Patient-Tailored Dementia Diagnosis with CNN-Based Brain MRI Classification
Abstract
Featured Application
Abstract
1. Introduction
1.1. Initial Considerations
1.2. Research Aims
- The primary objectives of this study were as follows:
- To demonstrate how CNN-based models can aid clinical decision-making for dementia patients;
- To utilize dementia-specific neuroimaging biomarkers to detect brain atrophy at the earliest possible stage and monitor disease progression from MCI to AD;
- To support the use of the DL tools for the development of personalized dementia care plans.
- The innovations of the proposed approach include the following:
- A lightweight 2D-CNN architecture That employs an optimized, low-complexity 2D-CNN model to balance diagnostic accuracy with reduced computational cost and memory usage;
- A single-slice classification strategy that implements a slice-wise classification approach using a single representative MRI slice per subject, minimizing data redundancy and model overfitting;
- An efficient training process that reduces training time significantly compared with volumetric 3D models, facilitating rapid experimentation and model adjustment;
- Robust performance on limited data, achieving high diagnostic accuracy despite limited labeled data, demonstrating strong generalization capabilities;
- Potential for real-time use, whereby the low computational burden and fast inference time make the model suitable for integration into real-time clinical research frameworks.
2. Materials and Methods
2.1. Data Collection and Preliminary Analysis
2.1.1. Data Sources
- AD—individuals diagnosed with AD and exhibiting typical signs and symptoms;
- CN—control group of CN subjects exhibiting no signs of dementia;
- MCI—individuals with subjective cognitive impairment of varying severity but without any other typical signs of dementia, whose daily activities remained mostly unaffected.
2.1.2. Data Preprocessing and Selection
2.2. CNN-Based Models for Diagnostic Support of Dementia
2.2.1. Used Tools and Software
2.2.2. Independently Developed CNN-Based Models
2.2.3. CNN-Based Models TL
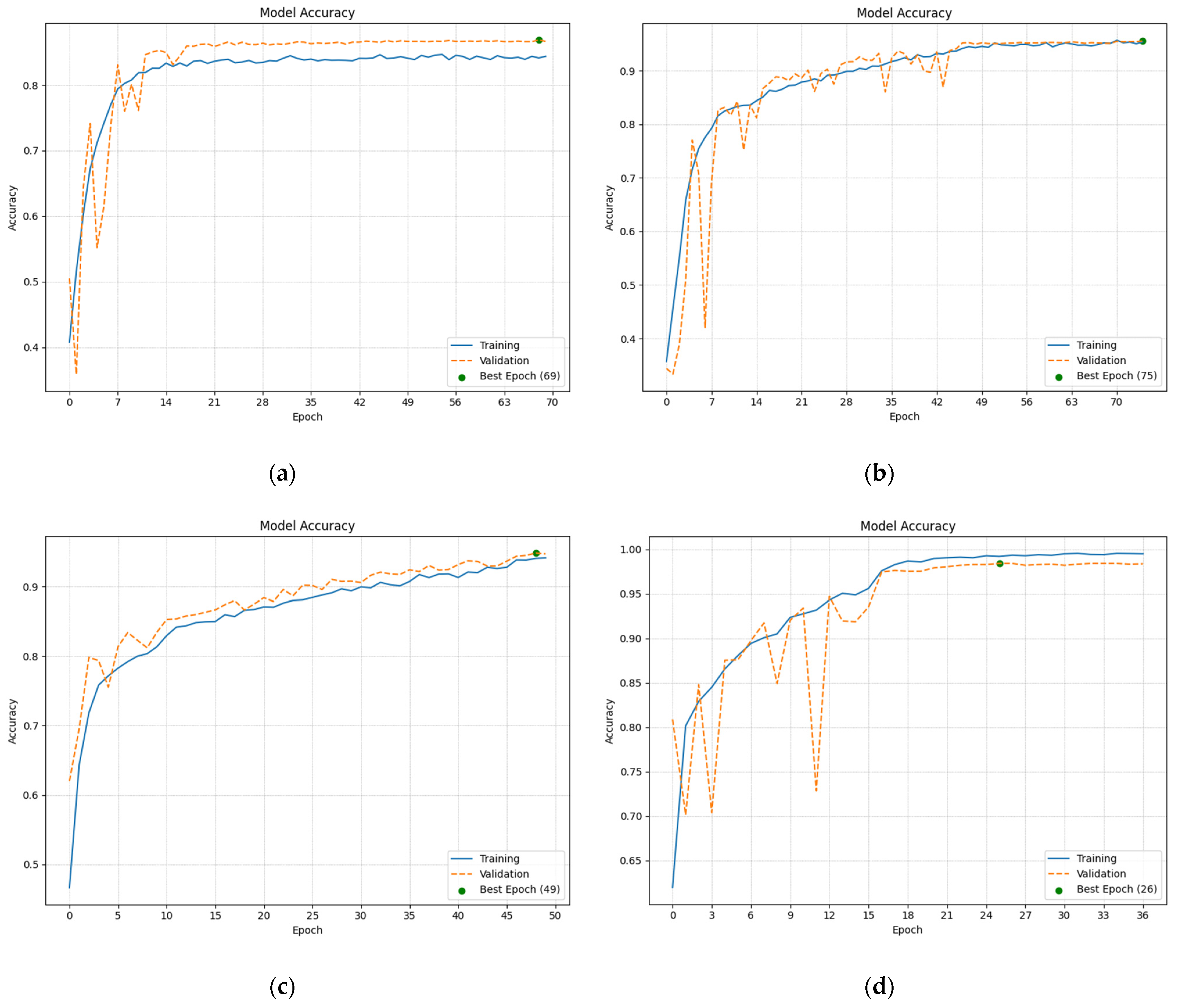
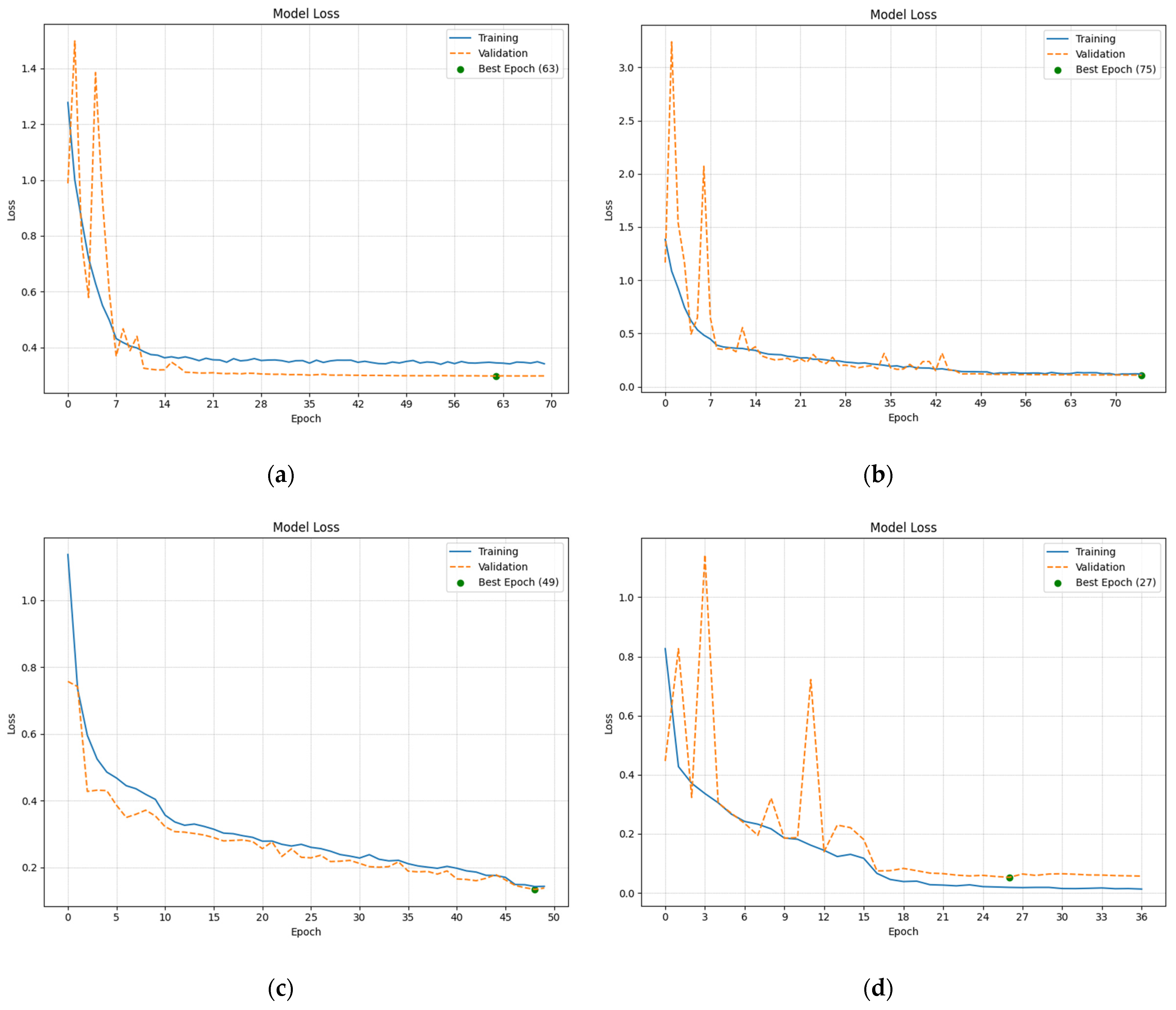
