef]
- Bateman, C.J.; McMahon, J.; Malpas, A.; de Ruiter, N.; Bell, S.; Butler, A.P.; Butler, P.H.; Renaud, P.F. Segmentation enhances material analysis in multi-energy CT: A simulation study. In Proceedings of the 2013 28th International Conference on Image and Vision Computing New Zealand (IVCNZ 2013), Wellington, New Zealand, 27–29 November 2013; pp. 190–195. [Google Scholar]
- Bateman, C.J. Methods for Material Discrimination in MARS Multi-Energy CT; University of Otago: Christchurch, New Zealand, 2015. [Google Scholar]
- Bateman, C.J.; Knight, D.; Brandwacht, B.; Mahon, J.M.; Healy, J.; Panta, R.; Aamir, R.; Rajendran, K.; Moghiseh, M.; Ramyar, M.; et al. MARS-MD: Rejection based image domain material decomposition. arXiv, 2018; arXiv:1802.05366. [Google Scholar]
- Barrett, J.F.; Keat, N. Artifacts in CT: Recognition and Avoidance1. Radiographics 2004, 24, 1679–1691. [Google Scholar] [CrossRef] [PubMed]
- Lin, P.; Beck, T.; Borras, C.; Cohen, G.; Jucius, R.; Kriz, R.; Nickoloff, E.; Rothenberg, L.; Strauss, K.; Villafana, T. Specification and Acceptance Testing of Computed Tomography Scanners; The American Association of Physicists in Medicine (AAPM): Alexandria, VA, USA, 1993. [Google Scholar]
- Davis, J.E. Event Pileup in Charge-Coupled Devices. Astrophys. J. 2001, 562, 575–582. [Google Scholar] [CrossRef]
- Taguchi, K.; Frey, E.C.; Wang, X.; Iwanczyk, J.S.; Barber, W.C. An analytical model of the effects of pulse pileup on the energy spectrum recorded by energy resolved photon counting X-ray detectors. Med. Phys. 2010, 37, 3957–3969. [Google Scholar] [CrossRef] [PubMed]
