Identification of Hybrid Okra Seeds Based on Near-Infrared Hyperspectral Imaging Technology
Abstract
1. Introduction
2. Materials and Methods
2.1. Okra Seed Samples Preparation
2.2. Near-Infrared Hyperspectral Imaging
2.3. Spectral Collection
2.4. Multivariate Data Analysis
2.5. Software Tools
3. Results and Discussion
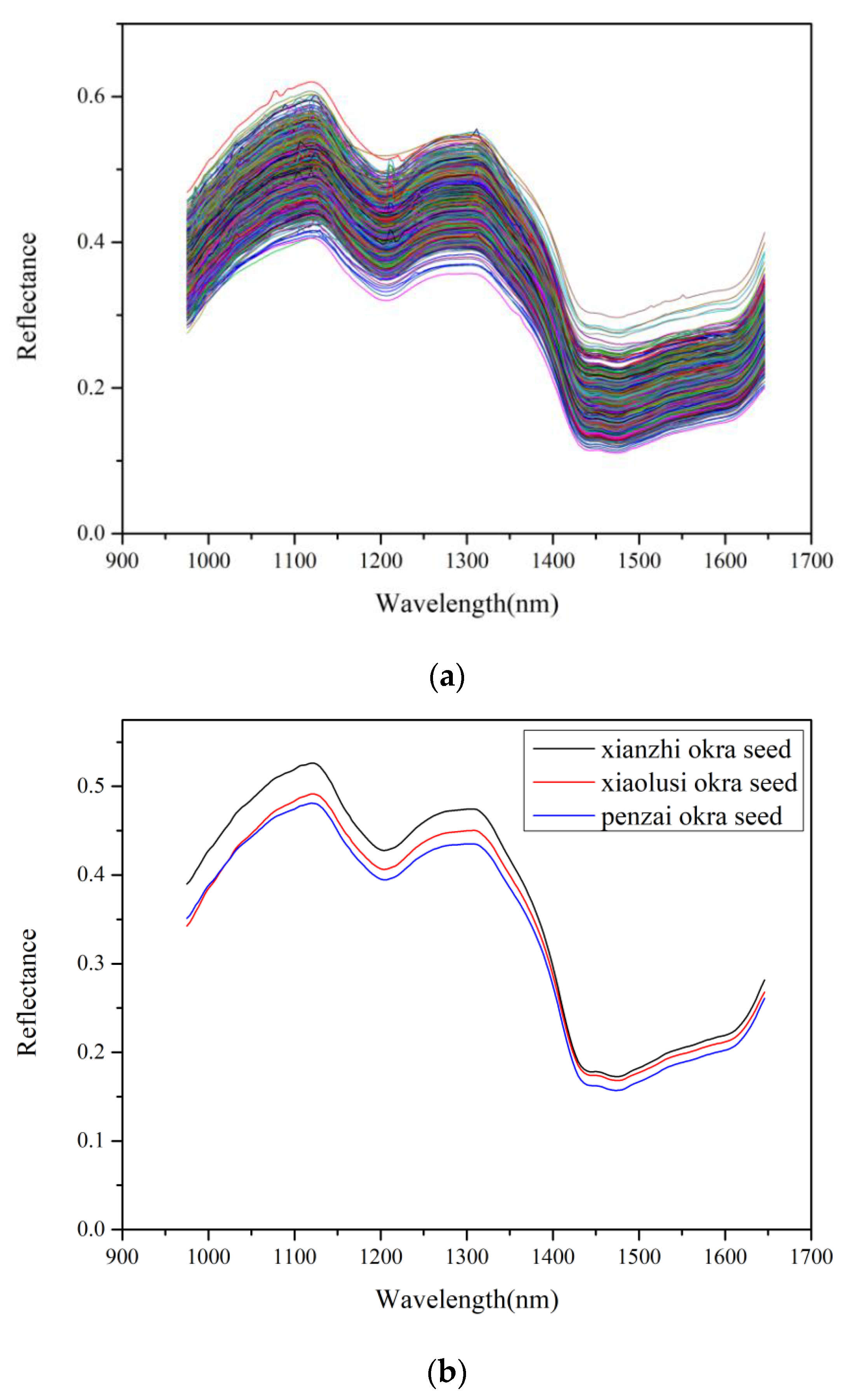
3.1. Spectroscopic Analysis
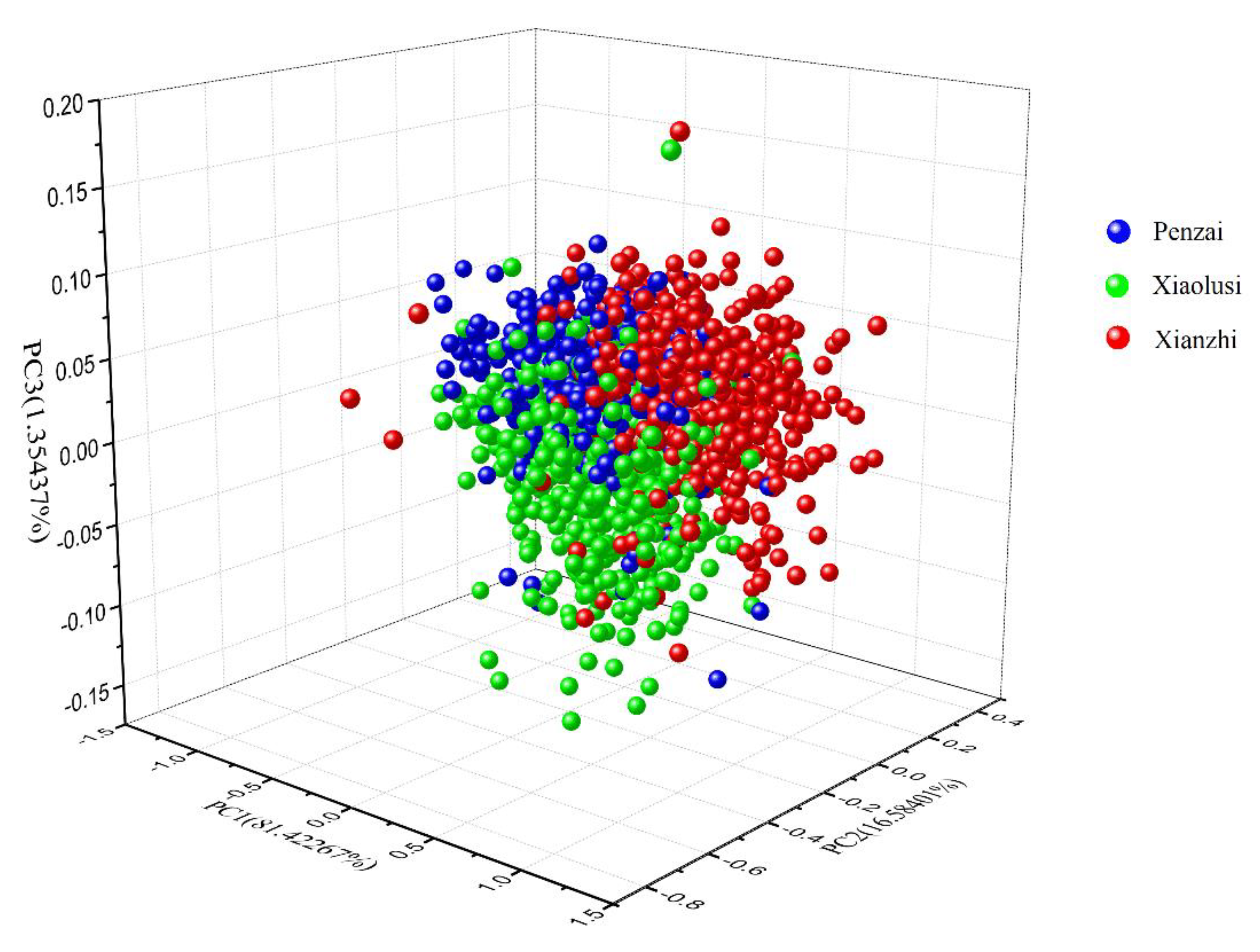
3.2. Principle Component Analysis of Spectral Data
3.3. Classification Results and Analysis by the Discrimination Models Based on the Full Spectrum
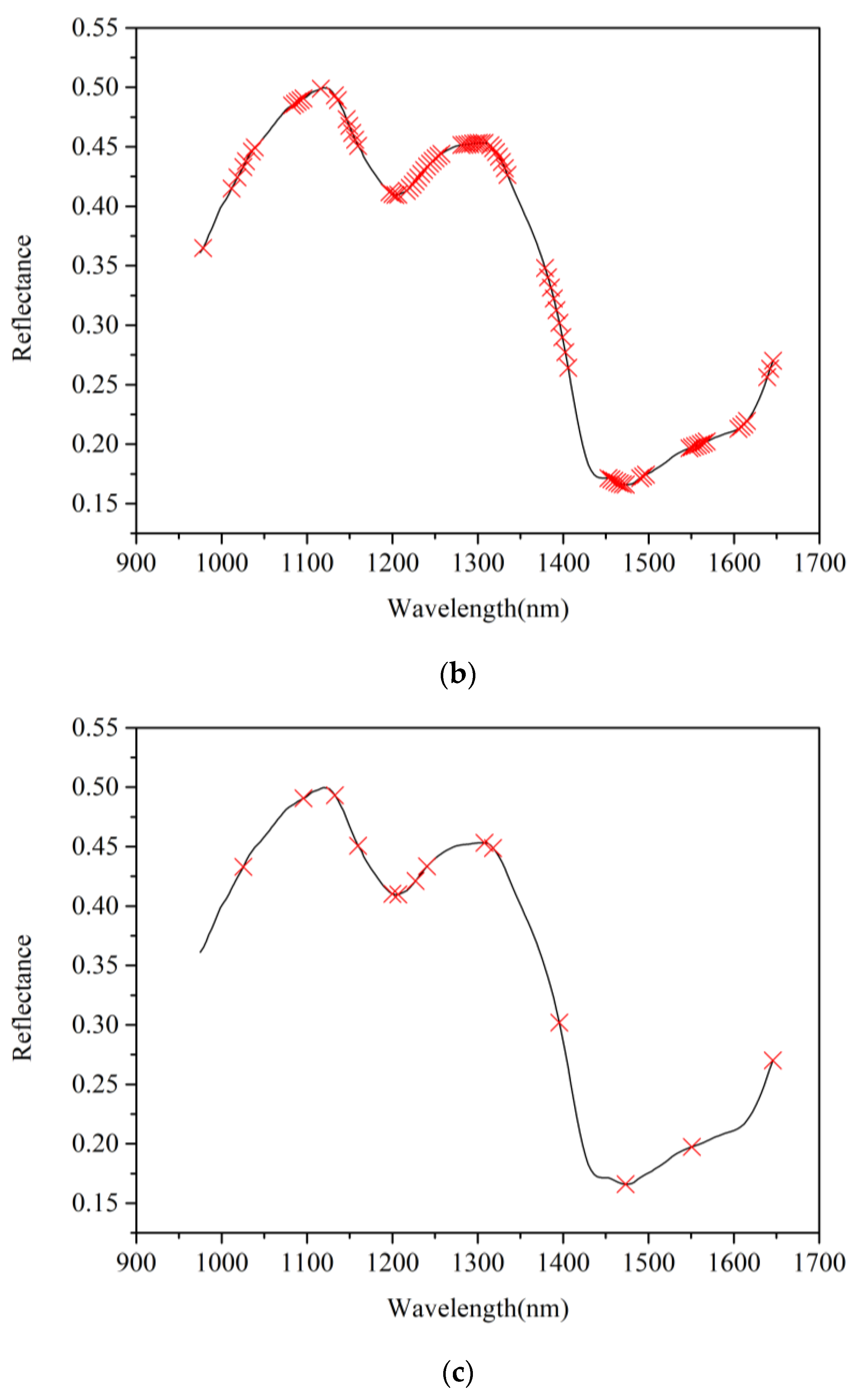
3.4. Selection of Effective Wavelengths
3.5. Classification Results and Analysis by the Discrimination Models Based on the Characteristic Band Spectrum
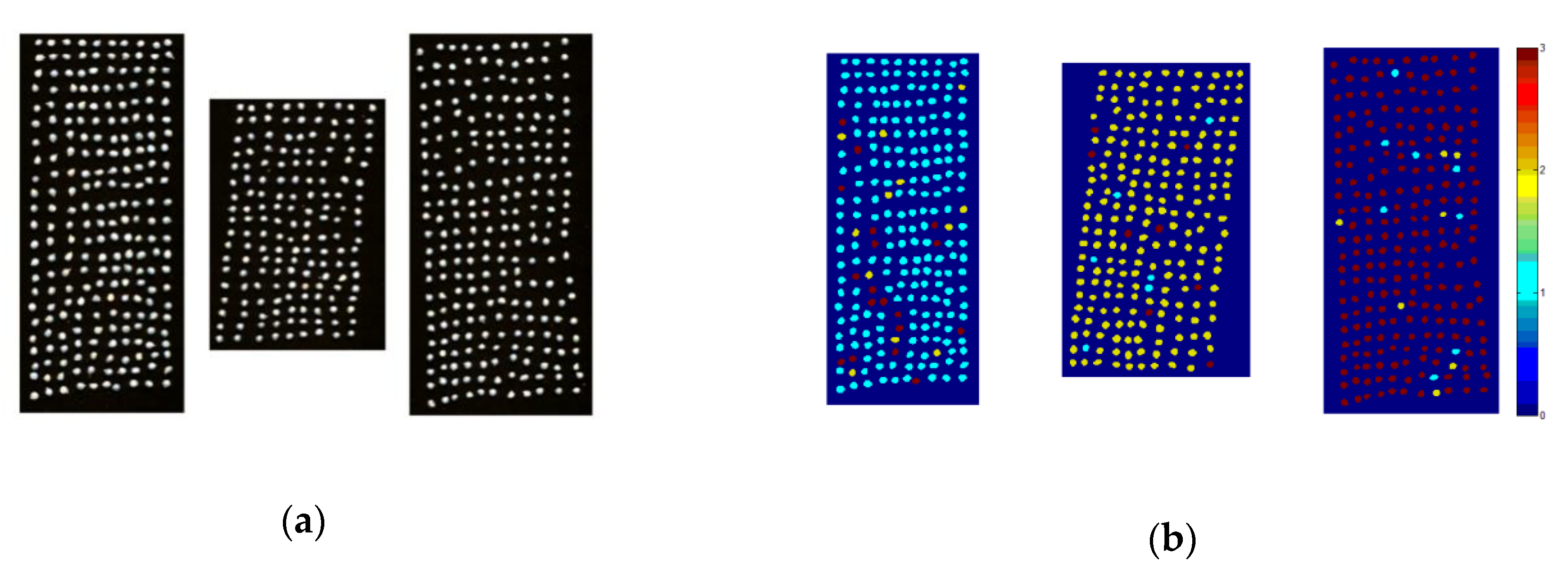
3.6. Visual Prediction of Okra Seeds
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kumar, P.S.; Sreeparvathy, S. Studies on heterosis in okra (Abelmoschus esculentus (L.) Moench). Electron. J. Plant Breed. 2010, 1, 1431–1433. [Google Scholar]
- Reddy, M.T.; Babu, K.H.; Ganesh, M.; Begum, H.; Babu, J.D. Exploitation of hybrid vigour for yield and its components in okra [Abelmoschus esculentus (L.) Moench]. Am. J. Agric. Sci. Technol. 2013, 1, 1–17. [Google Scholar] [CrossRef]
- Adelakun, O.E.; Oyelade, O.J.; Adeomowaye, B.I.; Adeyemi, I.A.; Van, D.V.M. Chemical composition and the antioxidative properties of nigerian okra seed (Abelmoschus esculentus Moench) flour. Food Chem. Toxic. 2009, 47, 1123–1126. [Google Scholar] [CrossRef]
