1. Introduction
Deformable registration of lung CT scans plays a pivotal role in real-world clinical workflows, including respiratory motion compensation [
1,
2], longitudinal disease monitoring [
3,
4,
5], and radiation therapy planning [
6,
7]. Unlike rigid registration methods [
8,
9,
10,
11] that assume static global transformations, deformable lung CT registration accounts for the complex non-linear anatomical changes induced by respiration, enabling precise spatial alignment across different breathing phases.
Traditionally, voxel-based registration has been the dominant paradigm for lung CT registration. These methods optimize the spatial transformation fields using intensity-based similarity metrics, such as mutual information [
12,
13] or normalized cross-correlation [
14]. While voxel-level registration provides dense deformation estimates, it suffers from critical limitations: (1) high computational and memory costs due to the dense volumetric nature of CT images [
15] and (2) sensitivity to the inconsistencies in intensity caused by artifacts, noise, and contrast variability [
7,
16].
Recent advances in geometric deep learning and 3D point cloud representations have enabled a new class of point-based deformable registration, offering computational efficiency and robustness to variations in intensity [
17,
18,
19]. These methods typically leverage sparse surface-based representations and geometric feature extraction to estimate dense deformation fields. However, they have not addressed the unique challenges of lung CT registration, where large, non-rigid respiratory deformations and fine-scale anatomical details must be jointly handled.
Against this backdrop, the recent release of the Lung250M dataset [
16] has introduced high-resolution lung CT point clouds across different respiratory phases, enabling foundational studies of point-based registration in lung CT scans. Unlike general registration tasks [
17,
18,
19], lung CT registration requires precise modeling of both the global anatomical structure (e.g., inter-lobar coherence) and local correspondence across complex structures such as bronchial trees and vascular bifurcations. In clinical terms, reliable registration of these structures is essential for tracking the disease progression in COPD and measuring the treatment response in pulmonary fibrosis or the accumulating radiation dose across respiratory cycles. These demands necessitate a multi-scale, context-aware framework that can simultaneously capture the long-range consistency and fine-level alignment.
In this work, we propose a novel point-based deformable lung CT registration framework designed to combine coarse-to-fine flow refinements with bi-directional geometric consistency. Specifically, our approach introduces three key innovations:
Geometric keypoint attention at coarse resolutions to enhance global-structure-aware feature matching across widely separated regions;
Context-aware flow refinement at finer levels via attention-based feature interactions, enabling precise modeling of subtle local deformations;
A bi-directional registration strategy that jointly estimates the forward and backward flow fields, enforcing transformation consistency and improving the anatomical plausibility.
By incorporating these components into a unified architecture, the proposed method achieves accurate, stable, and anatomically consistent alignment of lung CT point clouds, significantly improving over the existing point-based baselines [
17,
20]. Moreover, our method’s use of vascular structures as the keypoints aligns with clinically interpretable anatomical landmarks, offering better transparency and usability than fully voxel-based models.
3. The Proposed Approach
In this section, we first formalize our bi-directional objective for deformable lung CT registration (
Section 3.1), describe the multi-scale point feature extraction backbone (
Section 3.2), elaborate on geometric keypoint attention (
Section 3.3), detail the progressive refinement of the flow predictions with flow attention (
Section 3.4), and finally outline our comprehensive bi-directional training objective (
Section 3.5).
3.1. The Problem Setup
Given a pair of lung CT scans, the task of deformable lung CT registration aims to estimate a dense, non-rigid transformation that accurately aligns the anatomical structures between a fixed scan (source) and a moving scan (target). Formally, let and represent the 3D point clouds extracted from the source and target scans, respectively, where N denotes the number of sampled points. Our objective is to find a transformation , mapping each source point to its corresponding target location .
We parameterize this transformation as a displacement field, where each point-specific transformation function
is defined as
with
being the displacement vector to predict. Given a set of ground-truth correspondences
between the source and target points (precomputed using external tools such as corrField [
15]), the registration problem is formally expressed as minimizing
This optimization encourages the transformed source points
to closely match their ground-truth correspondences
, thereby achieving accurate anatomical alignment and maintaining spatial coherence.