- Aamir, R.; Anderson, N.G.; Butler, A.P.H.; Butler, P.H.; Lansley, S.P.; Doesburg, R.M.; Walsh, M.; Mohr, J.L. Characterization of Si and CdTe sensor layers in Medipix assemblies using a microfocus X-ray source. In Proceedings of the 2011 IEEE Nuclear Science Symposium and Medical Imaging Conference (NSS/MIC), Valencia, Spain, 23–29 October 2011; pp. 4766–4769. [Google Scholar]
- Wait, J.M.S.; Cody, D.; Jones, A.K.; Rong, J.; Baladandayuthapani, V.; Kappadath, S.C. Performance Evaluation of Material Decomposition With Rapid-Kilovoltage-Switching Dual-Energy CT and Implications for Assessing Bone Mineral Density. Am. J. Roentgenol. 2015, 204, 1234–1241. [Google Scholar] [CrossRef] [PubMed]
- Alessio, A.M.; MacDonald, L.R. Quantitative material characterization from multi-energy photon counting CT. Med. Phys. 2013, 40, 031108. [Google Scholar] [CrossRef] [PubMed]

| Imaging Protocols | Tube Voltage (kVp) | Tube Current (µA) | Exposure Time (ms) | External Filtration (mm) | Intrinsic Filtration (mm) | SDD (mm) | Energy Bins (keV) |
|---|---|---|---|---|---|---|---|
| Protocol-1 | 118 | 24 | 220/frame | 0.375 Brass | 1.8 (Al equivalent) | 250 | 30–45, 45–60, 60–78 and 78–118 |
| Protocol-2 | 118 | 30 | 130/frame | - | 1.8 (Al equivalent) | 250 | 27–33, 33–49, 49–60 and 60–118 |
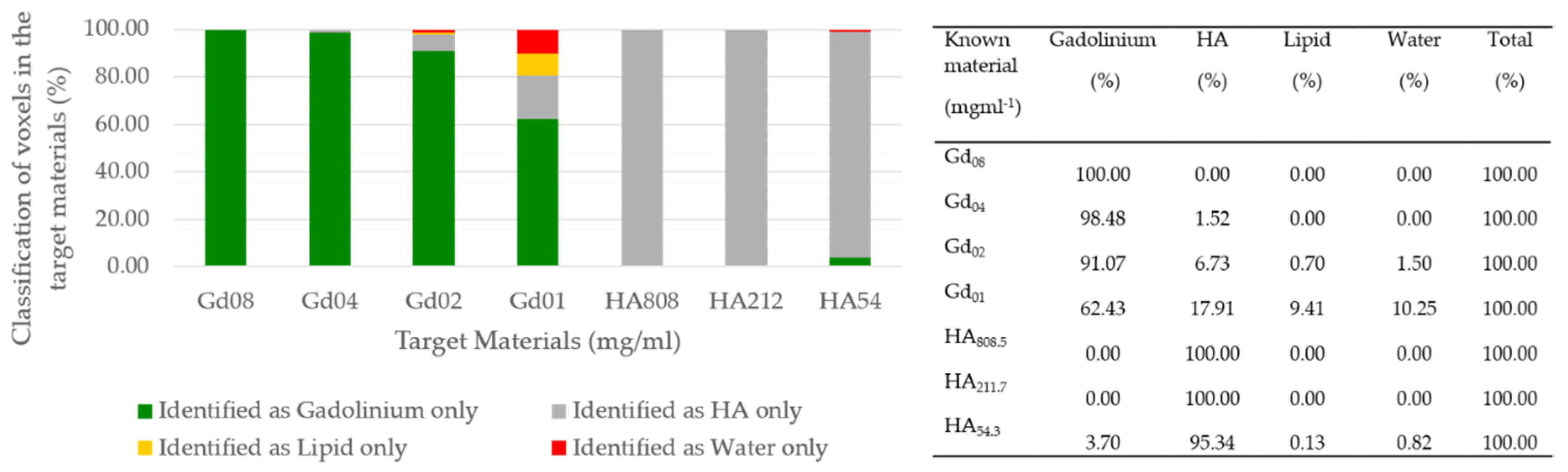
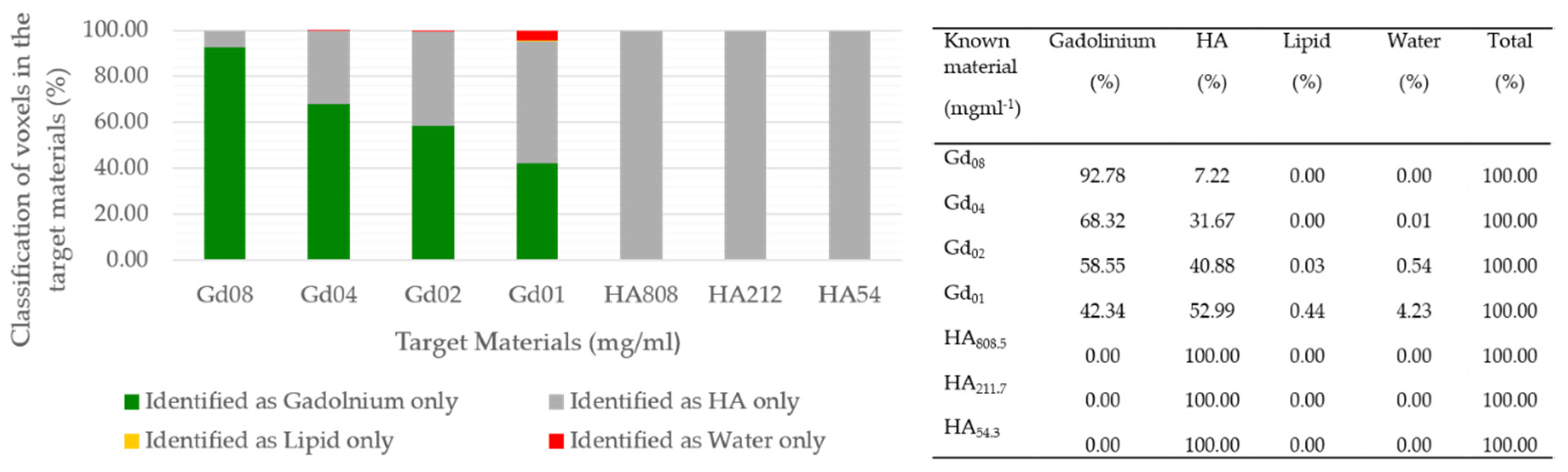
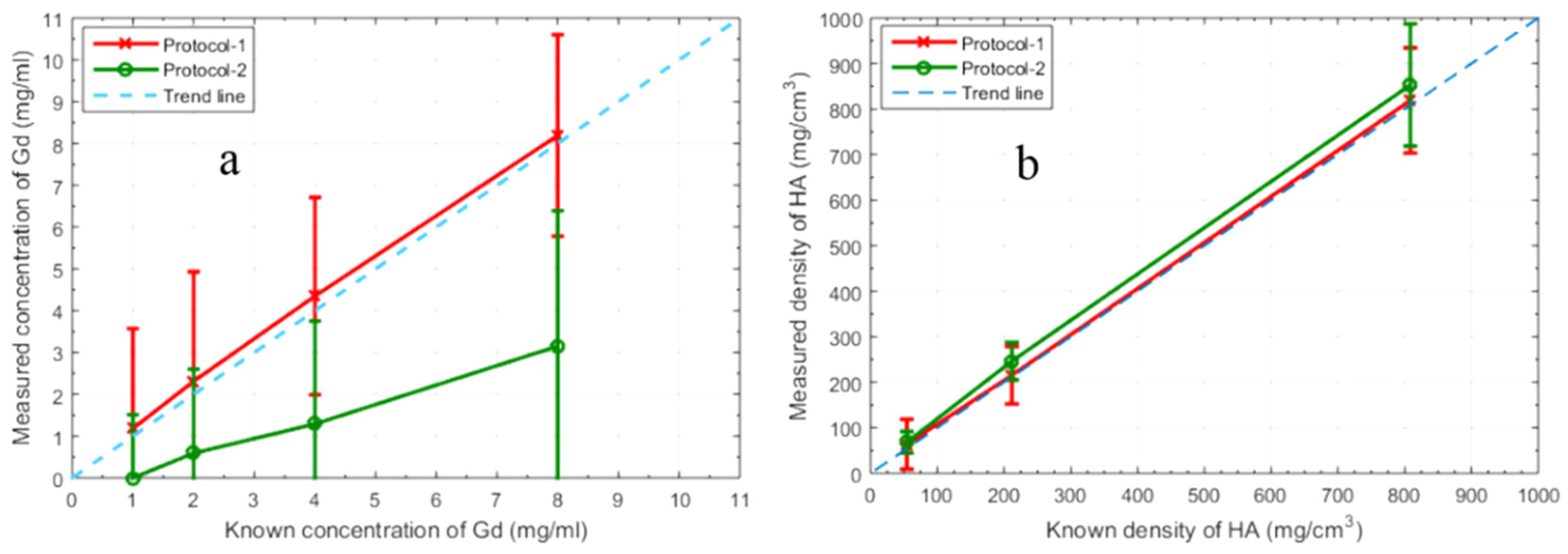
| Known Material (mg mL−1) | Protocol-1 Measured Material (mg mL−1) | Protocol-2 Measured Material (mg mL−1) | Protocol-1 Measured Misidentification (%) | Protocol-2 Measured Misidentification (%) |
