On Minimal Unique Induced Subgraph Queries
Abstract
Featured Application
Abstract
1. Introduction
- To our best knowledge, we are the first to propose MUIS query, which is a novel type of interesting and useful subgraph query. MUIS query enriches and develops graph data query and management methods;
- For the novel type of subgraph query, the formal definition is given and the properties are discussed in this paper;
- The EQA (Efficient Query Answering) algorithm is proposed to solve the MUIS query problem under the filtering-validation framework. In the EQA algorithm, BFS (Breadth First Search)-based candidate set generation strategy, matched vertices-based pruning strategy and query position-based subgraph isomorphism are proposed to improve the effectiveness and efficiency of MUIS query;
- Through comprehensive experiments on real datasets and synthetic datasets, EQA is demonstrated to outperform the state-of-the-art model to answer MUIS query. Influencing factors of the process speed are also verified by the experiments.
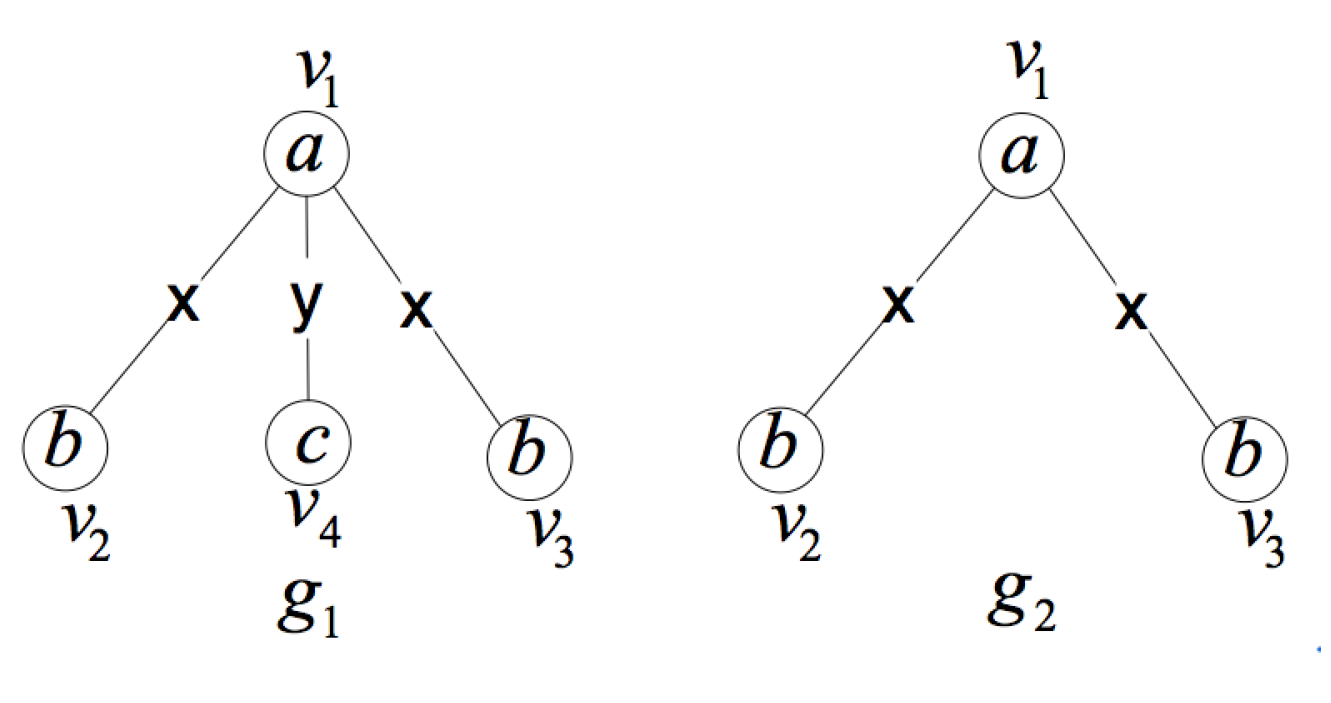
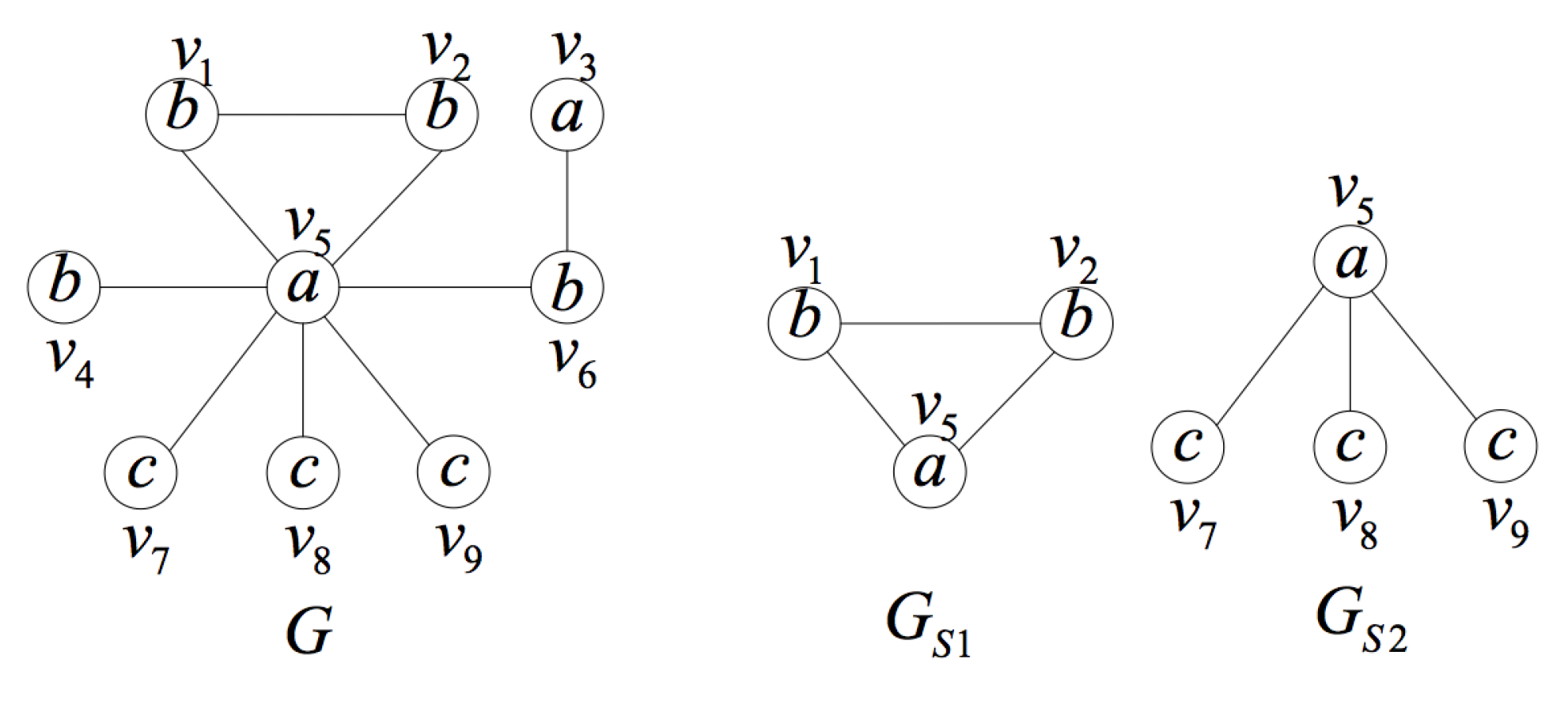
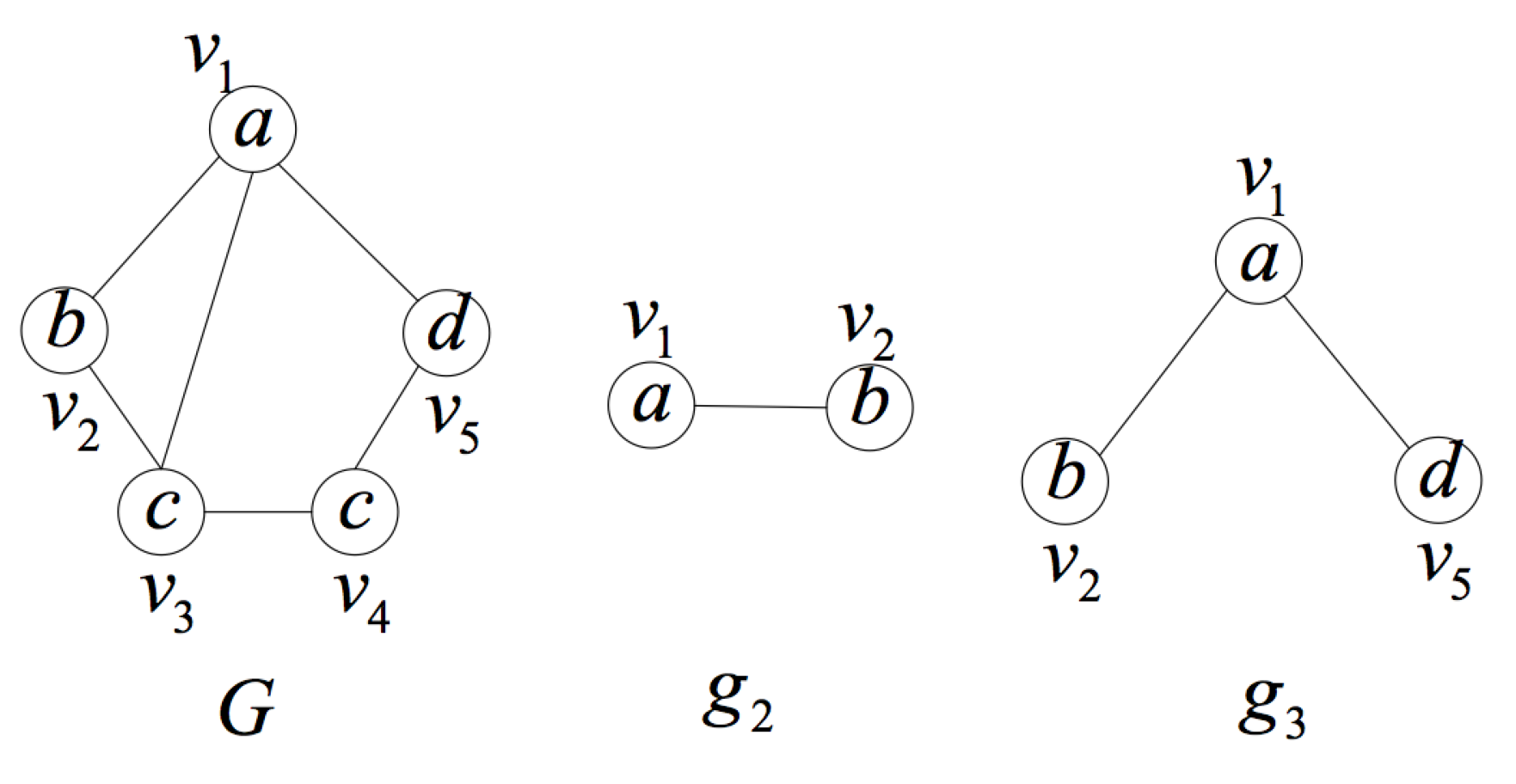
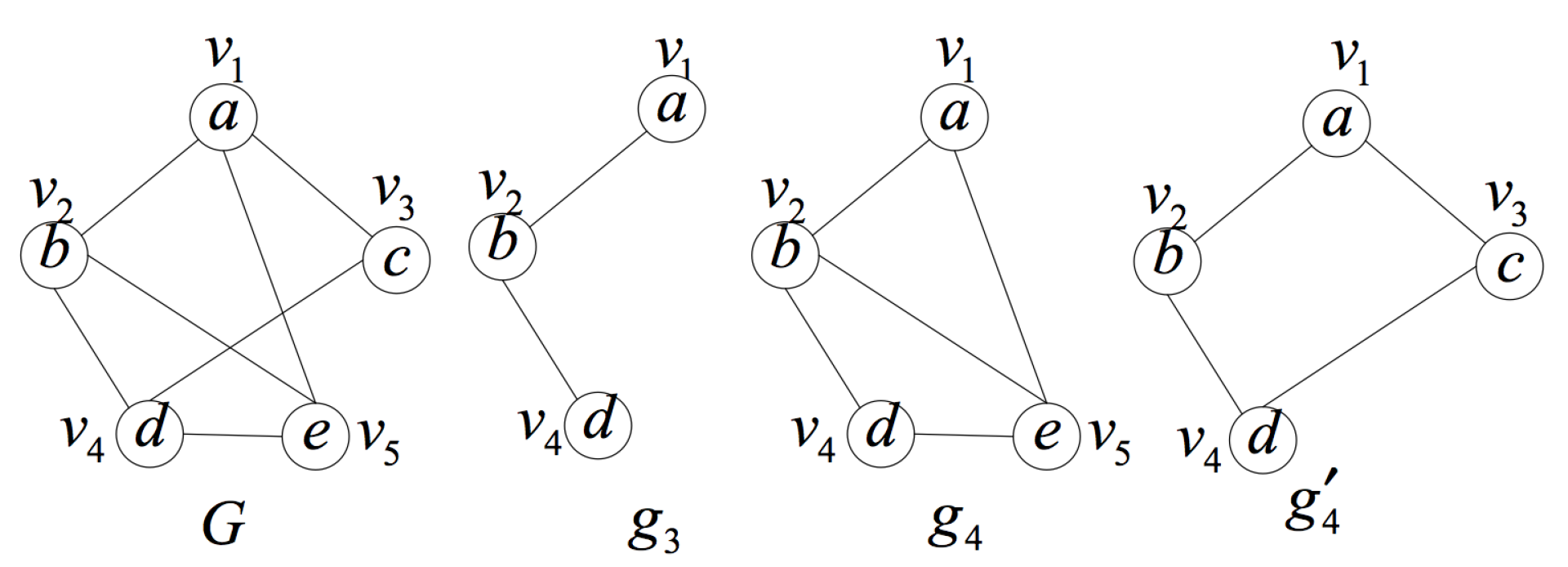
2. Formal Definition and Properties
- (1)
- ,
- (2)
- and ,
- (1)
- is a an induced subgraph of G (induced subgraph property).