In this work, we propose a learning-based approach to directly estimating the displacement field
from the input lung CT point clouds. Inspired by the point-based scene flow framework [
17], our architecture incorporates two key innovations. First, we introduce specialized attention mechanisms—geometric keypoint attention at the coarse scale and progressive flow attention at the fine scale—to capture both global structural relationships and local deformation cues better. Second, we employ a bi-directional flow estimation strategy, simultaneously predicting the forward flow (
, source-to-target) and the backward flow (
, target-to-source). Formally, the backward transformation is defined as
where
denotes the displacement vector for the target points. The backward flow is optimized by minimizing
This bi-directional formulation enables the network to leverage mutual consistency between the forward and backward transformations, resulting in more stable, accurate, and anatomically plausible registration outcomes.
Figure 1 provides an overview of our network design, and we detail each module in the subsequent sections.
3.2. Multi-Scale Feature Extraction
3.2.1. The PointConv Backbone
We extract hierarchical point features using PointConv layers [
38], which extend conventional convolutions to irregular 3D point sets. Formally, given an input point cloud
with the associated per-point features
, each PointConv layer aggregates the local features as follows:
where
denotes the local neighborhood of point
,
is an inverse density scale factor computed via kernel density estimation, and
represents the learnable convolutional weights conditioned on relative spatial coordinates. By stacking multiple PointConv blocks in a U-Net-style encoder–decoder [
35], we generate a coarse-to-fine hierarchy of informative point feature embeddings.
3.2.2. The Coarse-to-Fine Pipeline
To efficiently handle large deformations while preserving anatomical completeness, we progressively downsample the input point clouds using farthest point sampling (FPS) [
39], ensuring that the sampled points are broadly distributed across the lung’s geometry without over-concentrating on local regions. Since the original point clouds were extracted primarily from critical anatomical structures such as bronchial trees and vascular networks, FPS maintains essential structural coverage with high fidelity. This results in a hierarchical set of point clouds
, from the finest resolution (
) to the coarsest resolution (
), where each sampled point at the coarsest level encodes a broader anatomical region of the lung, facilitating global alignment. This multi-scale representation is critical for our attention-driven alignment strategy, where global coarse-level correspondences are first established and subsequently refined at higher resolutions.
3.3. Geometric Keypoint Attention
The geometric keypoint attention module captures the global structural correspondences prior to finer-scale refinement. At this stage, capturing the long-range geometric structure is essential for reducing the ambiguity of the correspondence under large respiratory motion, which motivates our use of a geometry-aware attention formulation. Consider the coarse-level feature embeddings
. The output attention-enhanced feature
is computed as an attention-weighted aggregation:
where the attention weights
measure the pairwise geometric relations among points:
Here,
represents the geometric embeddings, as proposed in GeoTransformer [
40], while
are learnable projection matrices for queries, keys, values, and geometric embeddings, respectively. Analogous computations are performed for the target cloud features
.
A similar attention mechanism is adopted for cross-attention, which explicitly models inter-cloud interactions by exchanging information between the source and target point clouds. Compared to self-attention, the key difference is that the query is computed from one point cloud (e.g., ), while the key and value are taken from the other (e.g., ). This directional change allows the network to attend across point clouds and capture alignment-relevant correspondences at the coarsest level.
To robustly capture both the intra-cloud structure and inter-cloud correspondence cues, the self- and cross-attention modules are alternated for iterations, resulting in enhanced features and . This interleaved design allows the network to iteratively refine the coarse-level representations by jointly reasoning about the global geometry and alignment consistency, which is particularly beneficial in reducing ambiguity in the correspondence under large deformations.
3.4. Progressive Flow Refinement
At each decoder level
l, we progressively refine the flow estimation by leveraging coarser predictions from level
, as shown in
Figure 2 (In the following sections, we focus on describing the forward flow
(source-to-target) for clarity and conciseness. The backward flow
(target-to-source) is computed in an analogous manner using the same architecture and refinement process.).
