Game Theory in Molecular Nanosensing System for Rapid Detection of Hg2+ in Aqueous Solutions
Abstract
1. Introduction
2. Experiment Materials and Methods
2.1. Materials and Reagents
2.2. Instruments
2.3. Synthesis of Cobalt Oxyhydroxide (CoOOH) Nanosheets
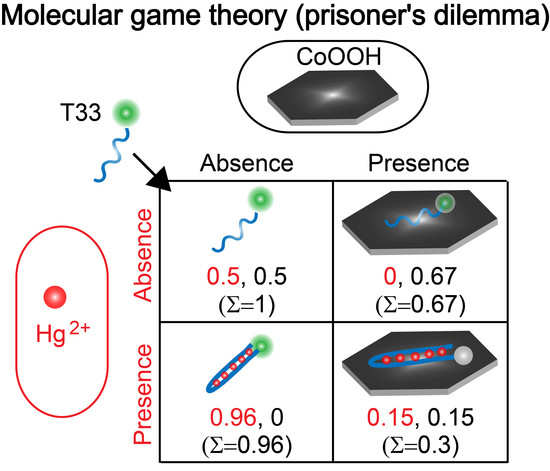
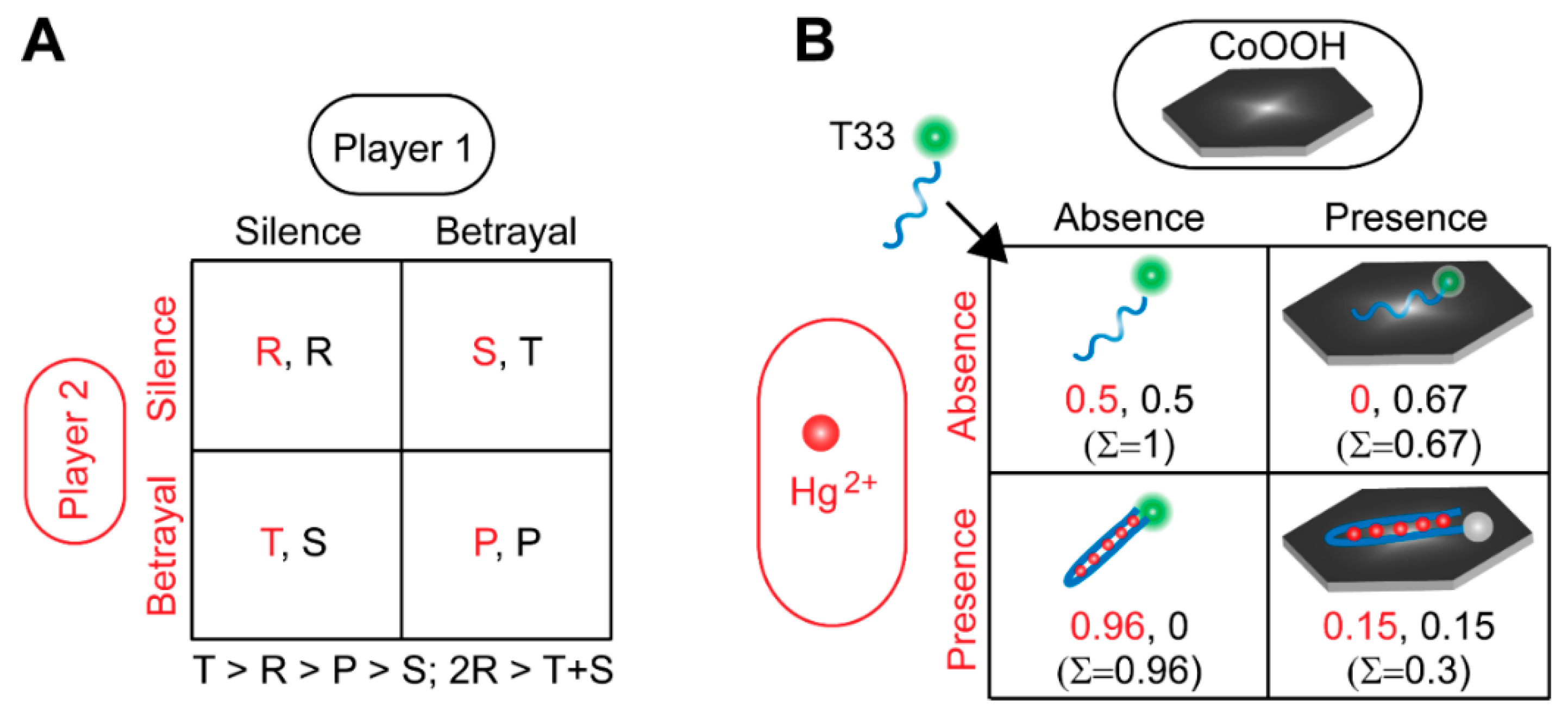
2.4. Construction and Demonstration of Molecular Game-Theoretical System and Application in Sensing Metal Ions
2.5. Real Sample Analysis
3. Results and Discussion
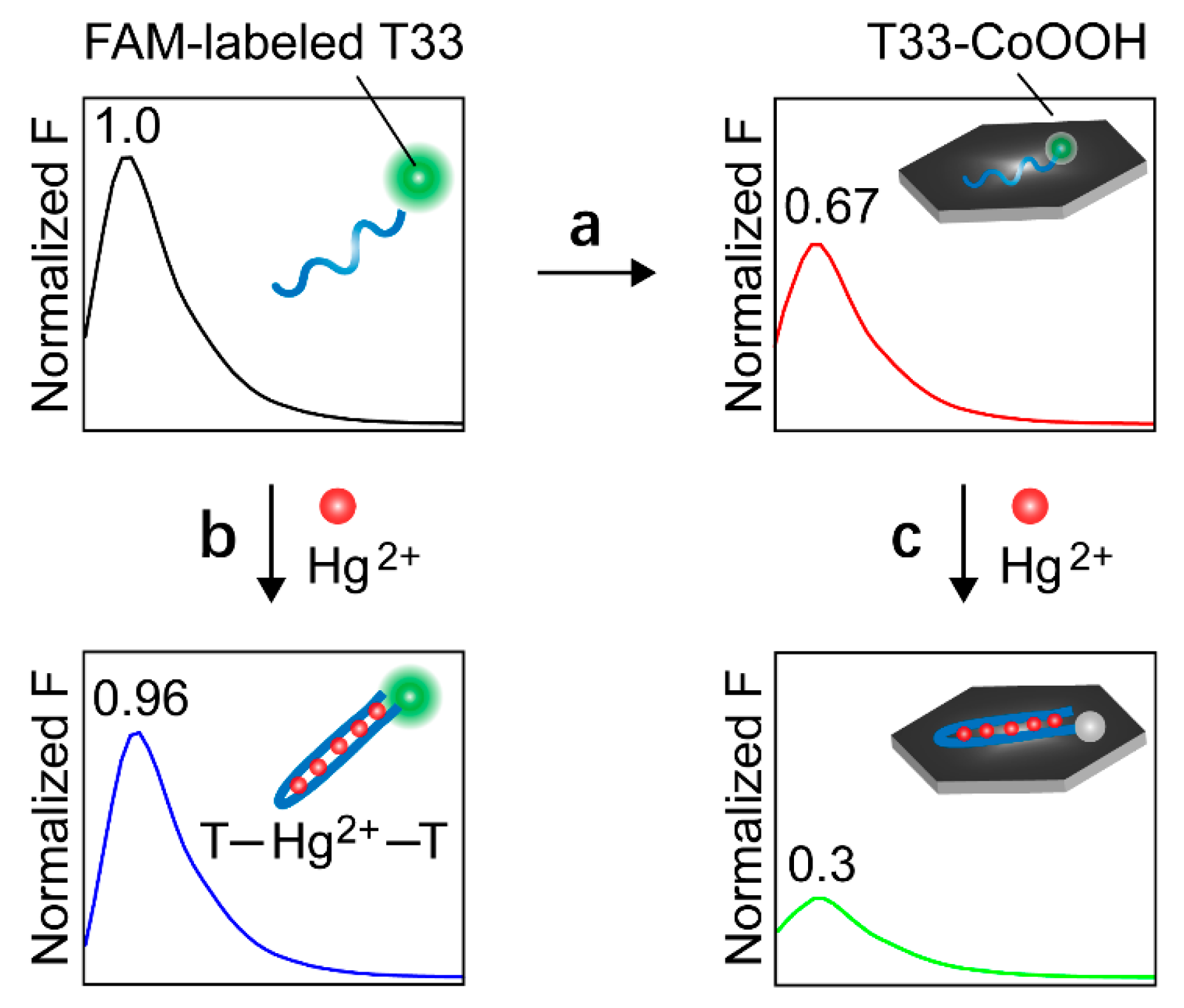
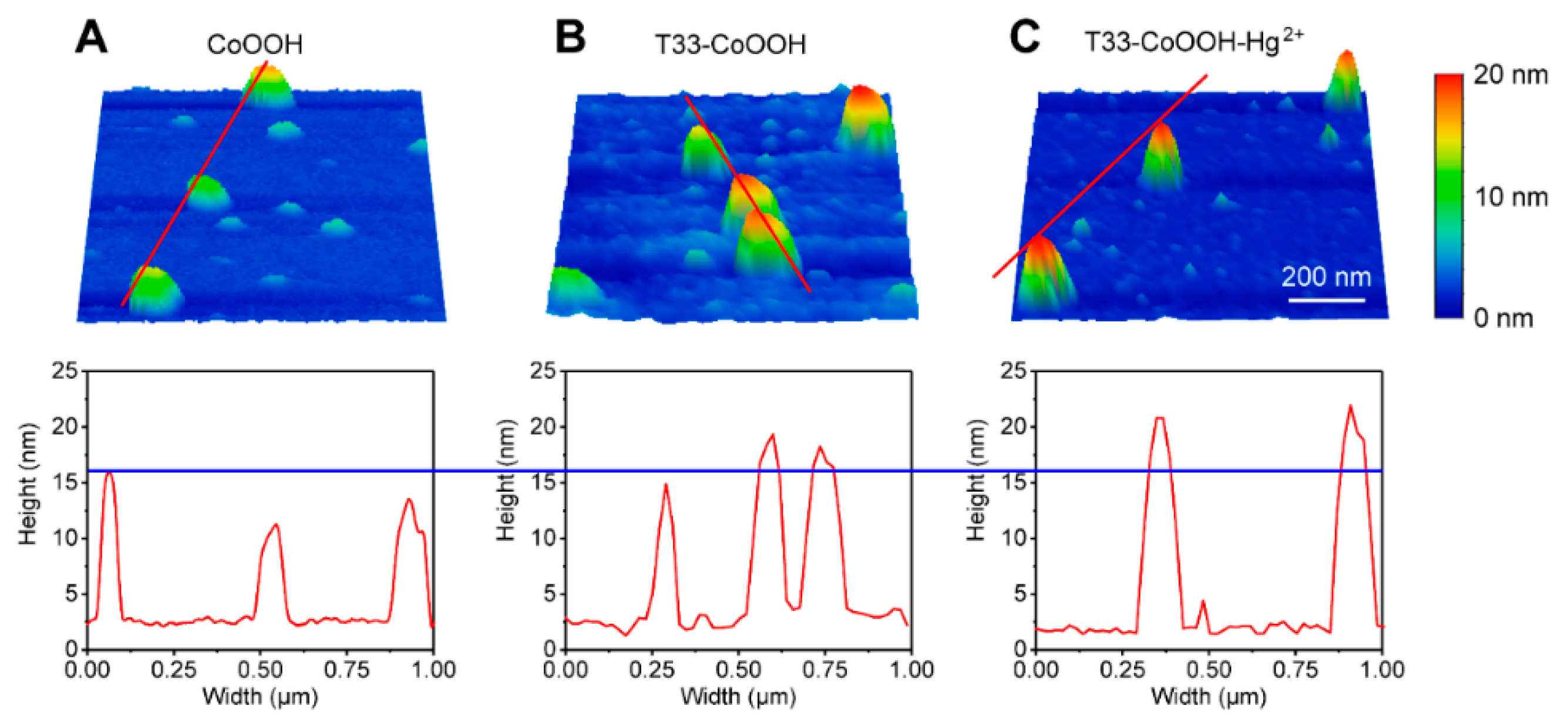
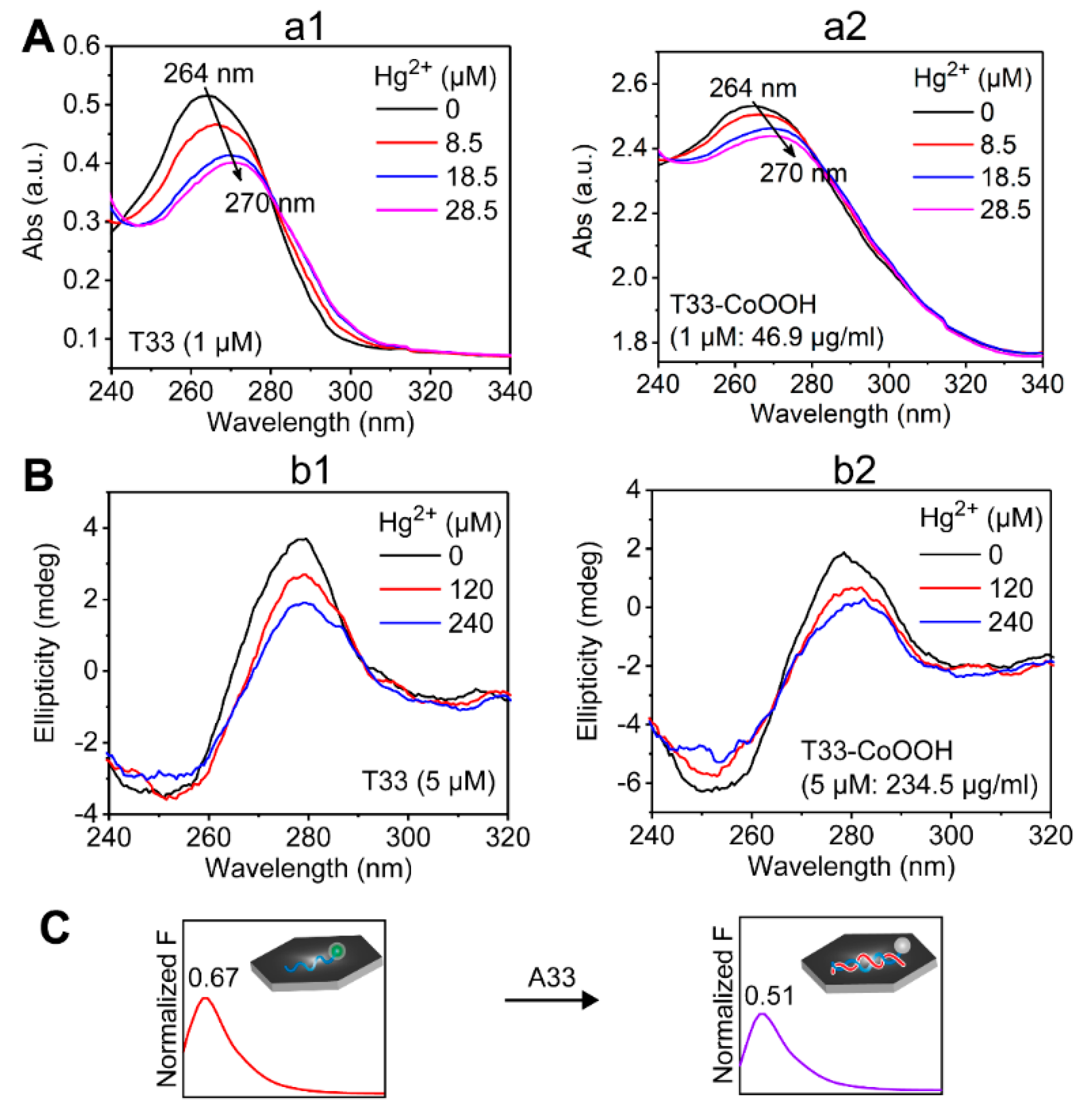
3.1. Construction and Characterization of Molecular Game-Theoretical System Based on Molecular Interactions
3.2. Demonstration of Molecular Game-Theoretical System
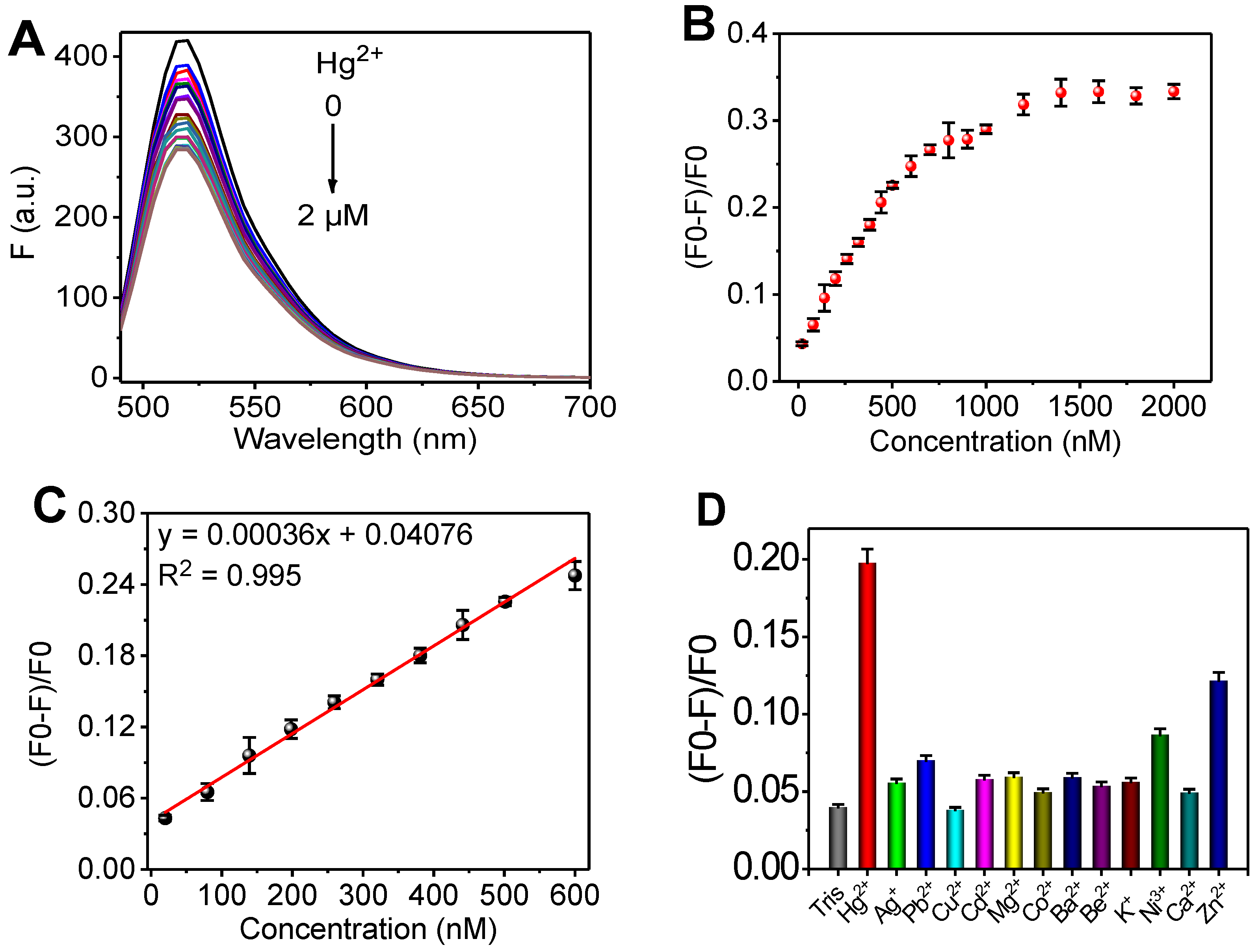
3.3. Molecular Game-Theoretical System for Sensing Hg2+
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Hummert, S.; Bohl, K.; Basanta, D.; Deutsch, A.; Werner, S.; Theissen, G.; Schroeter, A.; Schuster, S. Evolutionary game theory: Cells as players. Mol. Biosyst. 2014, 10, 3044–3065. [Google Scholar] [CrossRef]
- Sanfey, A.G. Social Decision-Making: Insights from Game Theory and Neuroscience. Science 2007, 318, 598–602. [Google Scholar] [CrossRef]
- Nowak, M.A.; Sasaki, A.; Taylor, C.; Fudenberg, D. Emergence of cooperation and evolutionary stability in finite populations. Nature 2004, 428, 646–650. [Google Scholar] [CrossRef]