- Arapitsas, P. Identification and quantification of polyphenolic compounds from okra seeds and skins. Food Chem. 2008, 110, 1041–1045. [Google Scholar] [CrossRef] [PubMed]
- Xia, F.; Zhong, Y.; Li, M.; Chang, Q.; Liao, Y.; Liu, X.; Pan, R. Antioxidant and anti-fatigue constituents of okra. Nutrients 2015, 7, 8846–8858. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Yu, W.; Li, Y.; Prasad, N.; Tang, Z. Antioxidant activity of extract and its major constituents from okra seed on rat hepatocytes injured by carbon tetrachloride. Biomed. Res. Int. 2015, 2014, 341291. [Google Scholar] [CrossRef] [PubMed]
- Maciel, G.M.; Luz, J.M.; Campos, S.F.; Finzi, R.R.; Azevedo, B.N.; Maciel, G.M.; Luz, J.M.; Campos, S.F.; Finzi, R.R.; Azevedo, B.N. Heterosis in okra hybrids obtained by hybridization of two methods: Traditional and experimental. Hort. Bras. 2017, 35, 119–123. [Google Scholar] [CrossRef]
- Seth, T.; Chattopadhyay, A.; Chatterjee, S.; Dutta, S.; Singh, B. Selecting parental lines among cultivated and wild species of okra for hybridization aiming at YVMV disease resistance. J. Agric. Sci. Technol. 2016, 18, 751–762. [Google Scholar]
- Das, S.; Chattopadhyay, A.; Dutta, S.; Chattopadhyay, S.B.; Hazra, P. Breeding okra for higher productivity and yellow vein mosaic tolerance. Int. J. Veg. Sci. 2013, 19, 58–77. [Google Scholar] [CrossRef]
- Yin, W.; Zhang, C.; Zhu, H.; Zhao, Y.; He, Y. Application of near-infrared hyperspectral imaging to discriminate different geographical origins of chinese wolfberries. PLoS ONE 2017, 12, e0180534. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Pulido, F.J.; Barbin, D.F.; Sun, D.W.; Gordillo, B.; González-Miret, M.L.; Heredia, F.J. Grape seed characterization by NIR hyperspectral imaging. Postharvest Biol. Technol. 2013, 76, 74–82. [Google Scholar] [CrossRef]