2.2.4. Analysis of the Training and Validation
3. Results
3.1. Evaluation of the Diagnostic Ability of the CNN-Based Models
3.2. Proposed CDSS for Dementia Grad-CAM
4. Discussion
4.1. Study Limitations
4.2. Added Value
4.3. Future Development Potential
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- What Is Dementia? Symptoms, Types, and Diagnosis. Available online: https://www.nia.nih.gov/health/alzheimers-and-dementia/what-dementia-symptoms-types-and-diagnosis (accessed on 5 March 2025).
- Dementia. Available online: https://www.who.int/news-room/fact-sheets/detail/dementia (accessed on 5 March 2025).
- How Is Alzheimer’s Disease Treated? Available online: https://www.nia.nih.gov/health/alzheimers-treatment/how-alzheimers-disease-treated (accessed on 5 March 2025).
- What Is Dementia? Available online: https://www.alz.org/alzheimers-dementia/what-is-dementia (accessed on 5 March 2025).
- Coupé, P.; Manjón, J.V.; Lanuza, E.; Catheline, G. Lifespan Changes of the Human Brain in Alzheimer’s Disease. Sci. Rep. 2019, 9, 3998. [Google Scholar] [CrossRef] [PubMed]
- Dementia. Symptoms & Causes. Available online: https://www.mayoclinic.org/diseases-conditions/dementia/symptoms-causes/syc-20352013 (accessed on 5 March 2025).
- Preclinical, Prodromal, and Dementia Stages of Alzheimer’s Disease. Available online: https://practicalneurology.com/articles/2019-june/preclinical-prodromal-and-dementia-stages-ofalzheimers-disease (accessed on 5 March 2025).
- Chen, Y.; Qi, Y.; Hu, Y.; Qiu, X.; Qiu, T.; Li, S.; Liu, M.; Jia, Q.; Sun, B.; Liu, C.; et al. Integrated Cerebellar Radiomic-network Model for Predicting Mild Cognitive Impairment in Alzheimer’s Disease. Alzheimer’s Dement. 2025, 21, e14361. [Google Scholar] [CrossRef]
- Oh, K.; Heo, D.-W.; Mulyadi, A.W.; Jung, W.; Kang, E.; Lee, K.H.; Suk, H.-I. A Quantitatively Interpretable Model for Alzheimer’s Disease Prediction Using Deep Counterfactuals. NeuroImage 2025, 309, 121077. [Google Scholar] [CrossRef]
- Blanco, K.; Salcidua, S.; Orellana, P.; Sauma-Pérez, T.; León, T.; Steinmetz, L.C.L.; Ibañez, A.; Duran-Aniotz, C.; De La Cruz, R. Systematic Review: Fluid Biomarkers and Machine Learning Methods to Improve the Diagnosis from Mild Cognitive Impairment to Alzheimer’s Disease. Alzheimer’s Res. Ther. 2023, 15, 176. [Google Scholar] [CrossRef]