- Yeates, J.A.M.; Hilbe, C.; Zwick, M.; Nowak, M.A.; Lehman, N. Dynamics of prebiotic RNA reproduction illuminated by chemical game theory. Proc. Natl. Acad. Sci. USA 2016, 113, 5030–5035. [Google Scholar] [CrossRef]
- Turner, P.E.; Chao, L. Prisoner’s dilemma in an RNA virus. Nature 1999, 398, 441–443. [Google Scholar] [CrossRef]
- Bohl, K.; Hummert, S.; Werner, S.; Basanta, D.; Deutsch, A.; Schuster, S.; Theissen, G.; Schroeter, A. Evolutionary game theory: Molecules as players. Mol. Biosyst. 2014, 10, 3066–3074. [Google Scholar] [CrossRef]
- Vaidya, N.; Manapat, M.L.; Chen, I.A.; Xulvi-Brunet, R.; Hayden, E.J.; Lehman, N. Spontaneous network formation among cooperative RNA replicators. Nature 2012, 491, 72–77. [Google Scholar] [CrossRef]
- Feynman, R.P. There’s plenty of room at the bottom: An invitation to enter a new field of physics. In Handbook of Nanoscience, Engineering, and Technology, 3rd ed.; CRC Press: Boca Raton, FL, USA, 2012; pp. 26–35. [Google Scholar]
- Nurse, P. Life, logic and information. Nature 2008, 454, 424–426. [Google Scholar] [CrossRef]
- Ausländer, D.; Ausländer, S.; Pierrat, X.; Hellmann, L.; Rachid, L.; Fussenegger, M. Programmable full-adder computations in communicating three-dimensional cell cultures. Nat. Methods 2017, 15, 57–60. [Google Scholar] [CrossRef]
- Green, A.A.; Kim, J.; Ma, D.; Silver, P.A.; Collins, J.J.; Yin, P. Complex cellular logic computation using ribocomputing devices. Nature 2017, 548, 117–121. [Google Scholar] [CrossRef]