- Sun, J.; Jiang, S.; Mao, H.; Wu, X.; Li, Q. Classification of black beans using visible and near infrared hyperspectral imaging. Int. J. Food Prop. 2016, 19, 1687–1695. [Google Scholar] [CrossRef]
- Huang, M.; Tang, J.; Yang, B.; Zhu, Q. Classification of maize seeds of different years based on hyperspectral imaging and model updating. Comput. Electron. Agric. 2016, 122, 139–145. [Google Scholar] [CrossRef]
- Serranti, S.; Cesare, D.; Marini, F.; Bonifazi, G. Classification of oat and goat kernels using nir hyperspectral imaging. Talanta 2013, 103, 276–284. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Hong, H.; You, Z.; Cheng, F. Spectral and image integrated analysis of hyperspectral data for waxy corn seed variety classification. Sensors 2015, 15, 15578–15594. [Google Scholar] [CrossRef] [PubMed]
- Gowen, A.A.; O’Donnell, C.P.; Cullen, P.J.; Downey., G.; Frias, J.M. Hyperspectral imaging—An emerging process analytical tool for food quality and safety control. Trends Food Sci. Technol. 2007, 18, 590–598. [Google Scholar] [CrossRef]
- Zhang, C.; Wang, Q.; Liu, F.; He, Y.; Xiao, Y. Rapid and non-destructive measurement of spinach pigments content during storage using hyperspectral imaging with chemometrics. Measurement 2017, 97, 149–155. [Google Scholar] [CrossRef]
