Catabolic Fingerprinting and Diversity of Bacteria in Mollic Gleysol Contaminated with Petroleum Substances
Abstract
:1. Introduction
2. Materials and Methods
2.1. Soil Characteristics and Experimental Design
2.2. Soil DNA Extraction and Molecular Identification of Bacteria
2.3. Community Level Physiological Profiling (CLPP) Analysis Using Biolog EcoPlates
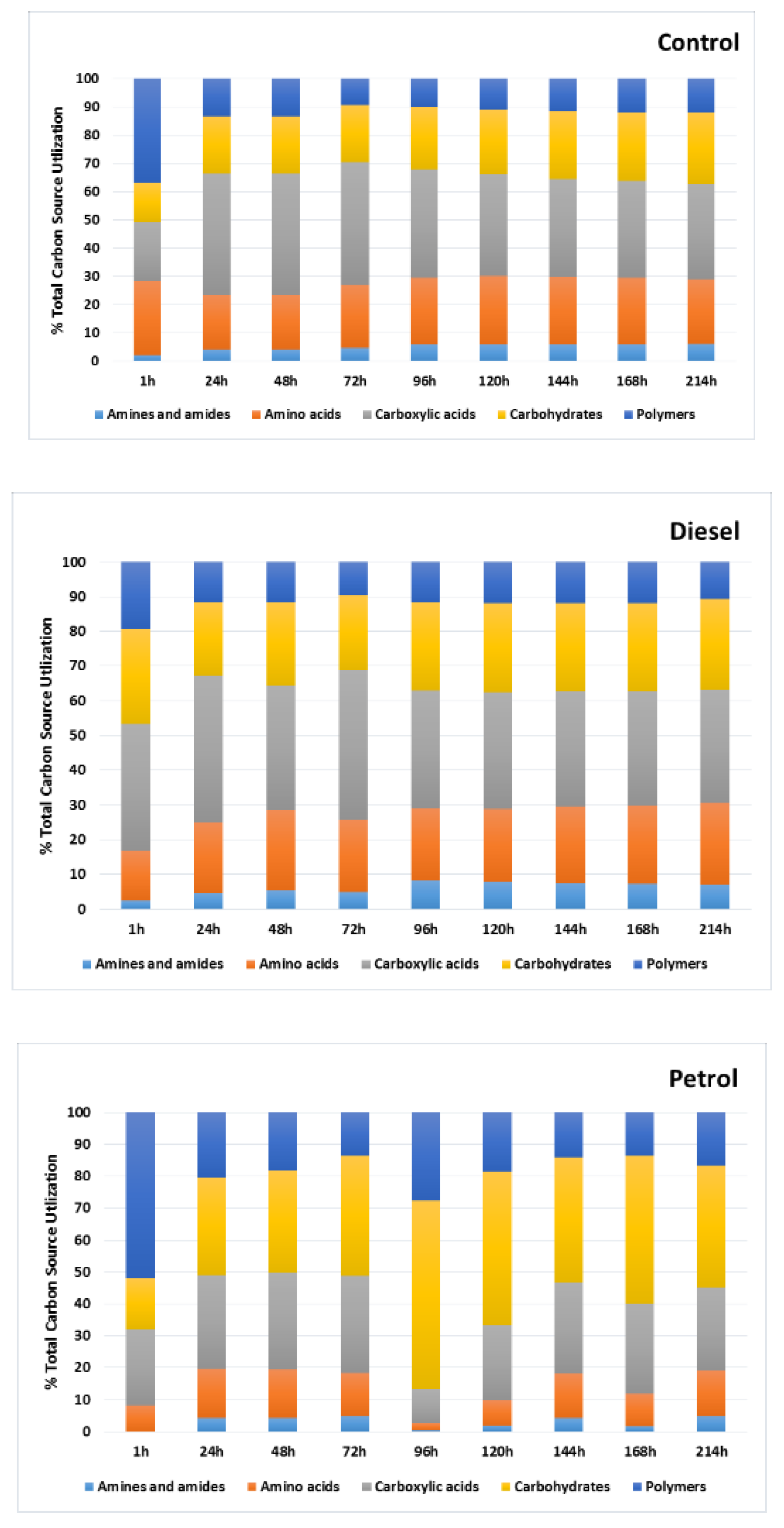
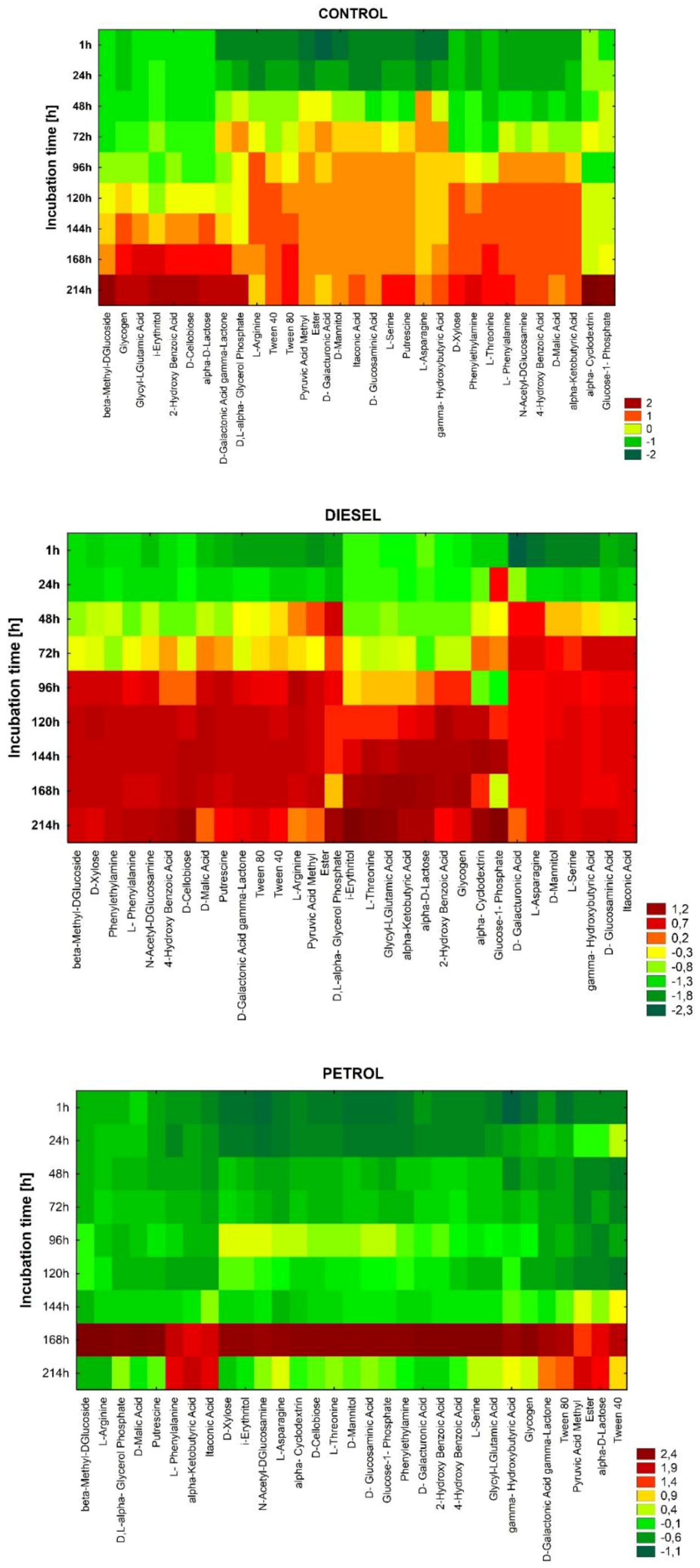
3. Results
3.1. Catabolic Diversity of Autochthonic Bacteria Inhabiting Soil Contaminated with Car Fuels
3.2. Genetic Diversity of Autochthonic Bacteria Inhabiting Soil Contaminated with Car Fuels
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Fernández-Luqueño, F.; Encinas, C.V.; Marsch, R.; Martínez-Suárez, C.; Vázquez-Núñez, E.; Dendooven, L. Microbial communities to mitigate contamination of PAHs in soil- possibilities and challenges: A review. Environ. Sci. Pollut. Res. 2011, 18, 12–30. [Google Scholar] [CrossRef] [PubMed]
- Alrumman, S.A.; Standing, D.B.; Paton, G.I. Effect of hydrocarbon contamination on soil microbial community and enzyme activity. J. King Saud Univ.-Sci. 2015, 27, 31–41. [Google Scholar] [CrossRef] [Green Version]
- Varjani, S.J. Microbial degradation of petroleum hydrocarbons. Biores. Technol. 2017, 223, 277–286. [Google Scholar] [CrossRef] [PubMed]
- Chikere, C.B.; Okpokwasili, G.C.; Chikere, B.O. Monitoring of microbial hydrocarbon remediation in the soil. 3 Biotech 2011, 1, 117–138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kucharski, J.; Tomkiel, M.; Boros, E.; Wyszkowska, J. The effect of soil contamination with diesel oil and petrol on the nitrification process. J. Elementol. 2010, 15, 111–118. [Google Scholar] [CrossRef]
- Wyszkowska, J.; Borowik, A.; Kucharski, J. Response of Avena sativa, microorganisms and enzymes to contamination of soil with diesel oil. Plant Soil. Environ. 2015, 61, 483–488. [Google Scholar] [CrossRef]
- Borowik, A.; Wyszkowska, J.; Kucharski, J. Functional diversity of fungal communities in soil contaminated with diesel oil. Front. Microbiol. 2017, 8, 1862. [Google Scholar] [CrossRef] [PubMed]
- Varjani, S.J.; Gnansounou, E. Microbial dynamics in petroleum oilfields and their relationship with physiological properties of petroleum oil reservoirs. Biores. Technol. 2017, 245, 1258–1265. [Google Scholar] [CrossRef] [PubMed]