- Erbas-Cakmak, S.; Kolemen, S.; Sedgwick, A.C.; Gunnlaugsson, T.; James, T.D.; Yoon, J.; Akkaya, E.U. Molecular logic gates: The past, present and future. Chem. Soc. Rev. 2018, 47, 2228–2248. [Google Scholar] [CrossRef]
- Szaciłowski, K. Digital Information Processing in Molecular Systems. Chem. Rev. 2008, 108, 3481–3548. [Google Scholar] [CrossRef]
- Cherry, K.M.; Qian, L. Scaling up molecular pattern recognition with DNA-based winner-take-all neural networks. Nature 2018, 559, 370–376. [Google Scholar] [CrossRef]
- Qian, L.; Winfree, E.; Bruck, J. Neural network computation with DNA strand displacement cascades. Nature 2011, 475, 368–372. [Google Scholar] [CrossRef]
- Boukis, A.C.; Reiter, K.; Froelich, M.; Hofheinz, D.; Meier, M.A.R. Multicomponent reactions provide key molecules for secret communication. Nat. Commun. 2018, 9, 1439. [Google Scholar] [CrossRef]
- Lustgarten, O.; Motiei, L.; Margulies, D. User Authorization at the Molecular Scale. ChemPhysChem 2017, 18, 1678–1687. [Google Scholar] [CrossRef]
- Huang, W.T.; Chen, L.X.; Lei, J.L.; Luo, H.Q.; Li, N.B. Molecular neuron: From sensing to logic computation, information encoding, and encryption. Sens. Actuator B-Chem. 2017, 239, 704–710. [Google Scholar] [CrossRef]
- Andréasson, J.; Pischel, U. Molecules for security measures: From keypad locks to advanced communication protocols. Chem. Soc. Rev. 2018, 47, 2266–2279. [Google Scholar] [CrossRef]
- Chatterjee, G.; Dalchau, N.; Muscat, R.A.; Phillips, A.; Seelig, G. A spatially localized architecture for fast and modular DNA computing. Nat. Nanotechnol. 2017, 12, 920–927. [Google Scholar] [CrossRef]
