1. Introduction
Ultrasound is one of the most widely used diagnostic techniques due to its numerous advantages, including cost-effectiveness, safety (being radiation-free), and the ability to be performed in real time [
1]. Additionally, ultrasound demonstrates significant versatility, being applicable to almost any part of the body except for bones, lungs, and sections of the intestine. However, performing an ultrasound scan correctly necessitates substantial training and years of experience. This requirement creates a considerable workload for specialists and complicates the repeatability of diagnostic examinations. Consequently, the expertise and skill of the sonographer profoundly influences the performance of an examination. This leads to substantial inter-operator variability stemming from inconsistent probe positioning and acquisition angles, which can in turn impact the clarity and diagnostic consistency of echocardiographic images. Recent advancements in artificial intelligence techniques have enabled the exploration of both robotic and assisted ultrasound applications to address these critical issues [
2].
In the robotic scenario, a robotic arm performs ultrasound examinations under the guidance of artificial intelligence algorithms. These algorithms manage various functions, including planning the scanning path, adjusting the arm’s movement in space and the probe’s pressure on the scanned body area, and completing the scan. Robotic ultrasound systems can be classified into three categories based on their level of autonomy: teleoperated [
3], semi-autonomous [
4], and fully autonomous [
5]. Teleoperated systems involve robotic arms piloted by an operator, aiming to reduce or prevent musculoskeletal disorders in physicians and facilitate remote diagnosis. Semi-autonomous systems autonomously perform some tasks, such as positioning the probe in the body region of interest, but still require operator intervention to conduct the examination. Fully autonomous systems can independently plan and execute the ultrasound scan, acquiring the necessary images without sonographer intervention and allowing the physician to review the examination subsequently. By contrast, in the assisted ultrasound scenario a sonographer performs the examination while being guided by an algorithm in their movement of the probe.
Although robotic ultrasound remains primarily within the research domain, some applications of assisted ultrasound have already reached the market, including [
6], which validated an AI-based real-time guidance system enabling novice operators to obtain diagnostic-quality echocardiographic images with minimal training, illustrating the potential of assisted ultrasound solutions to broaden access and streamline workflow in clinical settings. In particular, echocardiography requires precise probe placement and immediate feedback to capture complex cardiac dynamics, making it especially poised to benefit from these innovations.
Notable advancements in the echocardiographic domain have employed deep learning algorithms to guide non-expert operators. To reduce inter-operator variability, the use of deep learning algorithms in echocardiographic examinations is proving promising for the selection of echocardiographic views, image quality control, and improved real-time probe guidance [
7]. Among deep learning algorithms, CNNs and their variants have shown excellent performance in recognizing and segmenting cardiac structures [
8]. The use of CNNs allows for the quantification of cardiac parameters and identification of pathological patterns with accuracy comparable to that of clinical experts [
9]. Models developed for the classification of cardiac views differ in the number of classifiable views, but have shown accuracy levels of up to 98% [
10] for five cardiac views [
11] and fifteen cardiac views [
12]. An important distinction must be made between classification performed on videos and on static images; because the heart is a moving organ, it is necessary to identify transitions between different views and consider changes in the image due to the opening and closing of the heart valves. Some studies have explored the use of recurrent neural networks (RNNs) and generative adversarial networks (GANs); the former can enable real-time image classification and provide guidance for correct probe alignment [
13,
14], while the latter have been used to adapt the quality of the ultrasound image to a user-defined level without the need for matched pairs of low- and high-quality images [
15]. Using a GAN, [
16] achieved accuracy greater than 90% in both classification and segmentation of cardiac views for the diagnosis of left ventricular hypertrophy. To improve performance, architectures that simultaneously use spatial and temporal information have recently been explored, including time-distributed and two-stream networks [
14].
Despite significant advances in algorithms, there are numerous critical issues that need to be addressed in the development of assisted ultrasound applications that can be easily used by operators. The most relevant aspect is the need to acquire, label, and archive large datasets [
9]. This activity should be carried out through multi-center studies and with different equipment, which is contrary to usual practice [
11]. In addition, the use of specific datasets can improve the accuracy of automated diagnosis and promote greater integration of artificial intelligence into clinical practice for patients with congenital and structural heart disease [
17]. Another significant limitation is poor image quality due to background noise and limited acoustic windows [
18] as well as the movement of anatomical structures, which requires considering the temporal evolution of the image [
12]. This aspect is linked to the poor interpretability of deep learning algorithms [
7,
8] and may leads to more cautious adoption of these technologies by healthcare professionals [
10]. The computing power required to train such algorithms is another aspect that must be properly assessed, as it is necessary to find a balance between image resolution and computational load [
16].
For assisted probe positioning, one of the most important issues is the tracking method [
13], which can be implemented with cameras or IMUs. The use of cameras can lead to privacy issues and inflexible setups, while tracking with a single IMU is complex due to signal noise that prevents the correct reconstruction of the trajectory. One method that has been explored to obtain a generic trajectory of an object using a single IMU is to break down the acquired trajectory into basic trajectories, then pair these with predefined geometric models [
19]. Another promising technique appears to be exploiting the continuous wavelet transform to correct drift and obtain a more accurate position estimate [
20]. A further approach involves error compensation to correct drift accumulation through a reset mechanism. However, these approaches cannot compensate for poor sensor quality or avoid the need for accurate initial calibration [
21].