- Qiu, Z.; Chen, J.; Zhao, Y.; Zhu, S.; He, Y.; Zhang, C. Variety identification of single rice seed using hyperspectral imaging combined with convolutional neural network. Appl. Sci. 2018, 8, 212. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhu, S.; Zhang, C.; Feng, X.; Feng, L.; He, Y. Application of hyperspectral imaging and chemometrics for variety classification of maize seeds. RSC Adv. 2018, 8, 1337–1345. [Google Scholar] [CrossRef]
- Feng, X.; Zhao, Y.; Zhang, C.; Cheng, P.; He, Y. Discrimination of transgenic maize kernel using nir hyperspectral imaging and multivariate data analysis. Sensors 2017, 17, 1894. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, S.; Knapič, M.; Žibrat, U.; Deleuran, L.C.; Gislum, R. Single seed near-infrared hyperspectral imaging in determining tomato (Solanum lycopersicum L.) seed quality in association with multivariate data analysis. Sens. Actuators B Chem. 2016, 237, 1027–1034. [Google Scholar] [CrossRef]
- Gao, J.; Li, X.; Zhu, F.; He, Y. Application of hyperspectral imaging technology to discriminate different geographical origins of jatropha curcas l. Seeds. Comput. Electron. Agric. 2013, 99, 186–193. [Google Scholar] [CrossRef]
- Zhang, S.; Jie, D.; Zhang, H. NIR spectroscopy identification of persimmon varieties based on pca-svm. Comput. Comput. Technol. Agric. IV 2010, 345, 118–123. [Google Scholar]
- Shao, Y.; Yuan, L.; Jiang, L.; Jian, P.; Yong, H.; Dou, X. Identification of pesticide varieties by detecting characteristics of chlorella pyrenoidosa using visible/near infrared hyperspectral imaging and raman microspectroscopy technology. Water Res. 2016, 104, 432–440. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Jiang, H.; Liu, F.; He, Y. Application of near-infrared hyperspectral imaging with variable selection methods to determine and visualize caffeine content of coffee beans. Food Bioprocess Technol. 2017, 10, 1–9. [Google Scholar] [CrossRef]
- Burges, C.J.C. A tutorial on support vector machines for pattern recognition. Data Min. Knowl. Discov. 1998, 2, 121–167. [Google Scholar] [CrossRef]
- Wu, D.; He, Y.; Feng, S.; Sun, D.W. Study on infrared spectroscopy technique for fast measurement of protein content in milk powder based on LS-SVM. J. Food Eng. 2008, 84, 124–131. [Google Scholar] [CrossRef]
- Düzyaman, E. Phenotypic diversity within a collection of distinct okra (Abelmoschus esculentus) cultivars derived from turkish land races. Genet. Resour. Crop Evol. 2005, 52, 1019–1030. [Google Scholar] [CrossRef]
- Williams, P.J.; Kucheryavskiy, S. Classification of maize kernels using NIR hyperspectral imaging. Food Chem. 2016, 209, 131–138. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Fei, L.; Yong, H.; Li, X. Application of hyperspectral imaging and chemometric calibrations for variety discrimination of maize seeds. Sensors 2012, 12, 17234–17246. [Google Scholar] [CrossRef] [PubMed]
- Noh, H.K.; Lu, R. Hyperspectral laser-induced fluorescence imaging for assessing apple fruit quality. Postharvest Biol. Technol. 2007, 43, 193–201. [Google Scholar] [CrossRef]
- Zhang, J.; Rivard, B.; Rogge, D.M. The successive projection algorithm (SPA), an algorithm with a spatial constraint for the automatic search of endmembers in hyperspectral data. Sensors 2008, 8, 1321–1342. [Google Scholar] [CrossRef] [PubMed]