- Mohammadi-Sichani, M.M.; Assadi, M.M.; Farazmand, A.; Kianirad, M.; Ahadi, A.; Ghahderijani, H.H. Bioremediation of soil contaminated crude oil by Agaricomycetes. J. Environ. Health. Sci. Eng. 2017, 15, 8. [Google Scholar] [CrossRef] [PubMed]
- Moubasher, H.A.; Hegazy, A.K.; Mohamed, N.H.; Moustafa, Y.M.; Kabiel, H.F.; Hamad, A.A. Phytoremediation of soils polluted with crude petroleum oil using Bassia scoparia and its associated rhizosphere microorganisms. Int. Biodeter. Biodegr. 2015, 98, 113–120. [Google Scholar] [CrossRef]
- Hatami, E.; Abbaspour, A.; Dorostkar, V. Phytoremediation of a petroleum-polluted soil by native plant species in Lorenstan Province, Iran. Environ. Sci. Pollut. Res. 2018, 1–8. [Google Scholar] [CrossRef]
- Kaczyńska, G.; Borowik, A.; Wyszkowska, J. Soil dehydrogenases as an indicator of contamination of the environment with petroleum products. Water Air Soil Pollut. 2015, 226, 372. [Google Scholar] [CrossRef] [PubMed]
- Gałązka, A.; Król, M.; Perzyński, A. The efficiency of rhizosphere bioremediation with Azospirillum sp. and Pseudomonas stutzeri in soils freshly contaminated with PAHs and diesel fuel. Pol. J. Environ. Stud. 2012, 21, 345–353. [Google Scholar]
- Gałązka, A.; Gałązka, R. Phytoremediation of polycyclic aromatic hydrocarbons in soils artificially polluted using plant-associated-endophytic bacteria and Dactylis glomerata as the bioremediation plant. Pol. J. Microbiol. 2015, 64, 241–252. [Google Scholar] [CrossRef] [PubMed]
- Suja, F.; Rahim, F.; Taha, M.T.; Hambali, N.; Razali, R.; Khalid, A.; Hamzah, A. Effects of local microbial bioaugmentation and biostimulation on the bioremediation of total petroleum hydrocarbons (TPH) in crude oil contaminated soil based on laboratory and field observations. Int. Biodeter. Biodegr. 2014, 90, 115–122. [Google Scholar] [CrossRef]
- Stefanowicz, A. The Biolog Plate technique as a tool in ecological studies of microbial communities. Pol. J. Environ. Stud. 2006, 15, 669–676. [Google Scholar]
- Borowik, A.; Wyszkowska, J.; Wyszkowski, M. Resistance of aerobic microorganisms and soil enzyme response to soil contamination with Ekodiesel Ultra fuel. Environ. Sci. Pollut. Res. 2017, 24, 24346–24363. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grant, R.J.; Muckian, L.M.; Clipson, N.J.W.; Doyle, E.M. Microbial community changes during the bioremediation of creosote contaminated soil. Lett. Appl. Microbiol. 2007, 44, 293–300. [Google Scholar] [CrossRef] [PubMed]