- Schuster, S.; Kreft, J.U.; Schroeter, A.; Pfeiffer, T. Use of Game-Theoretical Methods in Biochemistry and Biophysics. J. Biol. Phys. 2008, 34, 1–17. [Google Scholar] [CrossRef]
- Liu, J.; Cao, Z.; Lu, Y. Functional Nucleic Acid Sensors. Chem. Rev. 2009, 109, 1948–1998. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.B.; Kong, R.M.; Lu, Y. Metal Ion Sensors Based on DNAzymes and Related DNA Molecules. Ann. Rev. Anal. Chem. 2011, 4, 105–128. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Wen, Y.; Xu, L.; Xu, Q.; Song, S.; Zuo, X.; Yan, J.; Zhang, W.; Liu, G. Development of mercury (II) ion biosensors based on mercury-specific oligonucleotide probes. Biosens. Bioelectron. 2016, 75, 433–445. [Google Scholar] [CrossRef] [PubMed]
- Park, K.S.; Jung, C.; Park, H.G. “Illusionary” Polymerase Activity Triggered by Metal Ions: Use for Molecular Logic-Gate Operations. Angew. Chem. Int. Ed. 2010, 49, 9757–9760. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Lu, Y. Rational Design of “Turn-On” Allosteric DNAzyme Catalytic Beacons for Aqueous Mercury Ions with Ultrahigh Sensitivity and Selectivity. Angew. Chem. Int. Ed. 2007, 46, 7587–7590. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Lu, L.; Kang, T.; Cheng, S. Intense charge transfer surface based on graphene and thymine–Hg(II)–thymine base pairs for detection of Hg2+. Biosens. Bioelectron. 2016, 77, 740–745. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Su, Y.; Li, J.; Li, D.; Zhang, J.; Song, S.; Zhao, Y.; Li, G.; Fan, C. Highly Sensitive Electrochemical Sensor for Mercury(II) Ions by Using a Mercury-Specific Oligonucleotide Probe and Gold Nanoparticle-Based Amplification. Anal. Chem. 2009, 81, 7660–7666. [Google Scholar] [CrossRef]