- (2)
- is unique in the set of induced subgraphs of G, that is to say, there is no other induced subgraph of graph G isomorphic to except itself (uniqueness property).
- (3)
- In G, there is no proper subgraph of satisfying (1) and (2) (smallest one property).
3. Related Works
3.1. Subgraph Matching Query
3.2. Frequent Subgraph Mining
3.3. Correlation Subgraph Query
3.4. Network Motif Discovery
4. The Proposed Model
4.1. The General Framework
| Algorithm 1 The general framework. |
Input: data graph, ; query position, q; Output: MUIS(q);
|
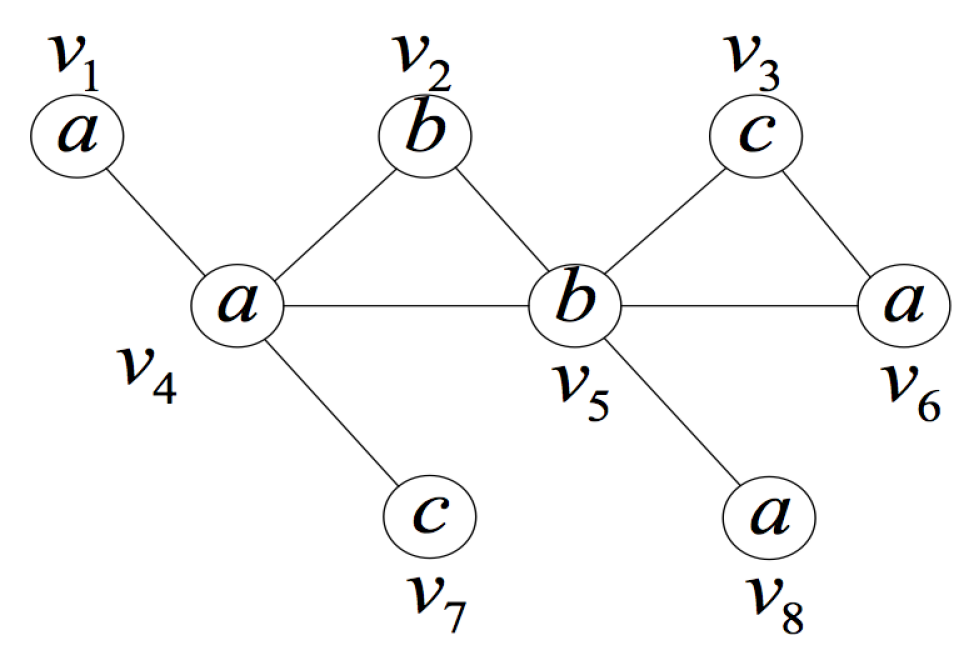
4.2. BFS-Based Candidate Set Generation Strategy
- (1)
- Search the induced subgraph space containing query position q in ascending order of the number of vertices. In particular, the first layer of the induced subgraph space is the query position self-constructed induced subgraph . Its importance will be explained in detail later.
- (2)
- Divide all the vertices of the data graph into two subsets. The two subsets are defined as follows:denotes the vertices already contained in the current induced subgraph. Note that it must have query position .denotes the vertices not contained in the current subgraph. Note that it must have .
- (3)
- When performing BFS for the -th layer induced subgraph from the i-th layer induced graph (containing i vertices), select a vertex v from the subset of the i-th layer induced subgraph. If vertex v is connected to any vertex of the subgraph, then add vertex v into the vertex set of the subgraph to generate a new induced subgraph (containing vertices).
- (4)
- We eliminate the generated duplicate induced subgraphs, which have been already obtained by other i-th layer induced subgraphs and vertices in , which can greatly reduce subsequent computational overhead.