- Nie, P.; Dong, T.; He, Y.; Xiao, S. Research on the effects of drying temperature on nitrogen detection of different soil types by near infrared sensors. Sensors 2018, 18, 391. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Xu, K.; Zhang, Y.; Sun, C.; He, Y. Optical determination of lead chrome green in green tea by fourier transform infrared (FT-IR) transmission spectroscopy. PLoS ONE 2017, 12, e0169430. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.; Xu, N.; Shao, Y.; He, Y. Using FT-IR spectroscopy technique to determine arginine content in fermented cordyceps sinensis mycelium. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2015, 149, 971. [Google Scholar] [CrossRef] [PubMed]
- Burns, D.A.; Ciurczak, E.W. Handbook of Near-Infrared Analysis; CRC Press: Boca Raton, FL, USA, 2008; pp. 211–230. [Google Scholar]
- Bryant, L.A.; Montecalvo, J.R.; Morey, K.S.; Loy, B. Processing, functional, and nutritional properties of okra seed products. J. Food Sci. 2010, 53, 810–816. [Google Scholar] [CrossRef]
- Agbo, E.A.; Nemlin, J.G.; Anvoh, B.K.; Gnakri, D. Characterisation of lipids in okra mature seeds. Int. J. Biol. Chem. Sci. 2010, 4, 184–192. [Google Scholar] [CrossRef]
- Kong, W.; Zhang, C.; Liu, F.; Nie, P.; He, Y. Rice seed cultivar identification using near-infrared hyperspectral imaging and multivariate data analysis. Sensors 2013, 13, 8916–8927. [Google Scholar] [CrossRef] [PubMed]

| Methods | Full Wavelength | ||
|---|---|---|---|