- Wolińska, A.; Kuźniar, A.; Szafranek-Nakonieczna, A.; Jastrzębska, N.; Roguska, E.; Stępniewska, Z. Biological activity of autochthonic bacterial community in oil-contaminated soils. Water Air Soil Pollut. 2016, 227, 130. [Google Scholar] [CrossRef] [PubMed]
- Khan, J.A.; Singh, S. Evaluation of oil degradation potential of micrococcus varians. Int. J. Appl. Biol. Pharm. Technol. 2011, 2, 75–80. [Google Scholar]
- Leilei, Z.; Mingxin, H.; Suiyi, Z. Enzymatic remediation of the polluted crude oil by Rhodococcus. Afr. J. Microbiol. Res. 2012, 6, 1213–1220. [Google Scholar] [CrossRef]
- Hamamura, N.; Ward, D.M.; Inskeep, W.P. Effects of petroleum mixture types on soil bacterial population dynamics associated with the biodegradation of hydrocarbons in soil environment. FEMS Microbiol. Ecol. 2013, 85, 168–178. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Xu, P.; Zeng, G.; Yang, C.; Huang, D.; Zhang, J. Bioremediation of soil contaminated with polycyclic aromatic hydrocarbons, petroleum, pesticides, chlorophenols and heavy metals by composting: Applications, microbes and future research needs. Biotechnol. Adv. 2015, 33, 745–755. [Google Scholar] [CrossRef] [PubMed]
- Rusin, M.; Gospodarek, J.; Nadgórska-Socha, A. The effect of petroleum-derived substances on the growth and chemical composition of Vicia faba L. Pol. J. Environ. Stud. 2015, 24, 2157–2166. [Google Scholar] [CrossRef]
- Palmroth, M.R.T.; Koskinen, P.E.P.; Kaksonen, A.H.; Münster, U.; Pichtel, J.; Puhakka, J.K. Metabolic and phylogenetic analysis of microbial communities during phytoremediation of soil contaminated with weathered hydrocarbons and heavy metals. Biodegradation 2007, 8, 769–782. [Google Scholar] [CrossRef] [PubMed]
- Wyszkowska, J.; Kucharski, J. Correlation between the number of cultivable microorganisms and soil contamination with diesel oil. Pol. J. Environ. Stud. 2005, 14, 347–356. [Google Scholar]
- Garland, J.L.; Millis, A.L. Classification and characterization of heterotrophic microbial communities on the basis of patterns of community-level sole-carbon-source utilization. Appl. Environ. Microbiol. 1991, 57, 2351–2359. [Google Scholar] [PubMed]
- Gałązka, A.; Gawryjołek, K.; Grządziel, J.; Frąc, M.; Księżak, J. Microbial community diversity and their interaction of soil under maize growth in different cultivation techniques. Plant Soil Environ. 2017, 63, 264–270. [Google Scholar] [CrossRef]