- Yoon, J.M.; Lim, C.Y.; Noh, H.; Nam, S.W.; Jun, S.Y.; Kim, M.J.; Song, M.Y.; Jang, H.; Kim, H.J.; Seo, S.W.; et al. Enhancing Foveal Avascular Zone Analysis for Alzheimer’s Diagnosis with AI Segmentation and Machine Learning Using Multiple Radiomic Features. Sci. Rep. 2024, 14, 1841. [Google Scholar] [CrossRef] [PubMed]
- Jytzler, J.A.; Lysdahlgaard, S. Radiomics Evaluation for the Early Detection of Alzheimer’s Dementia Using T1-Weighted MRI. Radiography 2024, 30, 1427–1433. [Google Scholar] [CrossRef]
- Schwarz, C.G.; Gunter, J.L.; Wiste, H.J.; Przybelski, S.A.; Weigand, S.D.; Ward, C.P.; Senjem, M.L.; Vemuri, P.; Murray, M.E.; Dickson, D.W.; et al. A Large-Scale Comparison of Cortical Thickness and Volume Methods for Measuring Alzheimer’s Disease Severity. NeuroImage Clin. 2016, 11, 802–812. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Pardoe, H.; Lichter, R.; Werden, E.; Raffelt, A.; Cumming, T.; Brodtmann, A. Cortical Thickness Estimation in Longitudinal Stroke Studies: A Comparison of 3 Measurement Methods. NeuroImage Clin. 2015, 8, 526–535. [Google Scholar] [CrossRef]
- Upadhyay, P.; Tomar, P.; Yadav, S.P. Advancements in Alzheimer’s Disease Classification Using Deep Learning Frameworks for Multimodal Neuroimaging: A Comprehensive Review. Comput. Electr. Eng. 2024, 120, 109796. [Google Scholar] [CrossRef]
- Wang, F.; Liang, Y.; Wang, Q.-W. Interpretable Machine Learning-Driven Biomarker Identification and Validation for Alzheimer’s Disease. Sci. Rep. 2024, 14, 30770. [Google Scholar] [CrossRef]
- Zhao, K.; Ding, Y.; Han, Y.; Fan, Y.; Alexander-Bloch, A.F.; Han, T.; Jin, D.; Liu, B.; Lu, J.; Song, C.; et al. Independent and Reproducible Hippocampal Radiomic Biomarkers for Multisite Alzheimer’s Disease: Diagnosis, Longitudinal Progress and Biological Basis. Sci. Bull. 2020, 65, 1103–1113. [Google Scholar] [CrossRef] [PubMed]
- Winchester, L.M.; Harshfield, E.L.; Shi, L.; Badhwar, A.; Khleifat, A.A.; Clarke, N.; Dehsarvi, A.; Lengyel, I.; Lourida, I.; Madan, C.R.; et al. Artificial Intelligence for Biomarker Discovery in Alzheimer’s Disease and Dementia. Alzheimer’s Dement. 2023, 19, 5860–5871. [Google Scholar] [CrossRef]