| Parameter | Calibration Set | Prediction Set | |
| PLS-DA | 9 | 83.36% | 82.59% |
| SVM | (256, 5.2780) | 99.31% | 93.62% |
| Methods | PLS-DA | SVM | ||
|---|---|---|---|---|
| SPA | Parameter | 8 | Parameter | (256, 27.8576) |
| Calibration Set | 79.74% | Calibration Set | 95.34% | |
| Prediction Set | 79.48% | Prediction Set | 91.55% | |
| CARS+SPA | Parameter | 9 | Parameter | (256, 48.5029) |
| Calibration Set | 81.47% | Calibration Set | 97.41% | |
| Prediction Set | 79.31% | Prediction Set | 92.24% | |
| CARS | Parameter | 9 | Parameter | (256, 9.1896) |
| Calibration Set | 84.40% | Calibration Set | 98.71% | |
| Prediction Set | 82.41% | Prediction Set | 94.83% | |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, J.; Feng, X.; Liu, X.; He, Y. Identification of Hybrid Okra Seeds Based on Near-Infrared Hyperspectral Imaging Technology. Appl. Sci. 2018, 8, 1793. https://doi.org/10.3390/app8101793
Zhang J, Feng X, Liu X, He Y. Identification of Hybrid Okra Seeds Based on Near-Infrared Hyperspectral Imaging Technology. Applied Sciences. 2018; 8(10):1793. https://doi.org/10.3390/app8101793
Chicago/Turabian StyleZhang, Jinnuo, Xuping Feng, Xiaodan Liu, and Yong He. 2018. "Identification of Hybrid Okra Seeds Based on Near-Infrared Hyperspectral Imaging Technology" Applied Sciences 8, no. 10: 1793. https://doi.org/10.3390/app8101793
APA StyleZhang, J., Feng, X., Liu, X., & He, Y. (2018). Identification of Hybrid Okra Seeds Based on Near-Infrared Hyperspectral Imaging Technology. Applied Sciences, 8(10), 1793. https://doi.org/10.3390/app8101793