In summary, existing solutions for assisted echocardiography often rely on large labeled datasets [
9,
11] or multiple sensors [
13,
19], and may still encounter issues with noise, drift, and interpretability, particularly when using single-IMU setups and AI-based image analysis [
7,
8]. While deep learning has achieved notable results in classifying echocardiographic views and monitoring image quality [
13,
14], practical barriers such as complex calibration steps [
20,
21], data scarcity [
9,
11], and computational overhead [
16] continue to persist.
The objective of the ongoing research activity reported in this paper is to develop a cognitive unit that enhances ultrasound imaging by assisting operators with real-time guidance and assessment. In this framework, ‘cognitive’ highlights the system’s planned ability to learn optimal probe manipulation by fusing real-time inertial sensor data with CNN-based analysis, ultimately providing adaptive and context-specific guidance to the operator. This unit aims to improve image quality and provide valuable feedback during the scanning process. Analysis of the proposed system’s recognition of elementary movements were carried out using the accelerometer signal. The integration of such a cognitive unit with commercial ultrasound scanners can potentially aid in reducing inter-operator variability, benefiting both trainees and general practitioners.
2. Materials and Methods
2.1. Hardware Setup
The selection of appropriate hardware for executing deep learning algorithms was guided by a thorough literature review focused on evaluating the performance of edge devices in artificial intelligence applications. Given the parallel development of software and the absence of specific algorithms to test, it was crucial to base hardware choices on established benchmarks and comparative analyses available in the literature.
The initial comparative analysis included devices commonly tested in image processing applications, such as the ASUS Tinker Edge R, Raspberry Pi 4, Google Coral Dev Board, NVIDIA Jetson Nano, and Arduino Nano 33 BLE. The evaluation considered inference speed and accuracy across various network models. Findings indicated that the Google Coral Dev Board demonstrated superior performance in continuous computational applications for models compatible with the TensorFlow Lite framework. The NVIDIA Jetson Nano ranked closely behind, offering greater versatility and the ability to train models using the onboard GPU [