- Chiang, C.K.; Huang, C.C.; Liu, C.W.; Chang, H.T. Oligonucleotide-based fluorescence probe for sensitive and selective detection of mercury (II) in aqueous solution. Anal. Chem. 2008, 80, 3716–3721. [Google Scholar] [CrossRef]
- Chen, G.H.; Chen, W.Y.; Yen, Y.C.; Wang, C.W.; Chang, H.T.; Chen, C.F. Detection of mercury (II) ions using colorimetric gold nanoparticles on paper-based analytical devices. Anal. Chem. 2014, 86, 6843–6849. [Google Scholar] [CrossRef]
- Xie, W.Y.; Huang, W.T.; Zhang, J.R.; Luo, H.Q.; Li, N.B. A triple-channel optical signal probe for Hg2+ detection based on acridine orange and aptamer-wrapped gold nanoparticles. J. Mater. Chem. 2012, 22, 11479–11482. [Google Scholar] [CrossRef]
- Zhang, J.R.; Huang, W.T.; Xie, W.Y.; Wen, T.; Luo, H.Q.; Li, N.B. Highly sensitive, selective, and rapid fluorescence Hg2+ sensor based on DNA duplexes of poly(dT) and graphene oxide. Analyst 2012, 137, 3300–3305. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, K.; Subramanian, K.; Murugan, K.; Dinakaran, K. Sensitive fluorescence detection of mercury(ii) in aqueous solution by the fluorescence quenching effect of MoS2 with DNA functionalized carbon dots. Analyst 2016, 141, 6344–6352. [Google Scholar] [CrossRef] [PubMed]