- Shi, M.; Feng, X.; Zhi, H.; Hou, L.; Feng, D. Machine Learning-based Radiomics in Neurodegenerative and Cerebrovascular Disease. MedComm 2024, 5, e778. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Huang, Y.; Zhang, X.; Yang, Q.; Guo, Y.; Xia, Y.; Peng, C.; Li, C. Research and Application Progress of Radiomics in Neurodegenerative Diseases. Meta-Radiology 2024, 2, 100068. [Google Scholar] [CrossRef]
- Peng, D.; Huang, W.; Liu, R.; Zhong, W. From Pixels to Prognosis: Radiomics and AI in Alzheimer’s Disease Management. Front. Neurol. 2025, 16, 1536463. [Google Scholar] [CrossRef]
- Shih, D.-H.; Wu, Y.-H.; Wu, T.-W.; Wang, Y.-K.; Shih, M.-H. Classifying Dementia Severity Using MRI Radiomics Analysis of the Hippocampus and Machine Learning. IEEE Access 2024, 12, 160030–160051. [Google Scholar] [CrossRef]
- Boeken, T.; Feydy, J.; Lecler, A.; Soyer, P.; Feydy, A.; Barat, M.; Duron, L. Artificial Intelligence in Diagnostic and Interventional Radiology: Where Are We Now? Diagn. Interv. Imaging 2023, 104, 1–5. [Google Scholar] [CrossRef]
- ur Rahman, J.; Hanif, M.; ur Rehman, O.; Haider, U.; Mian Qaisar, S.; Pławiak, P. Stages Prediction of Alzheimer’s Disease with Shallow 2D and 3D CNNs from Intelligently Selected Neuroimaging Data. Sci. Rep. 2025, 15, 9238. [Google Scholar] [CrossRef]
- Ali, M.U.; Kim, K.S.; Khalid, M.; Farrash, M.; Zafar, A.; Lee, S.W. Enhancing Alzheimer’s Disease Diagnosis and Staging: A Multistage CNN Framework Using MRI. Front. Psychiatry 2024, 15, 1395563. [Google Scholar] [CrossRef]
- Tripathy, S.K.; Nayak, R.K.; Gadupa, K.S.; Mishra, R.D.; Patel, A.K.; Satapathy, S.K.; Bhoi, A.K.; Barsocchi, P. Alzheimer’s Disease Detection via Multiscale Feature Modelling Using Improved Spatial Attention Guided Depth Separable CNN. Int. J. Comput. Intell. Syst. 2024, 17, 113. [Google Scholar] [CrossRef]
- Hussain, M.Z.; Shahzad, T.; Mehmood, S.; Akram, K.; Khan, M.A.; Tariq, M.U.; Ahmed, A. A Fine-Tuned Convolutional Neural Network Model for Accurate Alzheimer’s Disease Classification. Sci. Rep. 2025, 15, 11616. [Google Scholar] [CrossRef]