- Wolińska, A.; Frąc, M.; Oszust, K.; Szafranek-Nakonieczna, A.; Zielenkiewicz, U.; Stępniewska, Z. Microbial biodiversity of meadows under different modes of land use: Catabolic and genetic fingerprinting. World J. Microbiol. Biotechnol. 2017, 33, 154. [Google Scholar] [CrossRef] [PubMed]
- Frąc, M.; Oszust, K.; Lipiec, J. Community level physiological profiles (CLPP), characterization and microbial activity of soil amended with dairy sewage sludge. Sensors 2012, 12, 3253–3268. [Google Scholar] [CrossRef] [PubMed]
- Pohland, B.; Owen, B. Biolog EcoPlate standard methods. TAS Tech. Biul. Biol. 2009, 1, 1–3. [Google Scholar]
- Islam, M.R.; Chauhan, P.S.; Kim, Y.; Kim, M.; Sa, T. Community level functional diversity and enzyme activities in paddy soils under different long-term fertilizer management practices. Biol. Fertil. Soils 2011, 47, 99–604. [Google Scholar] [CrossRef]
- Labud, V.; Garcia, C.; Hernandez, T. Effect of hydrocarbon pollution on the microbial properties of a sandy and a clay soil. Chemosphere 2007, 66, 1863–1871. [Google Scholar] [CrossRef] [PubMed]
- Ziółkowska, A.; Wyszkowski, M. Toxicity of petroleum substances to microorganisms and plants. Ecol. Chem. Eng. 2010, 17, 73–82. [Google Scholar]
- Wolińska, A.; Rekosz-Burlaga, H.; Goryluk-Salmonowicz, A.; Błaszczyk, M.; Stępniewska, Z. Bacterial abundance and dehydrogenase activity in selected agricultural soils from Lublin region. Pol. J. Environ. Stud. 2015, 24, 2677–2682. [Google Scholar] [CrossRef]
- Łyszcz, M.; Gałązka, A. Selected molecular methods used in assessing the biodiversity of soil organisms. Post. Mikrobiol. 2016, 55, 309–319. [Google Scholar]
- Yousaf, S.; Andria, V.; Reichenauer, T.G.; Smalla, K.; Sessitsch, A. Phylogenetic and functional diversity of alkane degrading bacteria associated with Italian ryegrass (Lolium multiflorum) and Bridsfoot trefoil (Lotus corniculatus) in a petroleum oil-contaminated environment. J. Hazard. Mater. 2010, 184, 523–532. [Google Scholar] [CrossRef] [PubMed]
- White, J.; Gilbert, J.; Hill, G.; Hill, E.; Huse, S.M.; Weightman, A.J.; Mahenthiralingam, E. Culture-independent analysis of bacterial fuel contamination provides insight into the level of concordance with the standard industry practice of aerobic cultivation. Appl. Environ. Microbiol. 2011, 77, 4527–4538. [Google Scholar] [CrossRef] [PubMed]