- Xie, W.Y.; Huang, W.T.; Luo, H.Q.; Li, N.B. CTAB-capped Mn-doped ZnS quantum dots and label-free aptamer for room-temperature phosphorescence detection of mercury ions. Analyst 2012, 137, 4651–4653. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.R.; Huang, W.T.; Zeng, A.L.; Luo, H.Q.; Li, N.B. Ethynyl and pi-stacked thymine-Hg2+-thymine base pairs enhanced fluorescence quenching via photoinduced electron transfer and simple and sensitive mercury ion sensing. Biosens. Bioelectron. 2015, 64, 597–604. [Google Scholar] [CrossRef] [PubMed]
- Shankar, B.H.; Jayaram, D.T.; Ramaiah, D. A Reversible Dual Mode Chemodosimeter for the Detection of Cyanide Ions in Natural Sources. Chem. Asian J. 2014, 9, 1636–1642. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Li, Y.H.; Han, Y.Y.; Pan, W.; Zhang, T.T.; Tang, B. A Highly Selective and Instantaneous Nanoprobe for Detection and Imaging of Ascorbic Acid in Living Cells and in Vivo. Anal. Chem. 2014, 86, 3924–3930. [Google Scholar] [CrossRef] [PubMed]
- Cen, Y.; Yang, Y.; Yu, R.Q.; Chen, T.T.; Chu, X. A cobalt oxyhydroxide nanoflake-based nanoprobe for the sensitive fluorescence detection of T4 polynucleotide kinase activity and inhibition. Nanoscale 2016, 8, 8202–8209. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.Q.; Zhang, Z.; Liu, H.Q.; Wang, N.; Tang, J.L. Cobalt oxyhydroxide nanoflake based fluorescence sensing platform for label-free detection of DNA. Analyst 2016, 141, 4719–4724. [Google Scholar] [CrossRef]
- Miyake, Y.; Togashi, H.; Tashiro, M.; Yamaguchi, H.; Oda, S.; Kudo, M.; Tanaka, Y.; Kondo, Y.; Sawa, R.; Fujimoto, T.; et al. MercuryII-Mediated Formation of Thymine−HgII−Thymine Base Pairs in DNA Duplexes. J. Am. Chem. Soc. 2006, 128, 2172–2173. [Google Scholar] [CrossRef]