- Illakiya, T.; Ramamurthy, K.; Siddharth, M.V.; Mishra, R.; Udainiya, A. AHANet: Adaptive Hybrid Attention Network for Alzheimer’s Disease Classification Using Brain Magnetic Resonance Imaging. Bioengineering 2023, 10, 714. [Google Scholar] [CrossRef]
- Zhang, Y.; He, X.; Liu, Y.; Ong, C.Z.L.; Liu, Y.; Teng, Q. An End-to-End Multimodal 3D CNN Framework with Multi-Level Features for the Prediction of Mild Cognitive Impairment. Knowl.-Based Syst. 2023, 281, 111064. [Google Scholar] [CrossRef]
- Muksimova, S.; Umirzakova, S.; Iskhakova, N.; Khaitov, A.; Cho, Y.I. Advanced Convolutional Neural Network with Attention Mechanism for Alzheimer’s Disease Classification Using MRI. Comput. Biol. Med. 2025, 190, 110095. [Google Scholar] [CrossRef]
- Kang, W.; Lin, L.; Sun, S.; Wu, S. Three-Round Learning Strategy Based on 3D Deep Convolutional GANs for Alzheimer’s Disease Staging. Sci. Rep. 2023, 13, 5750. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Liu, S.; Cai, W.; Pujol, S.; Kikinis, R.; Feng, D. Early Diagnosis of Alzheimer’s Disease with Deep Learning. In Proceedings of the 2014 IEEE 11th International Symposium on Biomedical Imaging (ISBI), Beijing, China, 29 April–2 May 2014; pp. 1015–1018. [Google Scholar]
- Lian, C.; Liu, M.; Pan, Y.; Shen, D. Attention-Guided Hybrid Network for Dementia Diagnosis with Structural MR Images. IEEE Trans. Cybern. 2022, 52, 1992–2003. [Google Scholar] [CrossRef] [PubMed]
- Poloni, K.M.; Ferrari, R.J. Automated Detection, Selection and Classification of Hippocampal Landmark Points for the Diagnosis of Alzheimer’s Disease. Comput. Methods Programs Biomed. 2022, 214, 106581. [Google Scholar] [CrossRef]
- Cui, R.; Liu, M. Hippocampus Analysis by Combination of 3-D DenseNet and Shapes for Alzheimer’s Disease Diagnosis. IEEE J. Biomed. Health Inform. 2019, 23, 2099–2107. [Google Scholar] [CrossRef]
- Chen, Y.; Xia, Y. Iterative Sparse and Deep Learning for Accurate Diagnosis of Alzheimer’s Disease. Pattern Recognit. 2021, 116, 107944. [Google Scholar] [CrossRef]
- About ADNI. Available online: https://adni.loni.usc.edu/about/ (accessed on 5 March 2025).
- FreeSurferWiki. Available online: https://surfer.nmr.mgh.harvard.edu/fswiki/FreeSurferWiki (accessed on 5 March 2025).