22].
Further analysis narrowed the focus to GPU-equipped devices. The literature indicated that the NVIDIA Jetson Nano exhibited better image processing performance compared to the Jetson TX2, GTX 1060, and Tesla V100 in convolutional neural network applications [
23]. Within the NVIDIA Jetson series, the Jetson Orin Nano was identified as significantly outperforming both the Jetson Nano and Jetson AGX Xavier for video processing tasks using convolutional neural network models developed in PyTorch and optimized with NVIDIA’s Torch-TensorRT SDK [
24]. Based on these insights, the NVIDIA Jetson Orin Nano was selected for its robust performance.
Table 1 summarizes the main features of the NVIDIA Jetson Orin Nano.
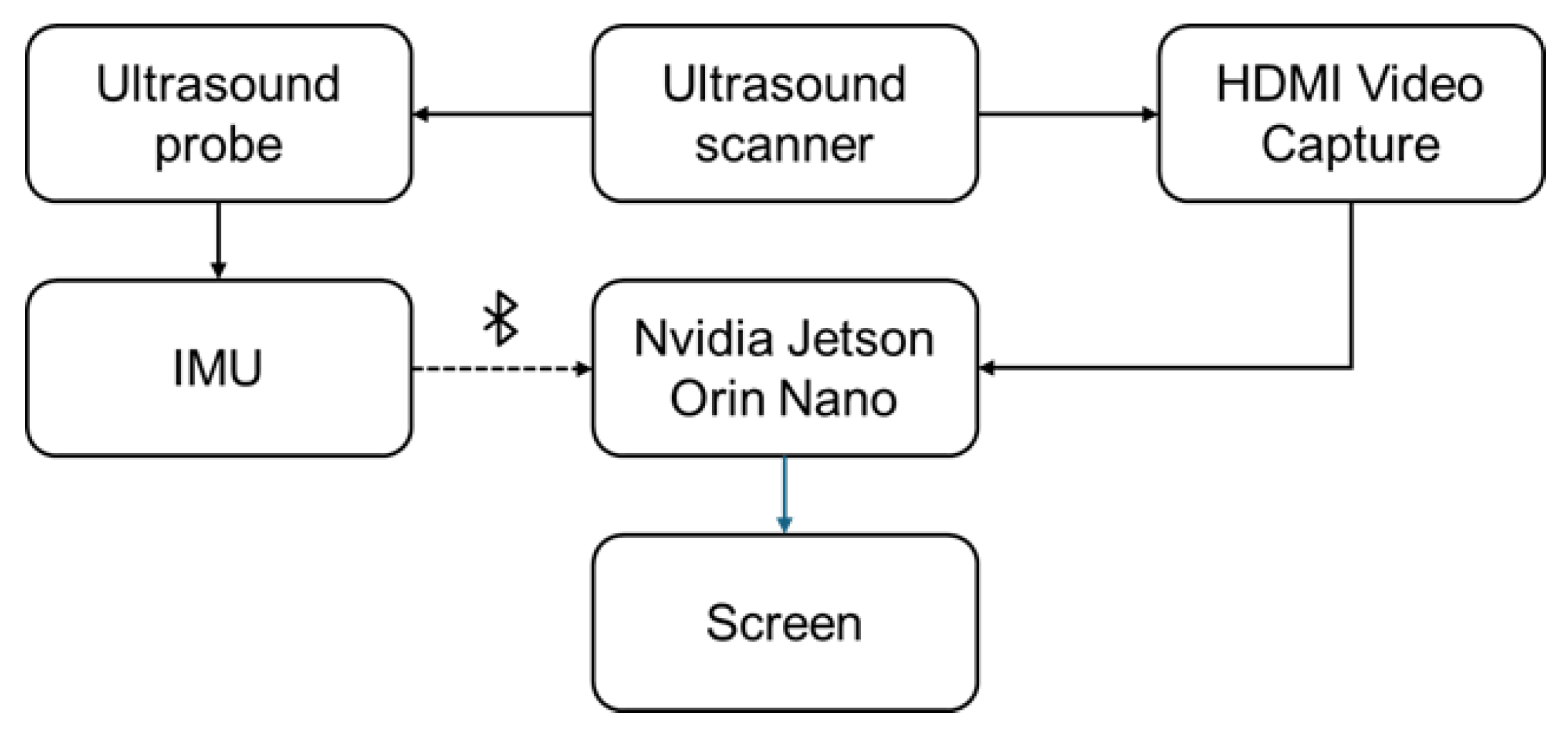
Figure 1 shows the experimental setup used for both ultrasound data collection and the inference phase. A small inertial measurement unit (IMU) was selected for attachment to the ultrasound probe in order to utilize the probe’s spatial position during inference.
After conducting tests on both online and offline ultrasound video processing through deep learning applications, we began to evaluate the performance of the hardware setup by developing a simple CNN model trained on the Camus dataset [
25], which includes echocardiographic sequences from 500 patients with varying image quality. Given the high variability in image acquisition and quality, the only preprocessing step we applied was resizing all images to 256 × 256 pixels to ensure input uniformity. This minimal preprocessing allows the model to remain robust across different input qualities, which is crucial for generalization in real-world clinical settings.
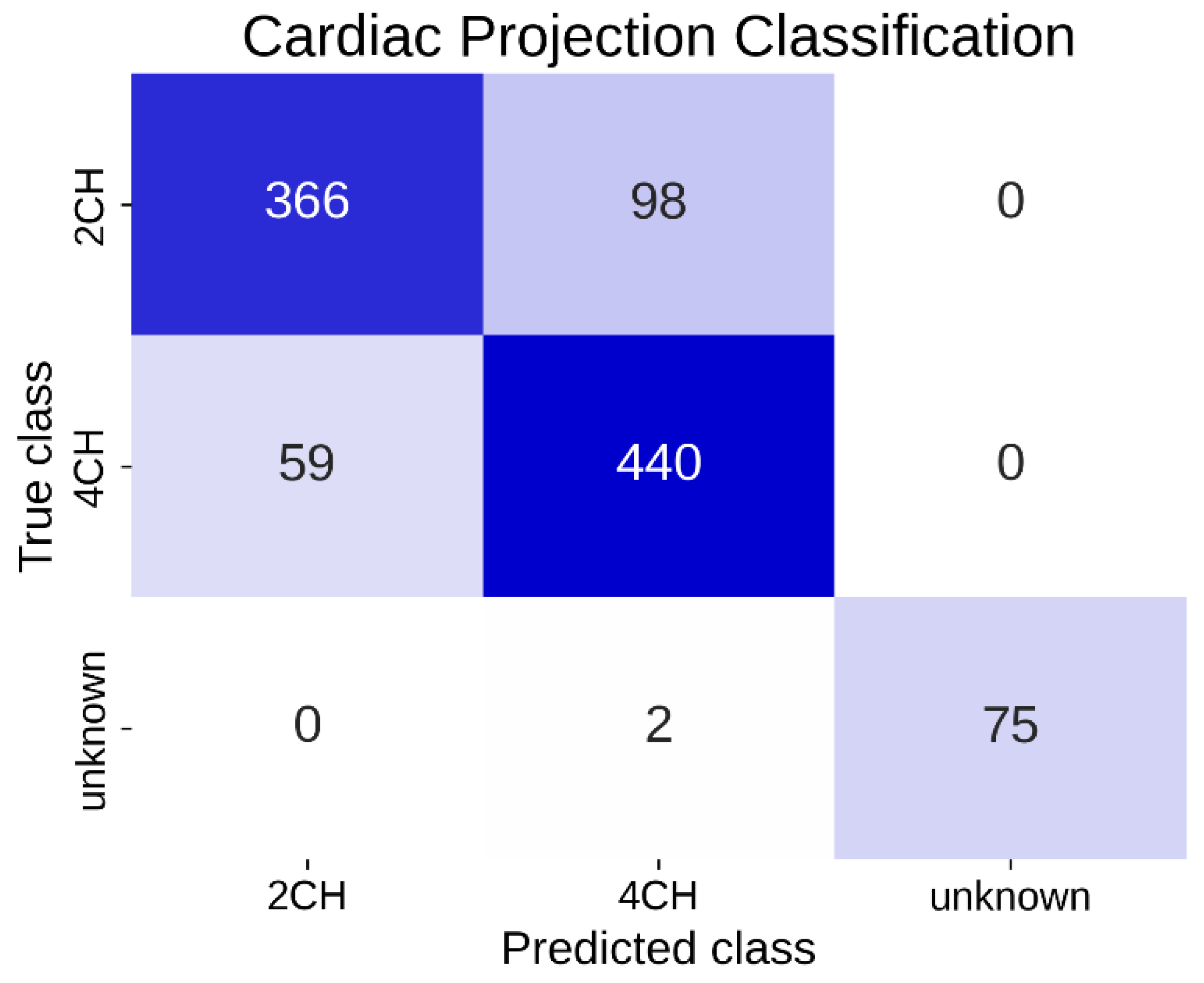
The neural network then classified echocardiographic images into “Apical projection with 2-chamber view (2CH)”, “Apical projection with 4-chamber view (4CH)”, and “Unknown”, with the latter including all non-classifiable images. This network was designed to perform classification of the observed cardiac projection automatically and in real time.