- Peng, M.; Zi, X.; Wang, Q. Bacterial community diversity of oil-contaminated soils assessed by high-throughput sequencing of 16S rRNA genes. Int. J. Environ. Res. Public Health 2015, 12, 12002–12015. [Google Scholar] [CrossRef] [PubMed]
- Martin-Sanchez, P.M.; Gorbushina, A.A.; Toepel, J. Quantification of microbial load in diesel storage tanks using culture- and qPCR-based approaches. Int. Biodeterior. Biodegrad. 2018, 126, 216–223. [Google Scholar] [CrossRef]
- Borowik, A.; Wyszkowska, J. Remediation of soil contaminated with diesel oil. J. Elem. 2018, 23, 767–788. [Google Scholar] [CrossRef]
- Saikia, R.R.; Deka, S.; Deka, M.; Banat, I.M. Isolation of biosurfactant producing Pseudomonas aeruginosa RS29 from oil-contaminated soil and evaluation of different nitrogen sources in biosurfactant production. Ann. Microbiol. 2012, 62, 753–763. [Google Scholar] [CrossRef]
- Pacwa-Płociniczak, M.; Płaza, G.A.; Poliwoda, A.; Piotrowska-Seget, Z. Characterization of hydrocarbon-degrading and biosurfactant producing Pseudomonas sp. P-1 strain as a potential tool for bioremediation of petroleum-contaminated soil. Environ. Sci. Pollut. Res. 2014, 21, 9385–9395. [Google Scholar] [CrossRef] [PubMed]
- Aparna, A.; Srinikethan, G.; Hedge, S. Effect of addition of biosurfactant produced by Pseudomonas ssp. on biodegradation of crude oil. Int. Proc. Chem. Biol. Environ. Eng. 2011, 6, 71–75. [Google Scholar]
| Parameter | Unit | Unleaded 95-Octane Petrol | Diesel |
|---|---|---|---|
| Carbon atoms number | – | C5–C12 | C9–C25 |
| Density | g cm−3 | 0.72–0.78 | 0.82–0.85 |
| PAH * content | % | 20 | 11 |
| Sulphur content | mg kg−1 | 10 | 10 |
| Sample ID | Bacterial Genera | Isolate Name in GenBank Database | Similarity (%) | Accession Number |
|---|---|---|---|---|
| CONTROL | Micrococcus | Micrococcus sp. CNJ719 PL04 | 97 | DQ448712 |
| Micrococcus sp. 76A3b | 97 | KJ744024 | ||
| Lysinobacterium | Lysinobacterium fusiformis | 99 | KY515463 | |
| Lysinobacterium sp. enrichment culture | 99 | JX992646 | ||
| Lysinobacterium sphaericus | 99 | KY673682 | ||
| Bacillus | Bacillus cereus | 99 | JN411317 | |
| Bacillus thuringiensis | 98 | KY368122 | ||
| Bacillus tovonensis | 98 | KY352881 | ||
| PETROL | Rhodococcus | Rhodococcus erythropolis strain HL-1 | 98 | KM670434 |
| Rhodococcus sp. A35 | 98 | FM986394 | ||
| Rhodococcus globerulus | 97 | AB828263 | ||
| Bacillus | Bacillus anthracis | 97 | KJ721202 | |