- Chang, E.L.; Simmers, C.; Knight, D.A. Cobalt Complexes as Antiviral and Antibacterial Agents. Pharmaceuticals (Basel) 2010, 3, 1711–1728. [Google Scholar] [CrossRef]
- Francisco, T.M.; Gee, W.J.; Shepherd, H.J.; Warren, M.R.; Shultz, D.A.; Raithby, P.R.; Pinheiro, C.B. Hard X-ray-Induced Valence Tautomeric Interconversion in Cobalt-o-Dioxolene Complexes. J. Phys. Chem. Lett. 2017, 8, 4774–4778. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.X.; Zhu, Q.Y.; Lu, J.Y.; Zhang, F.R.; Huang, W.T.; Ding, X.Z.; Xia, L.Q. Boolean logic tree of molecular self-assembly system based on cobalt oxyhydroxide nanoflakes for three-state logic computation, sensing and imaging of pyrophosphate in living cells and in vivo. Analyst 2018. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.S.; Liu, J.; Zhang, B.; Cheng, F.; Ruan, Y.J.; Ji, X.; Xu, K.; Chen, C.; Miao, L.; Jiang, J.J. Stabilizing the oxygen vacancies and promoting water-oxidation kinetics in cobalt oxides by lower valence-state doping. Nano Energy 2018, 53, 144–151. [Google Scholar] [CrossRef]
- Wang, J.W.; Kuo, Y.M. Synthesis of Nanosized Zinc-Doped Cobalt Oxyhydroxide Parties by a Dropping Method and Their Carbon Monoxide Gas Sensing Properties. J. Nanomater. 2013, 136145. [Google Scholar] [CrossRef]
- Aoki, S.; Kimura, E. Zinc-nucleic acid interaction. Chem. Rev. 2004, 104, 769–787. [Google Scholar] [CrossRef] [PubMed]