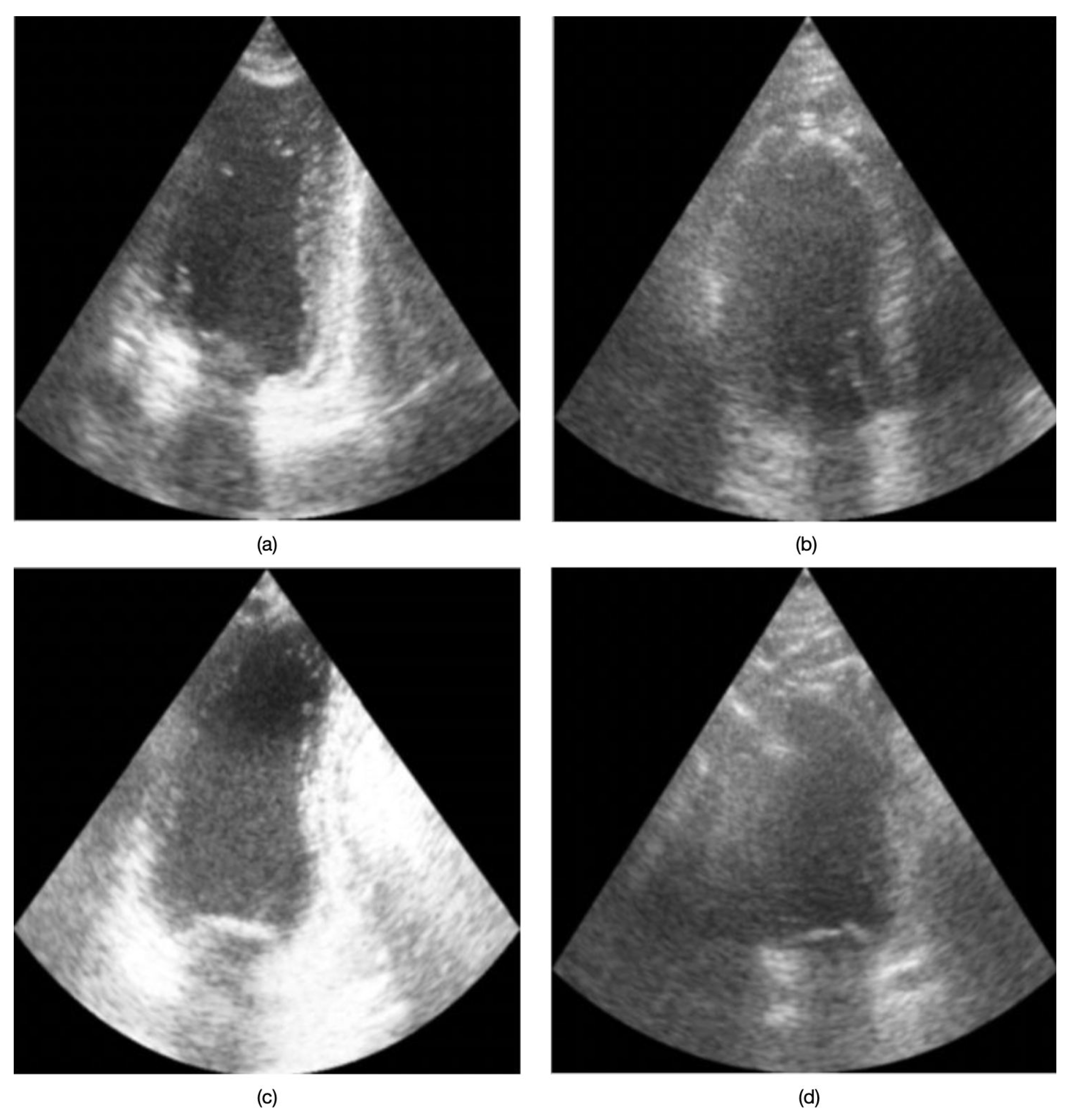
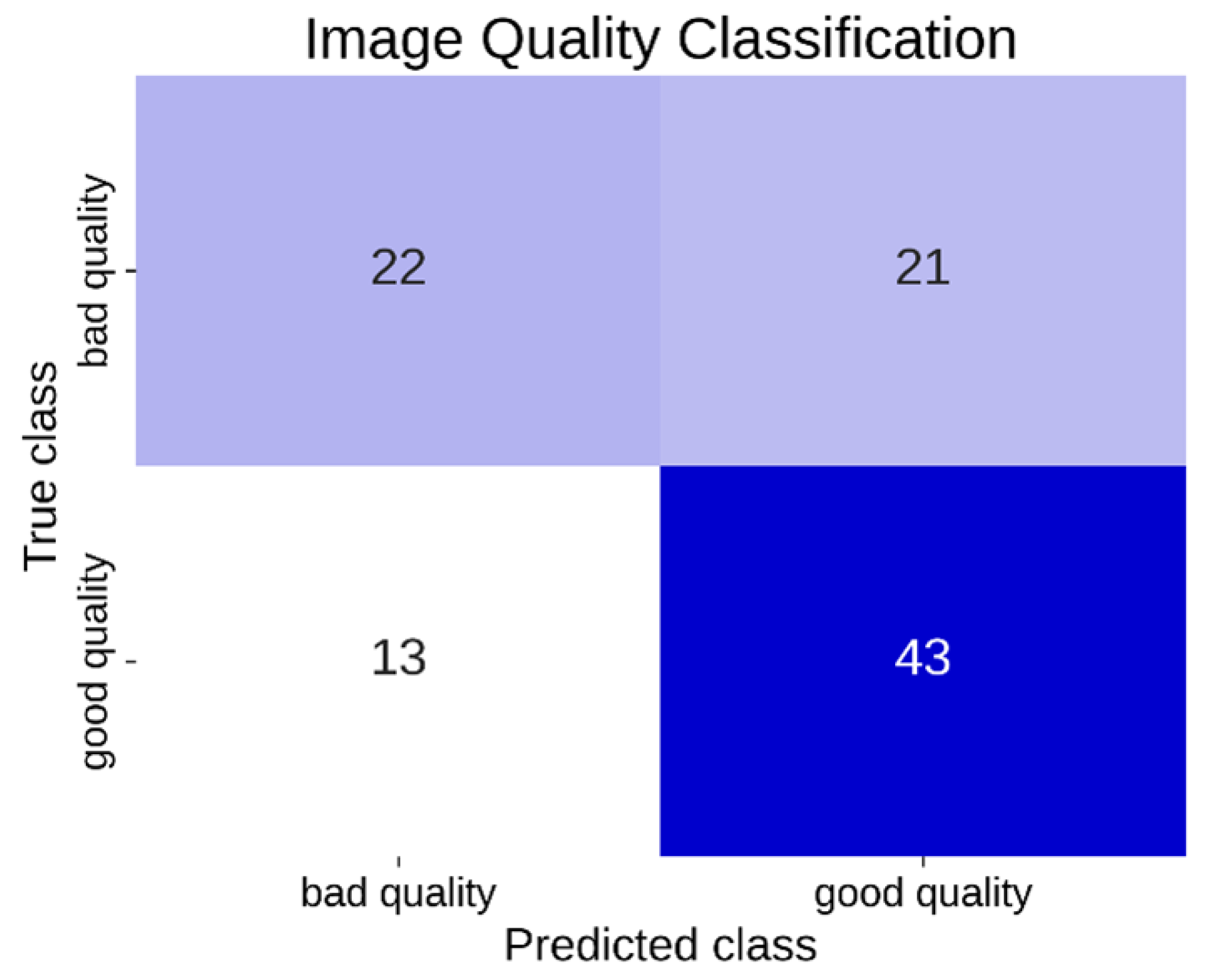
A second CNN was trained on the same dataset to perform binary classification of image quality, distinguishing between “good quality” and “bad/poor quality” for each frame. In this study, we define “good quality” images as those displaying clear anatomical structures, adequate contrast, and minimal artifacts, whereas “bad/poor quality” images exhibit significant noise, low contrast, or insufficient structural visibility.
Figure 2 presents example echocardiographic images acquired in both four-chamber (4CH) and two-chamber (2CH) views under varying quality conditions. No data from the inertial sensor were taken into account during the training and inference phases of the two models.