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar] [CrossRef]
- Chollet, F. Xception: Deep Learning with Depthwise Separable Convolutions. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 1800–1807. [Google Scholar] [CrossRef]
- Howard, A.G.; Zhu, M.; Chen, B.; Kalenichenko, D.; Wang, W.; Weyand, T.; Andreetto, M.; Adam, H. MobileNets: Efficient Convolutional Neural Networks for Mobile Vision Applications. arXiv 2017. [Google Scholar] [CrossRef]
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. arXiv 2015. [Google Scholar] [CrossRef]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-CAM: Visual Explanations from Deep Networks via Gradient-Based Localization. Int. J. Comput. Vis. 2020, 128, 336–359. [Google Scholar] [CrossRef]
- Alonso-Fernandez, F.; Hernandez-Diaz, K.; Buades, J.M.; Tiwari, P.; Bigun, J. An Explainable Model-Agnostic Algorithm for CNN-Based Biometrics Verification. In Proceedings of the IEEE International Workshop on Information Forensics and Security (WIFS), Nürnberg, Germany, 4–7 December 2023; pp. 1–6. [Google Scholar] [CrossRef]
- Lundberg, S.M.; Lee, S.-I. A Unified Approach to Interpreting Model Predictions. In Proceedings of the 31st Conference on Neural Information Processing Systems (NIPS 2017), Long Beach, CA, USA, 4–9 December 2017; Available online: https://proceedings.neurips.cc/paper_files/paper/2017/file/8a20a8621978632d76c43dfd28b67767-Paper.pdf (accessed on 20 April 2025).
- Zheng, Q.; Wang, Z.; Zhou, J.; Lu, J. Shap-CAM: Visual Explanations for Convolutional Neural Networks Based on Shapley Value. In Proceedings of the Computer Vision—ECCV 2022; Avidan, S., Brostow, G., Cissé, M., Farinella, G.M., Hassner, T., Eds.; Springer Nature: Cham, Switzerland, 2022; pp. 459–474. [Google Scholar] [CrossRef]
- Mattson, E. Decoding AI Decisions: Interpreting MNIST CNN Models Using LIME. PureAI. 2023. Available online: https://open.substack.com/pub/pureai/p/decoding-ai-decisions-using-lime?utm_campaign=post&utm_medium=web (accessed on 13 April 2025).
- Taneja, A. How SHAP Represent CNN Predictions. Medium 2024. Available online: https://medium.com/@ataneja.itprof/how-shap-represent-cnn-predictions-8a5a730d98c0 (accessed on 13 April 2025).
- Bernasconi, A. CHAPTER 8—Structural Analysis Applied to Epilepsy. In Magnetic Resonance in Epilepsy, 2nd ed.; Kuzniecky, R.I., Jackson, G.D., Eds.; Academic Press: Burlington, NJ, USA, 2005; pp. 249–269. ISBN 978-0-12-431152-7. [Google Scholar]