- Sander, S.A.; Van Hall, A.K.; Morrow, J.R. Zn2+-Selective Switch of Duplex to Hairpin DNA. Inorg. Chem. 2015, 54, 3084–3086. [Google Scholar] [CrossRef] [PubMed]
| Sample | Added (nM) | Found (nM) | Recovery (%) | RSD (%) |
|---|---|---|---|---|
| Pond water | 84.00 | 90.96 | 108.29 | 0.59 |
| 155.00 | 162.64 | 104.93 | 1.15 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nie, N.F.; Zhang, X.X.; Fang, C.S.; Zhu, Q.Y.; Lu, J.Y.; Zhang, F.R.; Yao, Q.F.; Huang, W.T.; Ding, X.Z.; Xia, L.Q. Game Theory in Molecular Nanosensing System for Rapid Detection of Hg2+ in Aqueous Solutions. Appl. Sci. 2018, 8, 2530. https://doi.org/10.3390/app8122530
Nie NF, Zhang XX, Fang CS, Zhu QY, Lu JY, Zhang FR, Yao QF, Huang WT, Ding XZ, Xia LQ. Game Theory in Molecular Nanosensing System for Rapid Detection of Hg2+ in Aqueous Solutions. Applied Sciences. 2018; 8(12):2530. https://doi.org/10.3390/app8122530
Chicago/Turabian StyleNie, Nan Fang, Xin Xing Zhang, Chu Shan Fang, Qiu Yan Zhu, Jiao Yang Lu, Fu Rui Zhang, Qing Feng Yao, Wei Tao Huang, Xue Zhi Ding, and Li Qiu Xia. 2018. "Game Theory in Molecular Nanosensing System for Rapid Detection of Hg2+ in Aqueous Solutions" Applied Sciences 8, no. 12: 2530. https://doi.org/10.3390/app8122530
APA StyleNie, N. F., Zhang, X. X., Fang, C. S., Zhu, Q. Y., Lu, J. Y., Zhang, F. R., Yao, Q. F., Huang, W. T., Ding, X. Z., & Xia, L. Q. (2018). Game Theory in Molecular Nanosensing System for Rapid Detection of Hg2+ in Aqueous Solutions. Applied Sciences, 8(12), 2530. https://doi.org/10.3390/app8122530