3.4.1. Warping and Cost Volume Construction
The estimated flow
at the coarser level
is propagated to the finer level through inverse distance weighted interpolation, producing an initial flow estimate
at the resolution
l. We use the upsampled flow
to warp the target points:
bringing the target points into approximate alignment with the source. This reduces large residual displacements and stabilizes the subsequent refinement steps.
Next, we construct a cost volume encoding the local geometric and feature-level differences. For each source point
, we first identify its
k-nearest neighbors among the warped target points
. Then, leveraging the warped target features
, we compute the residual feature embeddings between the source features
and their corresponding neighboring warped target features. These residual embeddings are subsequently aggregated via PointConv layers [
38], resulting in the cost volume features
, which effectively encode the geometric proximity and the discrepancies in the local appearance to guide the flow refinement.
3.4.2. The Flow Estimator
The cost volume features
and source features
are concatenated to form the refinement input:
To enhance the flow estimation accuracy, we introduce a contextual attention module composed of alternating self-attention and cross-attention layers. At this stage, the goal is to refine the localized residual flow patterns based on the learned contextual properties. Specifically, we first employ standard multi-head self-attention (MHSA) within the source point cloud to capture the internal structural context:
where MHSA is defined as
with
as the learnable linear projection matrices and
h as the number of attention heads.
Subsequently, to explicitly incorporate the inter-cloud correspondence cues, we perform a cross-attention operation between these self-attended source features
and the corresponding warped target features
. This step aggregates relevant information from the warped target points to refine the source features:
where
are the query, key, and value projection matrices, respectively.
By iteratively alternating the self-attention (intra-cloud) and cross-attention (inter-cloud) layers for steps, we obtain refined embeddings that jointly leverage the internal point cloud context and the external correspondence information.
Finally, these enriched embeddings
are input into a residual flow prediction network (MLP) to estimate the residual displacements
:
The refined flow at level
l is then computed by adding the residual displacement to the initial estimate:
which is recursively applied at each decoder level until the finest resolution is reached, producing the final flow prediction
.
Note that the same refinement procedure is applied in the reverse direction to obtain the backward flows .
3.4.3. Post-Processing for Dense Deformation Estimation
Although our model estimates the displacements only at sparse vessel-derived points, a dense deformation field across the entire lung volume is required. To achieve this, we employ Thin Plate Spline (TPS) interpolation, a classical method in landmark-based registration. Specifically, given the predicted displacements at a sparse set of source points, this interpolates the transformation to the entire 3D space, allowing the displacement vectors for all of the voxels in the volume to be computed. This design is motivated by the anatomical coupling between the vascular structures and lung parenchyma deformation, ensuring that clinically relevant motion patterns are faithfully captured across the lung.
3.5. The Training Objective
Our training objective combines supervised bi-directional flow losses with the unsupervised cycle consistency loss to promote accurate, coherent, and invertible transformations. By simultaneously supervising both the forward and backward displacement predictions at multiple resolutions and additionally enforcing consistency between these two directions, the network is guided towards robust and anatomically plausible lung registrations.
3.5.1. Bi-Directional Multi-Scale Flow Loss
At each resolution level
l, we supervise the predicted displacement fields by comparing them against their corresponding ground-truth flows. Specifically, we define a bi-directional loss that penalizes deviations from the ground truth in both the forward (source-to-target) and backward (target-to-source) directions:
where
and
denote the predicted forward and backward displacement functions, respectively, while
and
represent their ground-truth counterparts. This symmetric formulation provides balanced supervision, encouraging consistent flow estimation in both directions.
3.5.2. The Cycle Consistency Loss
To enhance the geometric plausibility and reversibility of the predicted transformations further, we introduce an unsupervised cycle consistency loss. Let and be the sets of predicted forward and backward displacement functions, respectively. Additionally, let and denote the transformation functions that deform a point cloud by applying the corresponding per-point displacements; for instance, .