2.2. Neural Network Model and Data Capture
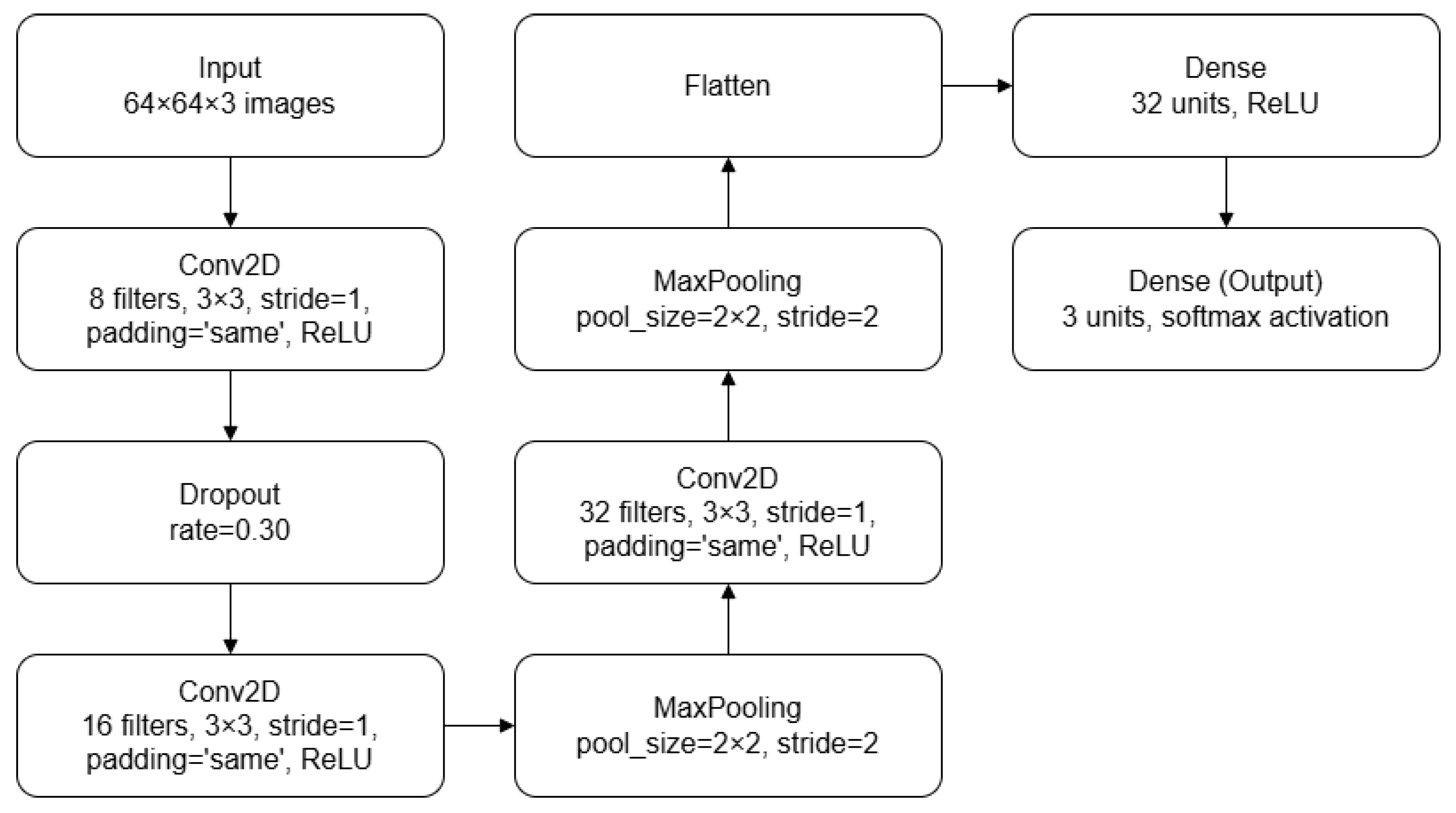
The CNN models used for cardiac projection classification (presented in
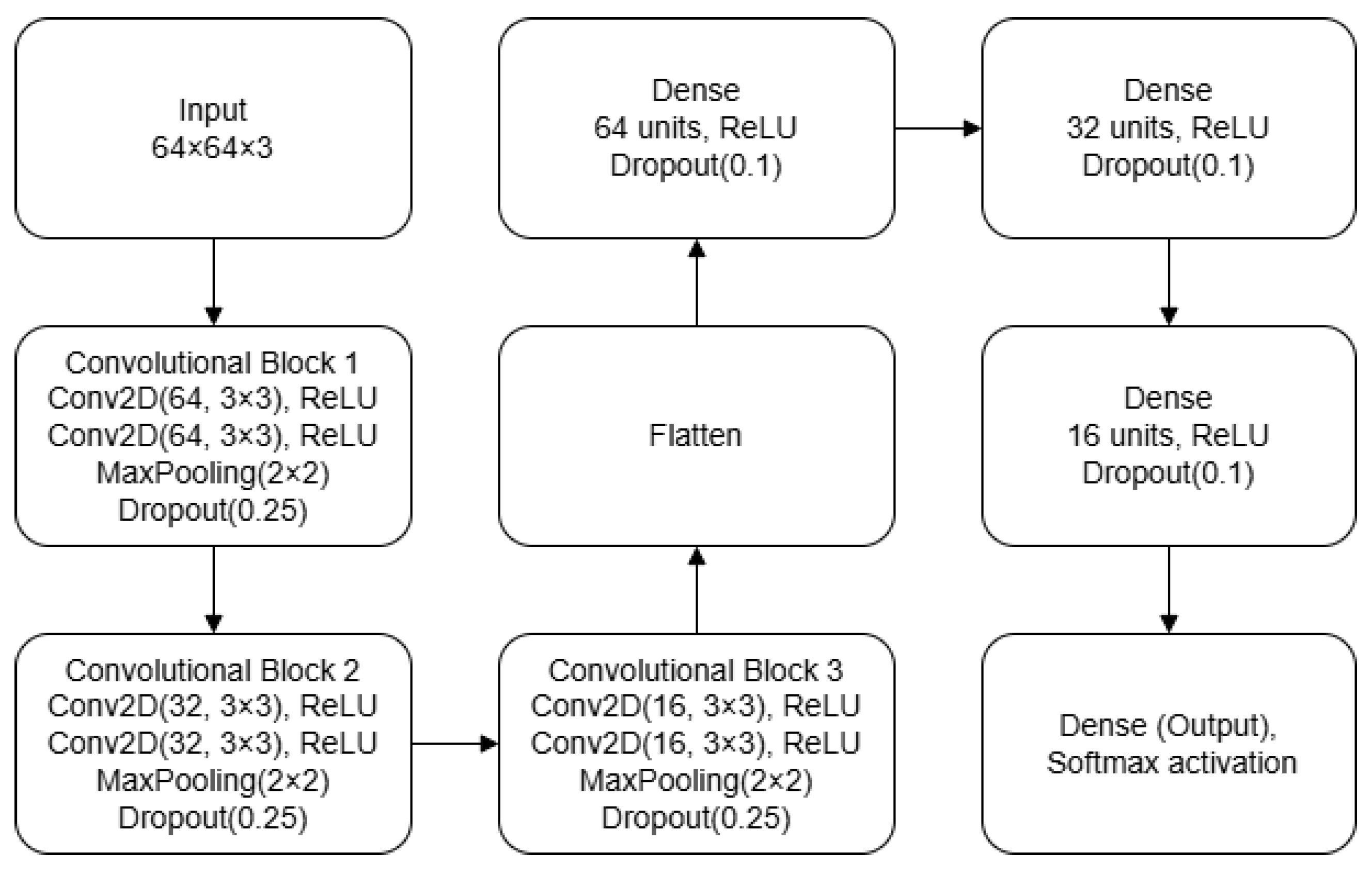
Figure 3) and quality classification (presented in
Figure 4) were both designed to balance computational complexity and generalization capacity, making them suitable for smaller datasets. The balance between computational complexity and generalization capacity was evaluated as a tradeoff between validation accuracy, inference time, and resource efficiency. The architecture comprises several layers, each of which performs specific operations to extract features and classify images. The original images were in NIfTI format and had different sizes; thus, they were converted into JPG format and resized to a resolution of
. This resolution was chosen as a compromise between preserving sufficient spatial information for classification and meeting the computational constraints of the deployment environment by keeping the inference computational cost lower. Each pixel is represented on 8 bits.
The first CNN model begins with a convolutional layer that applies eight filters of size
on each input image (
). This convolutional layer uses a stride of [1,1] and ‘same’ padding in order to maintain output dimensions equal to the input and preserve the spatial dimensions of the feature maps across convolutional layers. Following the convolution, a rectified linear unit (ReLU) activation function [
26] introduces nonlinearity into the model, which is essential for learning complex data relationships. To mitigate overfitting, a dropout layer with a 30% dropout rate is applied to randomly deactivate neurons during training. A dropout rate of 30% was selected through an iterative process in which different values were tested empirically. The second stage involves another convolutional layer, this time with 16 filters of size
, maintaining the same stride and padding settings. The ReLU activation function is used again to ensure nonlinearity. Subsequently, a max pooling layer with a pool size of [2,2] and stride of [2,2] reduces the spatial dimensions of the input while preserving the most significant features. This choice was based on its ability to highlight the strongest activations within each region, which is particularly effective in compact architectures.
In the third stage, the model includes a third convolutional layer with 32 filters of size , followed by a max pooling layer with an identical configuration to the previous one. This further reduces the spatial dimensions of the input, ensuring that the model focuses on the most prominent features. The output from the previous layers is then flattened into a one-dimensional vector to prepare it for the fully connected layers. The first fully connected layer consists of 32 units, with a ReLU activation function to introduce further nonlinearity. The final fully connected layer is the output layer. It comprises three units, corresponding to the number of classes in the classification problem. This layer uses a softmax activation function to assign probabilities to each class.