| Bacillus mycoides | 97 | KU877669 | ||
| Bacillus safensis | 99 | KY400282 | ||
| Bacillus pumilus | 99 | DQ837545 | ||
| Microbacterium | Microbacterium sp. IP5 | 98 | GU726546 | |
| Microbacterium oxydans | 97 | LN890176 | ||
| DIESEL | Rhodococcus | Rhodococcus erythropolis strain HS9 | 98 | AY168587 |
| Rhodococcus sp. DSD51Y | 98 | AB847903 | ||
| Arthrobacter | Arthrobacter sp. ES3-54 | 99 | AB496410 | |
| Arthrobacter humicola | 99 | NR041546 | ||
| Arthrobacter oxydans | 99 | KP235208 | ||
| Paenibacillus | Paenibacillus sp. 4-21 | 98 | HM537156 | |
| Bacillus sp. DB87 | 98 | HM566967 | ||
| Paenibacillus xylanexedens | 98 | KX156222 | ||
| Paenibacillus pabuli | 98 | JX293322 | ||
| Pseudomonas | Pseudomonas sp. | 99 | KR673340 | |
| Pseudomonas thiveralensis | 99 | KY457754 | ||
| Pseudomonas fluorescens | 99 | KT223372 |
| Bacterial Genera | Control | Unleaded 95-Octane Petrol | Diesel |
|---|---|---|---|
| Micrococcus | + | − | − |
| Lysinobacterium | + | − | − |
| Bacillus | + | + | − |
| Rhodococcus | − | + | + |
| Microbacterium | − | + | − |
| Arthrobacter | − | − | + |
| Paenibacillus | − | − | + |
| Pseudomonas | − | − | + |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wolińska, A.; Gałązka, A.; Kuźniar, A.; Goraj, W.; Jastrzębska, N.; Grządziel, J.; Stępniewska, Z. Catabolic Fingerprinting and Diversity of Bacteria in Mollic Gleysol Contaminated with Petroleum Substances. Appl. Sci. 2018, 8, 1970. https://doi.org/10.3390/app8101970
Wolińska A, Gałązka A, Kuźniar A, Goraj W, Jastrzębska N, Grządziel J, Stępniewska Z. Catabolic Fingerprinting and Diversity of Bacteria in Mollic Gleysol Contaminated with Petroleum Substances. Applied Sciences. 2018; 8(10):1970. https://doi.org/10.3390/app8101970
Chicago/Turabian StyleWolińska, Agnieszka, Anna Gałązka, Agnieszka Kuźniar, Weronika Goraj, Natalia Jastrzębska, Jarosław Grządziel, and Zofia Stępniewska. 2018. "Catabolic Fingerprinting and Diversity of Bacteria in Mollic Gleysol Contaminated with Petroleum Substances" Applied Sciences 8, no. 10: 1970. https://doi.org/10.3390/app8101970
APA StyleWolińska, A., Gałązka, A., Kuźniar, A., Goraj, W., Jastrzębska, N., Grządziel, J., & Stępniewska, Z. (2018). Catabolic Fingerprinting and Diversity of Bacteria in Mollic Gleysol Contaminated with Petroleum Substances. Applied Sciences, 8(10), 1970. https://doi.org/10.3390/app8101970