| Algorithm 2 BFS-based candidate set generation algorithm. |
Input: data graph, ; query position, q; Output: induced graphs, ;
|
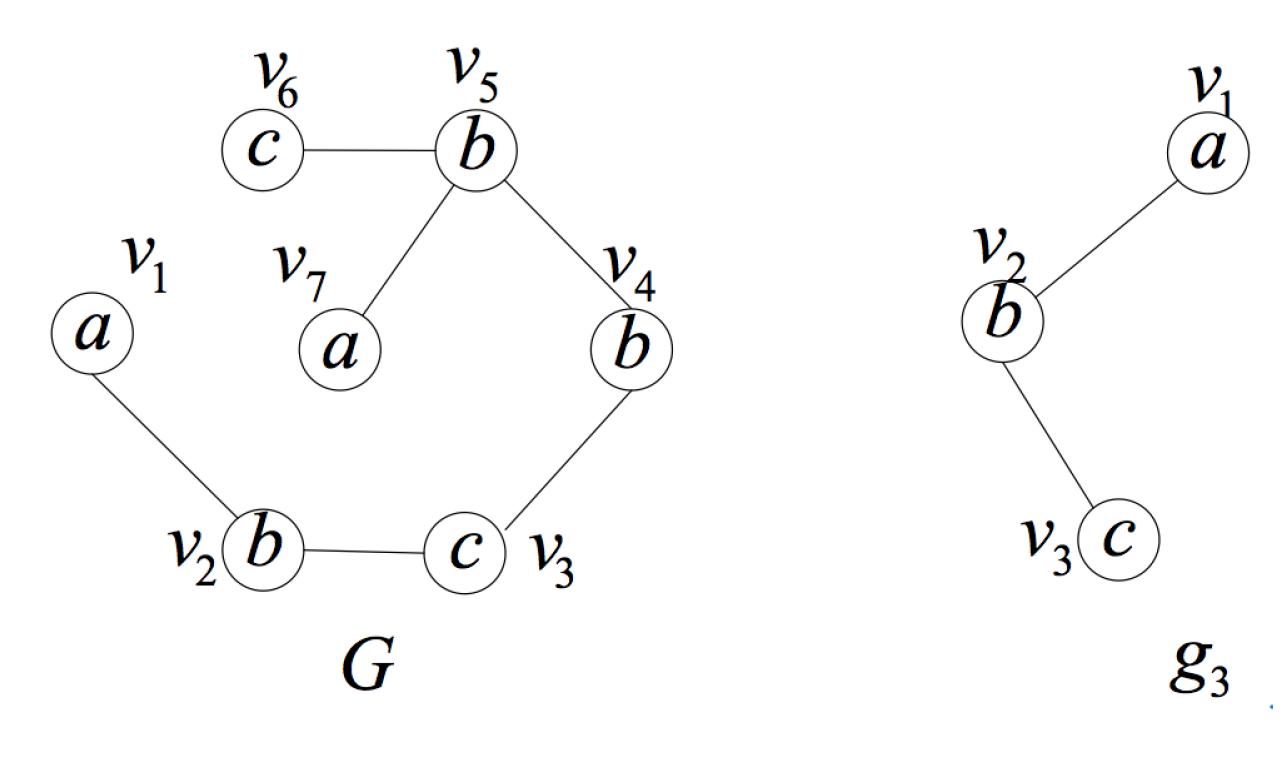
4.3. Matched Vertices-Based Pruning Strategy
- (1)
- During the isomorphism testing, some vertices in some subgraphs of data graph G are measured not to derive the subgraphs that are isomorphic to the induced subgraph. These vertices can be recorded for pruning.
- (2)
- Consider graph , query position and graph . In the case of corresponding to q, if all the subgraphs containing vertex of graph are not isomorphic to , then any hypergraph of graph and all subgraphs containing vertex of graph are not isomorphic in this case.
4.4. Query Position-Based Subgraph Isomorphism
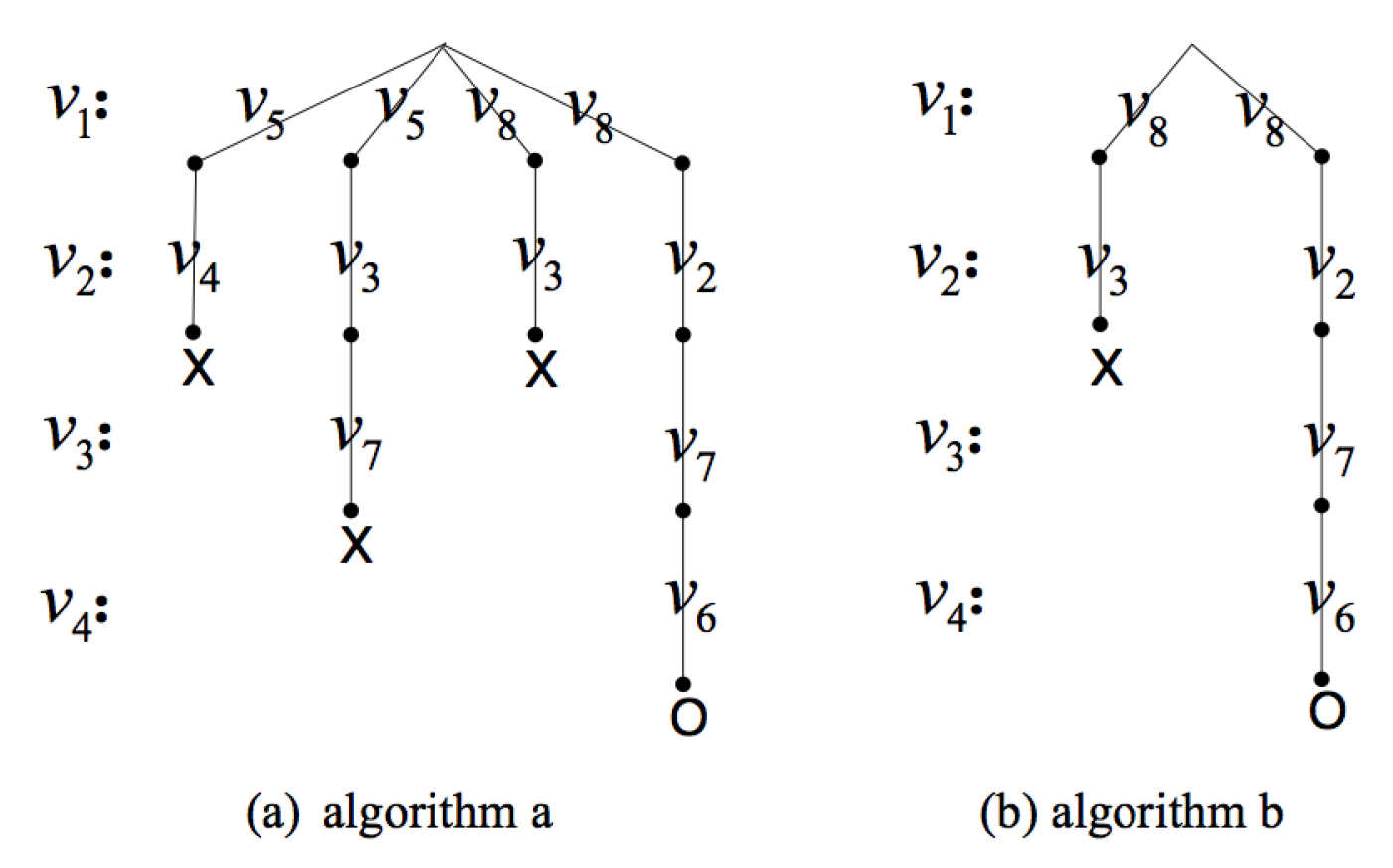
- (1)
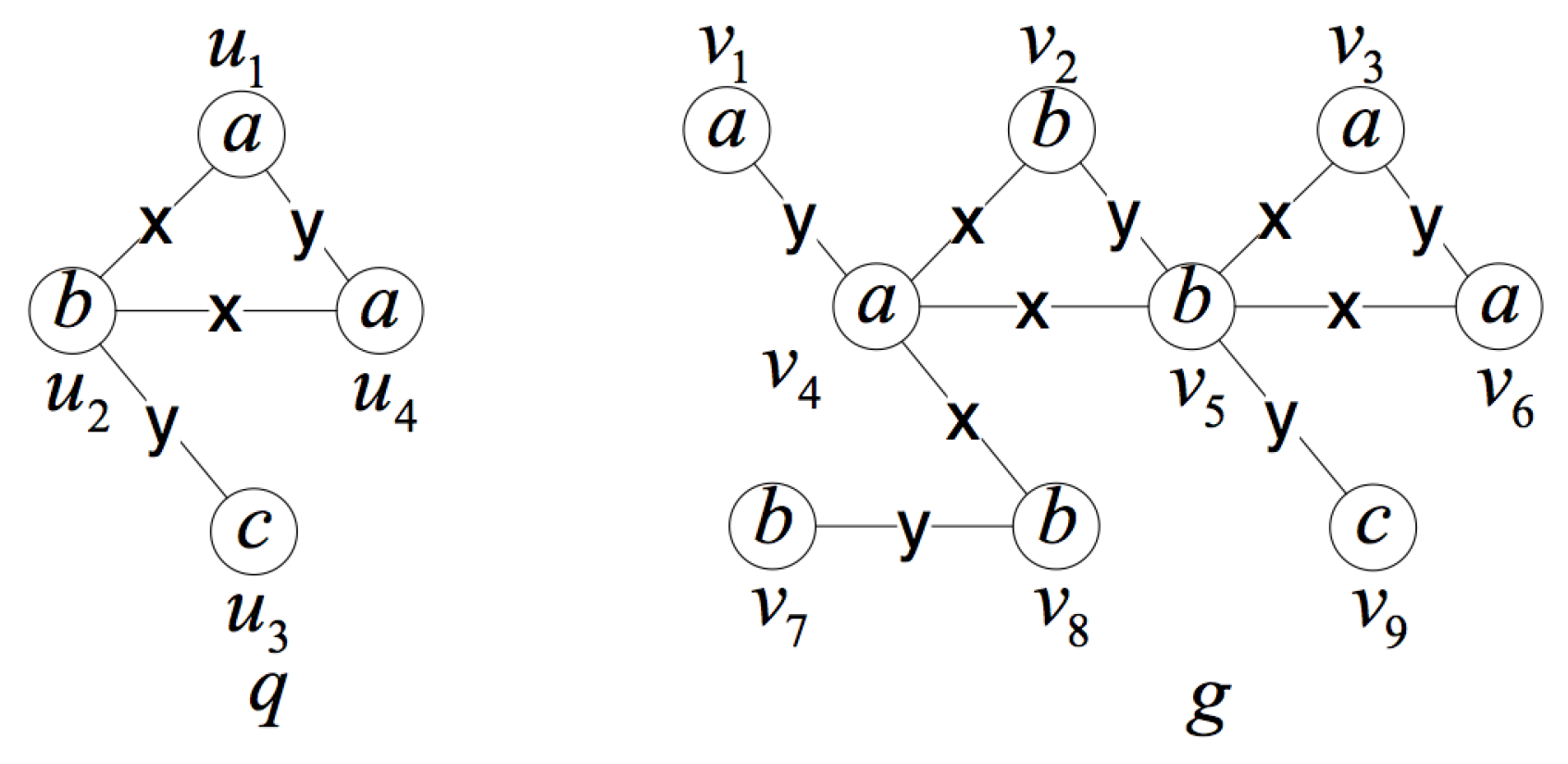
- Use the query position as the starting vertex of the isomorphism testingUsing the query position as the starting vertex of the isomorphism testing makes full use of the query position in the data graph, and it is the most important improvement in the isomorphism testing algorithm. When the subgraph isomorphism testing is performed on candidate subgraphs, matching the query position first can avoid invalid and extra isomorphism testing.Figure 9 is an illustration of the importance of matching the query position first. Considering the data graph G and the query position , is an induced subgraph of G containing the query position with three vertices. If we do not use as the first matching vertex, it will be matched in two directions. For the first direction, we can get matched pairs of vertices , and , where the isomorphic subgraph is itself. For the other direction, we can get matched pairs of vertices and and will stop for the non-matched vertices in and in G. We have to do more testing to judge whether is unique. When using as the first matching vertex, the second direction testing can be avoided.In fact, much local or partial matching can be avoided when using the query position as the starting vertex of the isomorphism testing, thus judging whether the induced subgraph is unique as soon as possible and improving the efficiency of verification process. In addition, we use the query rewrite method in [17] to rank the other vertices in the query graph and get a matching order according to the ranking value. In this way, we can reduce the candidate regions for performing subgraph isomorphism search and improve the efficiency.