The cycle consistency loss penalizes deviations after performing forward and backward transformations sequentially, promoting reversible mappings between point clouds:
where the Chamfer Distance (CD) between two point clouds
and
is defined as
This unsupervised regularization term ensures geometric coherence and encourages mutually consistent bi-directional flows.
3.5.3. The Final Training Objective
Our complete multi-scale training objective integrates these two losses across all resolution levels:
where the hyperparameter
controls the influence of the cycle consistency regularization and is empirically set to 0.1.
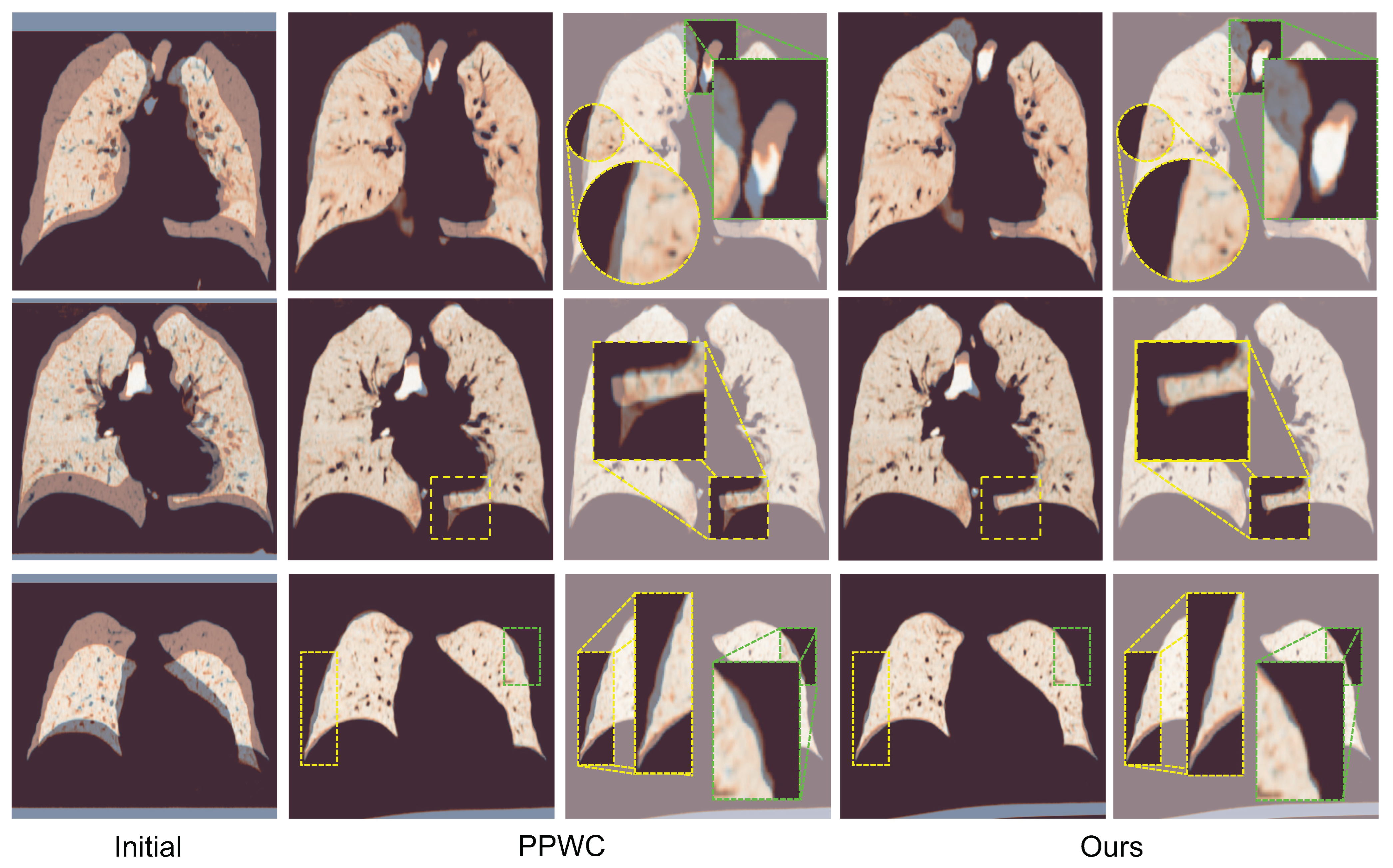
5. Discussion
Our results demonstrate that the proposed point-based deformable registration framework achieves a state-of-the-art performance on the Lung250M benchmark [
16]. By incorporating geometric keypoint attention at coarse resolutions, contextual flow refinement at finer scales, and bi-directional learning objectives, our method achieves a superior accuracy across all TRE metrics compared to that of the prior optimization-based and learning-based approaches. These findings validate our core hypothesis: that accurate lung CT registration requires joint modeling of the long-range structural consistency and localized deformation patterns, particularly given the large respiratory motion and anatomical variability inherent in this domain.