- Zoons, E.; Booij, J.; Nederveen, A.J.; Dijk, J.M.; Tijssen, M.A.J. Structural, Functional and Molecular Imaging of the Brain in Primary Focal Dystonia—A Review. NeuroImage 2011, 56, 1011–1020. [Google Scholar] [CrossRef]
- Whitwell, J.L. Voxel-Based Morphometry: An Automated Technique for Assessing Structural Changes in the Brain. J. Neurosci. 2009, 29, 9661–9664. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Wu, R.; Zeng, Y.; Qi, Z.; Ferraro, S.; Xu, L.; Zheng, X.; Li, J.; Fu, M.; Yao, S.; et al. Choice of Voxel-Based Morphometry Processing Pipeline Drives Variability in the Location of Neuroanatomical Brain Markers. Commun. Biol. 2022, 5, 913. [Google Scholar] [CrossRef]
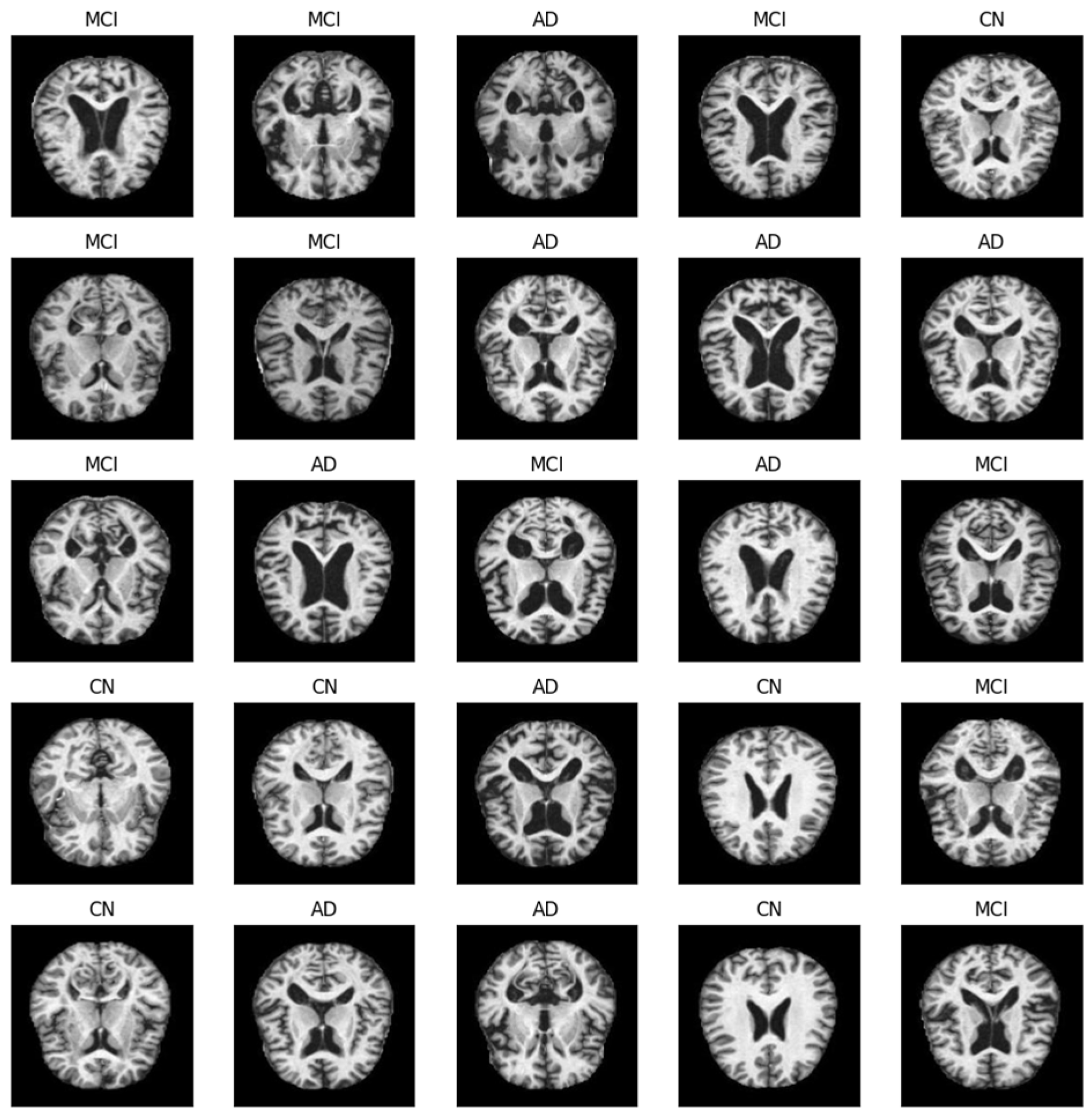




| Age | Gender | Research Group | |||
|---|---|---|---|---|---|
| Female | Male | CN | MCI | AD | |
| 40–49 | 1 | 0 | 1 | 0 | 0 |
| 50–59 | 78 | 32 | 54 | 30 | 26 |
| 60–69 | 327 | 250 | 302 | 187 | 88 |
| 70–79 | 478 | 563 | 440 | 377 | 224 |
| 80–89 | 154 | 237 | 116 | 150 | 125 |
| Above 89 | 13 | 12 | 6 | 7 | 12 |
| Model | No. of Conv2D Layers | No. of Filters in the Last Conv2D Layer | General Characteristics |
|---|---|---|---|
| Custom CNN 128 | 6 | 128 | Total params: 2,952,099 (11.26 MB) Trainable params: 2,951,075 (11.26 MB) Non-trainable params: 1024 (4.00 KB) |
| Custom CNN 256 | 8 | 256 | Total params: 10,646,307 (40.61 MB) Trainable params: 10,644,643 (40.61 MB) Non-trainable params: 1664 (6.50 KB) |
| Custom CNN 512 | 10 | 512 | Total params: 14,288,931 (54.51 MB) Trainable params: 14,285,475 (54.49 MB) Non-trainable params: 3456 (13.50 KB) |
| Custom CNN 1024 | 12 | 1024 | Total params: 28,848,675 (110.05 MB) Trainable params: 28,841,635 (110.02 MB) Non-trainable params: 7040 (27.50 KB) |
| Model | No. of Layers with Parameters (Depth) | General Characteristics |
|---|---|---|
| ResNet50-based | 107 | Total params: 26,079,747 (99.49 MB) Trainable params: 23,328,643 (88.99 MB) Non-trainable params: 2,751,104 (10.49 MB) |
| Xception-based | 81 | Total params: 23,353,515 (89.09 MB) Trainable params: 17,350,043 (66.19 MB) Non-trainable params: 6,003,472 (22.90 MB) |
| MobileNet-based | 55 | Total params: 4,541,251 (17.32 MB) Trainable params: 4,249,987 (16.21 MB) Non-trainable params: 291,264 (1.11 MB) |
| VGG16-based | 16 | Total params: 15,437,251 (58.89 MB) Trainable params: 13,701,507 (52.27 MB) Non-trainable params: 1,735,744 (6.62 MB) |
| Classification Quality Metrics | |||||||
|---|---|---|---|---|---|---|---|