- (2)
- Sorting the candidate vertices by degree for pruningFor vertex pairs in the candidate set P, u represents the vertex from induced subgraph , and the set of u can be denoted as , while v represents the vertex from data graph G, and the set of v can be denoted as .
| Algorithm 3 Query position-based subgraph isomorphism. |
Input: data graph, ; query position, q; induced graph, Output: Boolean variable to answer whether another induced graph in G isomorphic to graph , ;
Subroutine SubgraphSearch
|
4.5. Baseline and EQA Algorithms
5. Results
5.1. Experimental Performance Measurement
- (1)
- Average isomorphism timeThe running time of each algorithm in this paper contains two parts: the time of filtering and the time of verification. The time of filtering includes the time of searching and pruning in the induced subgraph space. Since the algorithms use the same method to search the induced subgraph space and the time of pruning is negligible relative to the time of searching, the filtering time was almost the same. Therefore, we adopted the verification time as the performance measurement. The verification time is the time of subgraph isomorphism testing.During the algorithms’ running process, the performance of the computer dynamically changes. Therefore, the average isomorphism time of five experiments was used as a criterion for evaluation. The average isomorphism time is represented by the symbol .
- (2)
- The times of calling the recursive functionCompared with the isomorphism time, the times of calling the recursive function were more stable and could better reflect the performance of the algorithms. As long as the data graph and query position were given, the times of calling recursive functions were the same for each algorithm and would not change when the PC hardware and software environment change.