Compared to prior point-based methods primarily developed for rigid registration or generic scene flow tasks [
17,
18], our approach is the first to specifically target deformable lung CT alignment using sparse point clouds, building on the recent Lung250M dataset [
16]. While voxel-based learning methods have achieved notable success in brain MRI and abdominal CT registration, their application to thoracic imaging remains limited by the computational costs and sensitivity to variations in intensity. Our method addresses these limitations by leveraging sparse geometric inputs and multi-scale feature interactions, offering a more efficient and robust alternative.
Moreover, the use of bi-directional training with cycle consistency constraints across all resolution levels contributes to the deformation stability by enforcing multi-scale coherence in the flow predictions. This regularization, in combination with a coarse-to-fine refinement architecture, enhances the robustness to noisy or imperfect training correspondences. The empirical consistency of our model’s performance across datasets further suggests resilience to residual noise in the pseudo-ground-truth labels without requiring additional denoising mechanisms.
From a clinical standpoint, the proposed framework offers practical benefits. Its efficiency enables rapid deployment across large-scale datasets, while its anatomical interpretability—grounded in vascular keypoints—aligns with radiologists’ mental models for thoracic anatomy. Such properties suggest its promising utility in workflows such as longitudinal disease monitoring and dose accumulation analyses in radiotherapy, where consistent, reliable registration is critical.
Despite these advantages, our framework also has limitations. As it operates on vessel-derived point clouds, the registration accuracy in regions without clear geometric landmarks—such as homogeneous parenchymal areas—is less directly evaluated. In addition, our training supervision relies on the pseudo-ground-truth correspondences generated by corrField [
15]. Although these correspondences have demonstrated high reliability, with reported mean TREs of 1.45 mm (test) and 2.67 mm (validation) against manual landmark annotations [
16], a certain level of noise is inevitable. Our model mitigates the potential supervision noise through bi-directional cycle consistency regularization, but an explicit analysis of the robustness to correspondence noise remains an area for future investigation.
Future work could explore hybrid architectures that fuse the voxel-level appearance with the point-based geometry, potentially enhancing the deformation estimations in texture-rich but landmark-sparse regions. Incorporating semantic priors such as airway labels or fissure segmentations may improve the regional interpretability and clinical usability further. Additionally, injecting controlled perturbations into the training correspondences would help quantify the model’s sensitivity and reinforce its reliability. Expanding this framework to handle full 4D CT sequences or to quantify the registration uncertainty also presents promising future directions.
In conclusion, we present a novel attention-based, bi-directional point cloud registration framework specifically designed for deformable lung CT alignment. Through architectural innovations and rigorous evaluations on a challenging large-scale dataset, we show that point-based learning, when tailored to the characteristics of respiratory deformation, can offer an accurate, efficient, and anatomically consistent registration performance. Beyond algorithmic improvements, our framework also provides interpretable and efficient deformation estimates that align with clinically meaningful structures, making it a strong candidate for deployment in medical image analysis workflows. We believe this work provides an important step toward more robust geometric modeling in medical image analysis.