| Model | Class | Precision | Recall | F1 Score | ACC | AUC | MCC |
| Custom CNN 128 | AD | 100% | 100% | 100% | 100% | 1 | 73.27% |
| CN | 68% | 85% | 76% | 85% | 0.83 | ||
| MCI | 80% | 60% | 69% | 60% | 0.76 | ||
| Average | 83% | 82% | 81% | 81.67% | 0.86 | ||
| Custom CNN 256 | AD | 100% | 100% | 100% | 100% | 1.00 | 88.33% |
| CN | 77% | 92% | 84% | 91.7% | 0.89 | ||
| MCI | 90% | 73% | 81% | 73.3% | 0.85 | ||
| Average | 89% | 88% | 88% | 88.33% | 0.91 | ||
| Custom CNN 512 | AD | 100% | 100% | 100% | 100% | 1 | 90.27% |
| CN | 85% | 97% | 91% | 96.7% | 0.94 | ||
| MCI | 96% | 83% | 89% | 83.3% | 0.91 | ||
| Average | 94% | 93% | 93% | 93.33% | 0.95 | ||
| Custom CNN 1024 | AD | 100% | 100% | 100% | 100% | 1 | 93.61% |
| CN | 88% | 100% | 94% | 100% | 0.97 | ||
| MCI | 100% | 87% | 93% | 86.7% | 0.93 | ||
| Average | 96% | 96% | 96% | 95.56% | 0.97 | ||
| ResNet50-based | AD | 100% | 100% | 100% | 100% | 1 | 91.73% |
| CN | 89% | 95% | 92% | 95% | 0.95 | ||
| MCI | 95% | 88% | 91% | 88.3% | 0.93 | ||
| Average | 95% | 94% | 94% | 94.44% | 0.96 | ||
| Xception-based | AD | 100% | 100% | 100% | 100% | 1 | 96.74% |
| CN | 94% | 100% | 97% | 100% | 0.98 | ||
| MCI | 100% | 93% | 97% | 93.3% | 0.97 | ||
| Average | 98% | 98% | 98% | 97.78% | 0.98 | ||
| MobileNet-based | AD | 100% | 100% | 100% | 100% | 1 | 97.54% |
| CN | 95% | 100% | 98% | 100% | 0.99 | ||
| MCI | 100% | 95% | 97% | 95% | 0.97 | ||
| Average | 98% | 98% | 98% | 98.33% | 0.99 | ||
| VGG16-based | AD | 100% | 100% | 100% | 100% | 1 | 100% |
| CN | 100% | 100% | 100% | 100% | 1 | ||
| MCI | 100% | 100% | 100% | 100% | 1 | ||
| Average | 100% | 100% | 100% | 100% | 1 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Knapińska, Z.; Mulawka, J. Patient-Tailored Dementia Diagnosis with CNN-Based Brain MRI Classification. Appl. Sci. 2025, 15, 4652. https://doi.org/10.3390/app15094652
Knapińska Z, Mulawka J. Patient-Tailored Dementia Diagnosis with CNN-Based Brain MRI Classification. Applied Sciences. 2025; 15(9):4652. https://doi.org/10.3390/app15094652
Chicago/Turabian StyleKnapińska, Zofia, and Jan Mulawka. 2025. "Patient-Tailored Dementia Diagnosis with CNN-Based Brain MRI Classification" Applied Sciences 15, no. 9: 4652. https://doi.org/10.3390/app15094652
APA StyleKnapińska, Z., & Mulawka, J. (2025). Patient-Tailored Dementia Diagnosis with CNN-Based Brain MRI Classification. Applied Sciences, 15(9), 4652. https://doi.org/10.3390/app15094652