The second CNN model is designed for image quality classification. It begins with two consecutive convolutional layers, each with 64 filters of size , using a stride of [1,1] and ‘same’ padding. Each convolution is followed by a ReLU activation, a max pooling layer with stride [2,2], and a 25% dropout layer. The second stage repeats this structure but with 32 filters per convolutional layer, while the third stage uses 16 filters. The feature maps are then flattened and passed through three fully connected layers of 64, 32, and 16 units, each followed by a ReLU activation and a 10% dropout. The final layer is a softmax output sized to the number of classes. The network is compiled using the Adam optimizer and the categorical cross-entropy loss function
Our choice of simpler models such as those described above was motivated by the limited availability of training data. More complex models such as ResNet [
27] and vision transformer (ViT) [
28] are more prone to overfitting and require extensive computational resources, which is not ideal for scenarios with smaller datasets. The modular structure of simple CNN models allows for easy integration of additional components such as long short-term memory (LSTM) layers [
29], allowing for further enhancements if necessary. In a real-time acquisition scenario when the probe is held steady in the correct position, LSTM layers can capture the temporal dependencies between consecutive frames, effectively modeling the cardiac motion cycle and potentially improving the temporal coherence of predictions. This flexibility and efficiency makes such models well suited for the targeted application. It is also important to note that until now there has been no need to quantize the neural network model.
Due to strict patient privacy regulations, scarce physician availability for labeling, and the time-consuming nature of data annotation, data collection represents a critical aspect of artificial intelligence model development. Available datasets are often insufficient to meet research objectives, necessitating the acquisition of new data in collaboration with clinical professionals. To streamline this process, a comprehensive data capture unit was implemented. This unit is capable of recording ultrasound screens, capturing the spatial position of the probe, and receiving anonymized data from the ultrasound scanner. It also facilitates secure transmission of the acquired data for further analysis. The data capture unit incorporates components that automatically receive, process, and anonymize data from the ultrasound scanner. A dedicated graphical user interface (GUI) was developed to support the data collection process.
Despite the automation provided by this system, labeling the data requires significant back-office effort from clinical staff members. It has been established that each ultrasound examination requires not only the DICOM files but also the videos of each ultrasound examination. For each frame of these videos, the corresponding timestamps are saved, allowing the samples of the signals acquired from the accelerometer and gyroscope to be associated with each frame. The goal is to be able to associate the variations in image quality used to label each frame with the movements performed by the ultrasonographer.
4. Discussion
This study introduces a novel approach to improving echocardiographic imaging by combining advanced convolutional neural networks with inertial motion data to guide operators and optimize image quality in real time. Our approach is novel in that it operates on an embedded device not bound to a specific ultrasound system, allowing onboard inference without high-end servers and facilitating integration into diverse clinical workflows. Furthermore, to simplify the hardware requirements and mitigate privacy concerns, we investigate the use of a single inertial sensor for motion tracking rather than employing traditional camera-based setups. Our system achieved an overall classification accuracy of over 85% in differentiating apical 2CH, apical 4CH, and unrelated views. In addition, it achieved 66% accuracy in distinguishing good-quality vs. poor-quality images on a limited dataset. Notably, we observed an average inference time of ∼13 ms per frame, allowing for a live on-screen classification overlay without disrupting the clinical workflow.
Our system addresses two key tasks in ultrasound practice: accurate identification of clinically relevant cardiac views, and automated assessment of frame quality. Evaluating image quality helps to ensure uniform acquisition standards and minimizes the need for repeat exams, enabling second opinions and consultations without further patient exposure. Simultaneously, identifying clinically relevant cardiac views is essential for acquiring reproducible measurements; our system’s automated guidance can significantly assist novice operators in mastering this crucial step. By developing and training two specialized neural networks (a multiclass model for distinguishing apical 2CH, apical 4CH, and unrelated images along with a binary model for evaluating image clarity), our system achieves notable performance on relatively small datasets. These results emphasize the effectiveness of streamlined architectures, which can provide robust feature extraction and high inference speeds in resource-limited clinical environments.
The hardware foundation built around the NVIDIA Jetson Orin Nano proved capable of handling real-time data processing, maintaining stable performance metrics and modest power usage during continuous operation. When compared with existing approaches that often rely on large labeled datasets or multiple sensors, our system is designed to minimize hardware complexity and data requirements by integrating a single low-cost IMU sensor with lightweight CNN models. By conducting preliminary tests with a low-cost IMU, we also found that the system can detect and categorize basic unidirectional translational movements along a single axis, achieving preliminary accuracy of around 75–90% in a controlled setting utilizing a robotic arm. Although integration with our CNN-based image analysis has not yet been carried out, the ongoing research plan is to collect synchronized IMU and ultrasound data from real-world scanning, enabling a fused approach that offers comprehensive real-time guidance.
Collaboration with sonographers and a structured data collection initiative will be essential for further refinement, as large and diverse datasets will allow the system to accommodate a wide range of patient anatomies, ultrasound machines, and operator skill levels. The dedicated data acquisition setup captures anonymized video streams, IMU signals, and clinical metadata, paving the way for deeper analyses of how operator actions correlate with ultrasound images. In addition to standardizing the acquisition process, this integrated solution could significantly streamline training, reduce inter-operator variability, and enable more consistent echocardiographic examinations.
Looking ahead, the system’s design supports additional enhancements. Incorporating advanced drift compensation algorithms for the IMU along with interpretability techniques could allow clinicians to more transparently understand the network’s outputs. By guiding operators towards optimal probe positioning and providing immediate quality feedback, this approach aims to mitigate common errors in ultrasound scanning and improve patient care. Ultimately, these innovations hold promise not only for accelerating workflows but also for expanding the reach of expert-level echocardiographic imaging across diverse clinical settings.