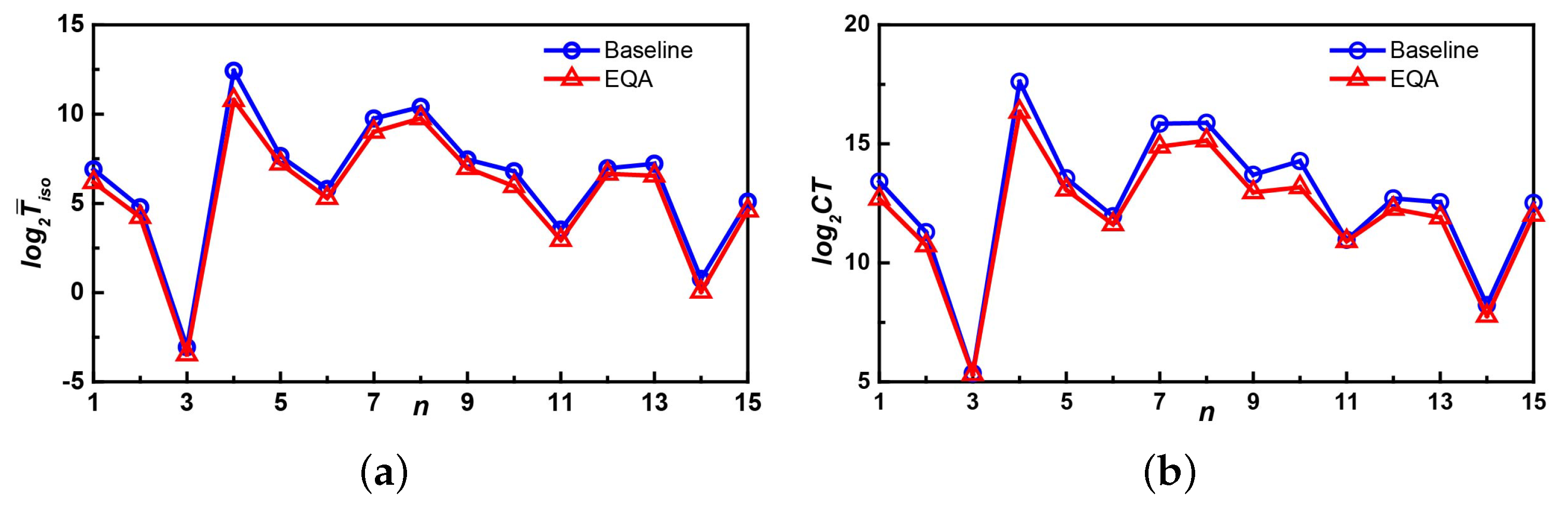
5.2. Experiment on the YEAST Dataset
5.3. Experiment on the HPRD Dataset
5.4. Experiment on the Synthetic Datasets
- (1)
- Experiments on the increasing number of edgesWe investigated the influence of graph size on EQA process speed first. Keep the same number of vertex labels and edge labels, and increase the number of edges. The number of vertices was 3000; the number of edge labels was set as five; the number of vertex labels was set as five; and the number of edges was set as 6000, 7000, 8000 and 9000 separately. The experimental results are shown in Table 3. As can be seen from the table, was increasing with the increasing number of edges, that is to say, the answering speed was decreasing. When the number of vertices in the graph was kept the same and the number of edges was increasing, the average degree of vertices was also increasing. Therefore, when searching the induced subgraph space in ascending order of the number of vertices, more candidate subgraphs would be generated in each layer, and more graphs would participate in the subgraph isomorphism testing; so the query time became longer, and the answering speed decreased.
- (2)
- Experiments on the increasing number of vertex labelsWe investigated the influence of vertex labels on EQA process speed subsequently. Keep the same size of graphs and the same number of edge labels, then increase the number of vertex labels. The graphs contained 3000 vertices and 8000 edges. The number of edge labels was set as five, and the number of vertex labels was set as 10, 30, 50 and 70 separately. The experimental results are shown in Table 4. As seen from the table, was decreasing with the increasing number of vertex labels, that is to say, the answering speed was increasing. Since both the number of vertices and the number of edges in each graph were the same, the number of candidate subgraphs generated had little difference, as well as the number of candidate subgraphs participating in the isomorphism testing. However, with the increasing number of vertex labels, there were more unique induced subgraphs on the dataset and a higher probability to obtain MUIS earlier. Therefore, was decreasing, and the answering speed was increasing.
- (3)
- Experiments on the increasing number of edge labelsWe investigated the influence of edge labels on EQA process speed at last. Keep the same size of graphs and the same number of vertex labels, then increase the number of edge labels. All the graphs contained 3000 vertices and 8000 edges. The number of vertex labels was set as five, and the number of edge labels was set as 10, 30, 50 and 70 separately. The experimental results are shown in Table 5. As seen from the table, was decreasing with the increasing number of edge labels, that is to say, the answering speed was increasing. Since the size of each graph was the same, the number of candidate subgraphs generated in each graph was also almost the same, that is to say, the number of candidate subgraphs participating in the isomorphism testing also had little difference. However, the number of unique induced subgraphs was increasing in each graph with the increasing number of edge labels, so there was a higher probability to obtain MUIS earlier. Therefore, was decreasing, and the answering speed was increasing.