|---|---|---|---|---|
| Gd08 | 8.19 | 3.15 | 0 | 7.22 |
| Gd04 | 4.35 | 1.29 | 1.52 | 31.68 |
| Gd02 | 2.31 | 0.59 | 8.93 | 41.45 |
| Gd01 | 1.18 | 0 | 37.57 | 57.66 |
| HA808.5 | 818.9 | 852.9 | 0 | 0 |
| HA211.7 | 215.4 | 246.4 | 0 | 0 |
| HA54.3 | 63.78 | 68.09 | 4.66 | 0 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Raja, A.Y.; Moghiseh, M.; Bateman, C.J.; De Ruiter, N.; Schon, B.; Schleich, N.; Woodfield, T.B.F.; Butler, A.P.H.; Anderson, N.G. Measuring Identification and Quantification Errors in Spectral CT Material Decomposition. Appl. Sci. 2018, 8, 467. https://doi.org/10.3390/app8030467
Raja AY, Moghiseh M, Bateman CJ, De Ruiter N, Schon B, Schleich N, Woodfield TBF, Butler APH, Anderson NG. Measuring Identification and Quantification Errors in Spectral CT Material Decomposition. Applied Sciences. 2018; 8(3):467. https://doi.org/10.3390/app8030467
Chicago/Turabian StyleRaja, Aamir Younis, Mahdieh Moghiseh, Christopher J. Bateman, Niels De Ruiter, Benjamin Schon, Nanette Schleich, Tim B. F. Woodfield, Anthony P. H. Butler, and Nigel G. Anderson. 2018. "Measuring Identification and Quantification Errors in Spectral CT Material Decomposition" Applied Sciences 8, no. 3: 467. https://doi.org/10.3390/app8030467
APA StyleRaja, A. Y., Moghiseh, M., Bateman, C. J., De Ruiter, N., Schon, B., Schleich, N., Woodfield, T. B. F., Butler, A. P. H., & Anderson, N. G. (2018). Measuring Identification and Quantification Errors in Spectral CT Material Decomposition. Applied Sciences, 8(3), 467. https://doi.org/10.3390/app8030467