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Zaslavskiy, M.; Bach, F.; Vert, J.P. Global alignment of protein–protein interaction networks by graph matching methods. Bioinformatics 2009, 25, 259–267. [Google Scholar] [CrossRef] [PubMed]
- Liao, C.S.; Lu, K.; Baym, M.; Singh, R.; Berge, B. IsoRankN: Spectral methods for global alignment of multiple protein networks. Bioinformatics 2009, 25, 253–258. [Google Scholar] [CrossRef] [PubMed]
- Couennea, F.; Jalluta, C.; Maschkea, B.; Tayakouta, M.; Breedveldb, P. Bond graph for dynamic modelling in chemical engineering. Chem. Eng. Process. 2008, 47, 1994–2003. [Google Scholar] [CrossRef]
- Khakzad, N.; Landucci, G.; Reniers, G. Application of Graph Theory to Cost-Effective Fire Protection of Chemical Plants During Domino Effects. Risk Anal. 2017, 37, 1652–1667. [Google Scholar] [CrossRef] [PubMed]
- Faloutsos, M. Detecting malware with graph-based methods: Traffic classification, botnets, and facebook scams. In Proceedings of the 22nd International Conference on World Wide Web, Rio De Janeiro, Brazil, 13–17 May 2013; pp. 495–496. [Google Scholar]
- Khan, K.U.; Alam, A.; Dolgorsuren, B.; Uddin, M.A.; Umair, M.; Sang, U.; Duong, V.T.; Xu, W.; Lee, Y.K. LPaMI: A Graph-Based Lifestyle Pattern Mining Application Using Personal Image Collections in Smartphones. Appl. Sci. 2017, 7, 1200. [Google Scholar] [CrossRef]
- Rezig, S.; Achour, Z.; Rezg, N.; Kammoun, M.A. Supervisory control based on minimal cuts and Petri net sub-controllers coordination. Int. J. Syst. Sci. 2015, 1–11. [Google Scholar] [CrossRef]
- Rezig, S.; Achour, Z.; Rezg, N. Control Synthesis Based on Theory of Regions with Minimal Reachability Graph Knowledge. IFAC-Pap. Online 2016, 49, 1383–1388. [Google Scholar] [CrossRef]
- Rezig, S.; Achour, Z.; Rezg, N. Theory of Regions for Control Synthesis without Computing Reachability Graph. Appl. Sci. 2017, 7, 270. [Google Scholar] [CrossRef]
- Fortin, S. The Graph Isomorphism Problem; Tech. Rep.; University of Alberta: Edmonton, AB, Canada, 1996. [Google Scholar]
- Yuan, Y.; Wang, G.; Chen, L.; Wang, H. Efficient subgraph similarity search on large probabilistic graph databases. In Proceedings of the VLDB Endowment, Istanbul, Turkey, 27–31 August 2012; pp. 800–811. [Google Scholar]
- Cook, S.A. The Complexity of Theorem-proving. In Proceedings of the Third Annual ACM Symposium on Theory of Computing, Shaker Heights, OH, USA, 3–5 May 1971; pp. 151–158. [Google Scholar]
- Shamir, R.; Tsur, D. Faster subtree isomorphism. J. Algorithms 1999, 33, 267–280. [Google Scholar] [CrossRef]
- Shasha, D.; Wang, J.; Giugn, R. Algorithmics and applications of tree and graph searching. In Proceedings of the Twenty-First ACM SIGMOD-SIGACT-SIGART Symposium on Principles of Database Systems, Madison, WI, USA, 3–5 June 2002; pp. 39–52. [Google Scholar]
- Cordella, L.P.; Foggia, P.; Sansone, C.; Vento, M. A (sub)graph isomorphism algorithm for matching large graphs. IEEE PAMI 2004, 26, 1367–1372. [Google Scholar] [CrossRef] [PubMed]
- Shang, H.; Zhang, Y.; Lin, X.; Yu, J.X. Taming verification hardness: An efficient algorithm for testing subgraph isomorphism. In Proceedings of the VLDB Endowment, Auckland, New Zealand, 23–28 August 2008; pp. 364–375. [Google Scholar]
- Han, W.S.; Lee, J.; Lee, J.H. TurboISO: Towards ultrafast and robust subgraph isomorphism search in large graph databases. In Proceedings of the 2013 ACM SIGMOD International Conference on Management of Data, New York, NY, USA, 22–27 June 2013; pp. 337–348. [Google Scholar]
- Zhao, X.; Xiao, C.; Lin, X.; Wang, W.; Ishikawa, Y. Efficient processing of graph similarity queries with edit distance constraints. VLDB J. 2013, 22, 727–752. [Google Scholar] [CrossRef]
- Zhao, X.; Xiao, C.; Lin, X.; Zhang, W.; Wang, Y. Efficient structure similarity searches: A partition-based approach. VLDB J. 2018, 27, 53–78. [Google Scholar] [CrossRef]
- Lin, W.; Xiao, X.; Ghinita, G. Large-scale frequent subgraph mining in mapreduce. In Proceedings of the IEEE 30th International Conference on Data Engineering, Chicago, IL, USA, 31 March–4 April 2014; pp. 844–855. [Google Scholar]
- Horváth, T.; Otaki, K.; Ramon, J. Efficient frequent connected induced subgraph mining in graphs of bounded tree-width. In Proceedings of the Joint European Conference on Machine Learning and Knowledge Discovery in Databases, Prague, Czech Republic, 23–27 September 2013; pp. 622–637. [Google Scholar]
- Qiao, F.; Zhang, X.; Li, P.; Ding, Z.; Jia, S.; Wang, H. A parallel approach for frequent subgraph mining in a single large graph using spark. J. Appl. Sci. 2018, 8, 230. [Google Scholar] [CrossRef]
- Inokuchi, A.; Washio, T.; Motoda, H. An apriori-based algorithm for mining frequent substructures from graph data. In Proceedings of the Joint European Conference on Machine Learning and Knowledge Discovery in Databases, Lyon, France, 13–16 September 2000; pp. 13–23. [Google Scholar]
- Kuramochi, M.; Karypis, G. Frequent subgraph discovery. In Proceedings of the 2001 IEEE International Conference on Data Mining, San Jose, CA, USA, 29 November–2 December 2001; pp. 313–320. [Google Scholar]
- Yan, X.; Han, J. Gspan: Graph-based substructure pattern mining. In Proceedings of the 2002 IEEE International Conference on Data Mining, Maebashi City, Japan, 9–12 December 2002; pp. 721–724. [Google Scholar]
- Huan, J.; Wang, W.; Prins, J. Efficient Mining of Frequent Subgraphs in the Presence of Isomorphism. In Proceedings of the 2003 IEEE International Conference on Data Mining, Melbourne, FL, USA, 19–22 November 2003. [Google Scholar]
- Nijssen, S.; Kok, J.N. A quickstart in frequent structure mining can make a difference. In Proceedings of the tenth ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, Seattle, WA, USA, 22–25 August 2004; pp. 647–652. [Google Scholar]
- Zou, L.; Chen, L.; Lu, Y. Top-K correlation sub-graph search in graph databases. In Proceedings of the 14th International Conference on Database Systems for Advanced Applications, Brisbane, Australia, 21–23 April 2009; pp. 168–185. [Google Scholar]
- Ke, Y.; Cheng, J.; Ng, W. Efficient correlation search from graph databases. IEEE Trans. Knowl. Data Eng. 2008, 20, 1601–1615. [Google Scholar]
- Ke, Y.; Cheng, J.; Ng, W. Correlation search in graph databases. In Proceedings of the 13th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Jose, CA, USA, 12–15 August 2007; pp. 390–399. [Google Scholar]
- Ke, Y.; Cheng, J.; Yu, J.X. Efficient discovery of frequent correlated subgraph pairs. In Proceedings of the Ninth IEEE International Conference on Data Mining, Miami, FL, USA, 6–9 December 2009; pp. 239–248. [Google Scholar]
- Ronen, M.; Rosenberg, R.; Shraiman, B.I.; Alon, U. Assigning numbers to the arrows: Parameterizing a gene regulation network by using accurate expression kinetics. Proc. Natl. Acad. Sci. USA 2002, 99, 10555–10560. [Google Scholar] [CrossRef] [PubMed]
- Grochow, J.A.; Kellis, M. Network motif discovery using subgraph enumeration and symmetry-breaking. In Proceedings of the Annual International Conference on Research in Computational Molecular Biology, Oakland, CA, USA, 21–25 April 2007; pp. 92–106. [Google Scholar]
- Ribeiro, P.; Silva, F. G-Tries: A data structure for storing and finding subgraphs. Data Min. Knowl. Dis. 2014, 28, 337–377. [Google Scholar] [CrossRef]
- Michale, G.; Giugno, R.; Ferro, A.; Mongiovi, M.; Shasha, D.; Pulvirenti, A. Fast Analytical Methods for Finding Significant Labeled Graph Motifs. Data Min. Knowl. Dis. 2018, 32, 504–531. [Google Scholar] [CrossRef]
- Mcgregor, J. Backtrack search algorithms and the maximal common subgraph problem. Softw. Pract. Exp. 1982, 12, 23–34. [Google Scholar] [CrossRef]
- Williams, D.W.; Huan, J.; Wang, W. Graph database indexing using structured graph decomposition. In Proceedings of the 23rd International Conference on Data Engineering, Istanbul, Turkey, 15–20 April 2007; pp. 976–985. [Google Scholar]
- Shokoufandeh, A.; Dickinson, S.J.; Siddiqi, K.; Zucker, S.W. Indexing using a spectral encoding of topological structure. In Proceedings of the 1999 Conference on Computer Vision and Pattern Recognition, Collins, CO, USA, 23–25 June 1999; pp. 491–497. [Google Scholar]
- Bu, D.; Zhao, Y.; Cai, L. Topological structure analysis of the protein-protein interaction network in budding yeast. Nucleic Acids Res. 2003, 31, 2443–2450. [Google Scholar] [CrossRef] [PubMed]
- Goel, R.; Muthusamy, B.; Pandey, A.; Prasad, T.S.K. Human protein reference database and human proteinpedia as discovery resources for molecular biotechnology. Mol. Biotechnol. 2011, 48, 87–95. [Google Scholar] [CrossRef] [PubMed]



| # | Query Position | Vertex # | MUIS |
|---|---|---|---|
| #1 | 265 | 4 | 257 265 267 991 |
| #2 | 321 | 5 | 81 208 321 522 1024 |
| #3 | 345 | 5 | 146 338 345 400 849 |
| #4 | 495 | 4 | 495 499 1525 1816 |
| #5 | 620 | 6 | 275 303 619 620 625 866 |
| #6 | 752 | 4 | 67 71 752 1040 |
| #7 | 899 | 8 | 186 895 896 891 898 899 900 1280 |
| #8 | 987 | 5 | 368 507 987 1201 1477 |
| #9 | 1501 | 4 | 144 429 1501 1678 |
| #10 | 1758 | 5 | 1483 1526 1724 1725 1758 |
| #11 | 1895 | 3 | 198 1576 1895 |
| #12 | 1984 | 5 | 17 1191 1508 1982 1984 |
| #13 | 2013 | 6 | 1515 1553 1559 1562 1563 2013 |
| #14 | 2236 | 4 | 1442 1617 1885 2236 |
| #15 | 2300 | 6 | 1005 1357 1517 2121 2122 2300 |
| # | Query Position | Vertex # | MUIS |
|---|---|---|---|
| #1 | 1890 | 4 | 100 568 723 1890 |
| #2 | 2155 | 3 | 134 2155 2157 |
| #3 | 2977 | 2 | 1098 2977 |
| #4 | 3434 | 4 | 236 2329 3434 6928 |
| #5 | 3789 | 3 | 1144 3789 4832 |
| #6 | 4334 | 3 | 69 4334 4677 |
| #7 | 4567 | 4 | 87 2734 4567 4569 |
| #8 | 5332 | 4 | 77 153 235 5332 |
| #9 | 5347 | 5 | 269 346 347 349 5347 |
| #10 | 5701 | 4 | 659 2255 5701 6596 |
| #11 | 5734 | 3 | 5734 5767 5768 |
| #12 | 6758 | 4 | 419 1282 1818 6758 |
| #13 | 7345 | 4 | 1686 3142 3840 7345 |
| #14 | 8434 | 3 | 127 2959 8434 |
| #15 | 9147 | 4 | 1457 1728 3277 9417 |
| Dataset # | Edge # | Vertex Label # | Edges Label # | |
|---|---|---|---|---|
| #1 | 6000 | 5 | 5 | 566,534 |
| #2 | 7000 | 5 | 5 | 616,321 |
| #3 | 8000 | 5 | 5 | 685,132 |
| #4 | 9000 | 5 | 5 | 763,026 |
| Dataset # | Edge # | Vertex Label # | Edges Label # | |
|---|---|---|---|---|
| #1 | 8000 | 10 | 5 | 666,345 |
| #2 | 8000 | 30 | 5 | 602,654 |
| #3 | 8000 | 50 | 5 | 538,935 |
| #4 | 8000 | 70 | 5 | 464,682 |
| Dataset # | Edge # | Vertex Label # | Edges Label # | |
|---|---|---|---|---|
| #1 | 8000 | 5 | 10 | 675,634 |
| #2 | 8000 | 5 | 30 | 610,635 |
| #3 | 8000 | 5 | 50 | 542,684 |
| #4 | 8000 | 5 | 70 | 476,325 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, L.; Zhao, X.; Ge, B.; Hu, S.; Xiao, W.; Shang, H.; Jing, Y. On Minimal Unique Induced Subgraph Queries. Appl. Sci. 2018, 8, 1798. https://doi.org/10.3390/app8101798
Jiang L, Zhao X, Ge B, Hu S, Xiao W, Shang H, Jing Y. On Minimal Unique Induced Subgraph Queries. Applied Sciences. 2018; 8(10):1798. https://doi.org/10.3390/app8101798
Chicago/Turabian StyleJiang, Lincheng, Xiang Zhao, Bin Ge, Shengze Hu, Weidong Xiao, Haichuan Shang, and Yumei Jing. 2018. "On Minimal Unique Induced Subgraph Queries" Applied Sciences 8, no. 10: 1798. https://doi.org/10.3390/app8101798
APA StyleJiang, L., Zhao, X., Ge, B., Hu, S., Xiao, W., Shang, H., & Jing, Y. (2018). On Minimal Unique Induced Subgraph Queries. Applied Sciences, 8(10), 1798. https://doi.org/10.3390/app8101798





