Abstract
Adrenal lesions are common findings in abdominal imaging, with adrenal adenomas being the most frequent type. Accurate detection of adrenal adenomas is essential to avoid unnecessary diagnostic procedures and treatments. However, conventional imaging-based evaluation relies heavily on the expertise of radiologists and can be complicated by pseudo-lesions, overlapping imaging features, and suboptimal imaging techniques. To address these challenges, we propose an end-to-end machine learning pipeline that integrates deep learning-based lesion detection (FCOS) with an ensemble classifier for adrenal lesion classification in MRI. Our pipeline operates directly on broader regions of interest, eliminating the need for manual lesion segmentation. Our method was evaluated on a multi-sequence MRI dataset comprising 206 adenomas and 45 non-adenomas. The pipeline achieved 87.45% accuracy, 87.33% specificity, and 87.63% recall for adenoma classification, demonstrating competitive performance compared to prior studies. The results highlight strong non-adenoma identification while maintaining robust adenoma detection. Future research should focus on dataset expansion, external validation, and comparison with radiologist performance to further validate clinical applicability.
1. Introduction
Adrenal lesions affect approximately 9% of the global population, with most cases detected incidentally during abdominal imaging—adrenal incidentalomas [1]. These lesions can be malignant, such as carcinomas or metastases, or benign, including adenomas and pheochromocytomas [2]. Among these, adrenal adenomas account for 50–80% of cases [3]. Typically, adrenal adenomas are non-functional, asymptomatic, and do not need treatment [4,5]. Their prevalence increases significantly with age, from around 0.14% in individuals aged 20–29 years to approximately 7% in those over 70 [6]. Distinguishing adenomas from potentially malignant lesions is vital to avoid unnecessary surgical procedures or overtreatment [7].
Diagnosis of adrenal lesions is a complex process often involving both biochemical and radiological evaluation [8]. Adrenal radiologic evaluation via conventional imaging is a challenging process that depends largely on the experience and knowledge of the radiologist [9]. Several pitfalls can result in the misdiagnosis of adrenal lesions, such as the presence of pseudo-lesions, overlap of imaging features of different lesions, or incorrect choice of the imaging technique [10]. Approximately 70–80% of adrenal adenomas are lipid-rich, making them easier to detect using imaging techniques sensitive to intracytoplasmic lipid content. However, lipid-poor adenomas pose a greater diagnostic challenge due to substantial overlap in appearance with malignant lesions [4]. Conversely, some benign and malignant lesions with high lipid content can mimic lipid-rich adenomas, further complicating accurate differentiation [11].
Traditional machine learning (ML) and deep learning (DL) have both made significant contributions to medical imaging analysis. Traditional machine learning methods rely on feature extraction, often requiring domain-specific expertise to identify relevant features in medical images [12,13]. In contrast, DL, particularly through convolutional neural networks (CNNs), automates feature extraction by learning hierarchical representations directly from raw image data. This automation has resulted in notable improvements in accuracy and efficiency for tasks such as segmentation, classification, and detection of various medical conditions [14,15,16,17]. However, DL faces data quality and availability challenges, as collecting sufficient data and creating high-quality annotations remain time-intensive tasks that require significant effort from medical experts [18]. In the context of adrenal lesion analysis, most applications to date have utilized legacy machine learning models that rely on radiomic features to classify adrenal lesions [9].
In this study, we aimed to develop a machine learning pipeline that integrates a DL model for adrenal lesion detection with a traditional ML model for classifying the radiomic features of the detected lesions. The object detection DL model was chosen for its ability to learn hierarchical features directly from raw magnetic resonance imaging (MRI) images, enabling precise and efficient lesion detection. Once detected, the identified lesions are analyzed through an ensemble learning model, which classifies them based on their radiomic features. This combination creates a robust end-to-end framework for adrenal lesion analysis, focusing on adenoma detection.
1.1. Deep Learning for Object Detection
Deep learning object detection models were initially developed for natural images, but have been successfully adapted for identifying and localizing different lesions in medical images [19,20].
These models comprise two main components: the backbone and the detector. The backbone, typically a convolutional network, such as Residual Network (ResNet) or VGG (the model from Visual Geometry Group), is responsible for extracting features from the input images. The detector, also known as the head, analyzes these features to classify and localize the objects within the defined classes. The object detectors can be categorized according to their anchor dependence or the number of stages in their architecture. Single-stage detectors, such as Fully Convolutional One-Stage Object Detection (FCOS) and RetinaNet use dense sampling to analyze the images in a single-shot fashion. In contrast, two-stage detectors, such as Region-based Convolutional Neural Networks (R-CNNs), have an initial stage where region proposals are computed that will then be classified in the second stage. Single-stage detectors are more efficient and the two-stage detectors tend to be more accurate. Regarding anchor dependence, anchor-based models, like RetinaNet or R-CNN, rely on predefined bounding boxes (anchors) for object detection. Meanwhile, anchor-free models, like FCOS, eliminate this dependency, simplifying the detection process. The performance of the object detection model is mainly affected by three challenges: intra-class variation, a high number of classes, and efficiency [21].
1.2. Radiomics
Radiomics involves the extraction of quantitative features from medical images, such as those from MRI or Computed Tomography (CT), to uncover patterns and insights not visible to the naked eye [9]. Before feature extraction, the region of interest (ROI) is segmented to focus the analysis and reduce the volume of data that needs processing, thereby enhancing computational efficiency. This process can be performed manually, which is considered the gold standard. In this approach, a medical expert segments the region of interest (ROI). This time-consuming task heavily relies on the level of expertise of the individual performing the segmentation. Furthermore, there are fully automated methods for ROI segmentation. However, they struggle in cases with lesions with indistinct borders and are highly dependent on the quality of the image. For that reason, semi-automatic methods are preferable. These methods have minimal user interaction for seed identification or manual correction [22]. After the ROI segmentation, several different features can be extracted. Commonly, these features are divided into four categories [22]:
- Shape-Based Features: numeric information respecting geometric characteristics, like shape and size.
- First-Order Features: distribution of voxel values without spatial information, generally histogram-based.
- Second-Order Features: “texture” features, focus on the spatial relationships between voxels with similar grey levels.
- High-Order Features: filters are used to extract patterns from the images. From the resultant images, first- and second-order features are extracted.
Traditional ML models are frequently used to classify radiomic features of regions of interest and have achieved high classification performance in different anatomical regions [9,23]. These models are used instead of conventional statistical methods as they perform better with large amounts of data [5].
1.3. Traditional Machine Learning and Ensemble Learning
Traditional machine learning has significantly progressed in the medical domain, particularly in analyzing clinical data and medical imaging. Traditional machine learning can effectively classify and predict outcomes based on complex datasets by employing algorithms such as decision trees, support vector machines, and artificial neural networks. These models can analyze vast amounts of data, identifying patterns that may not be apparent to human observers and enhancing diagnostic accuracy [12].
Ensemble learning is an approach in machine learning that aims to improve predictive performance by combining multiple models, known as base learners, to create a single, more accurate model. This technique leverages the diversity of the individual models, which may have different strengths and weaknesses, to enhance overall performance. Ensemble learning methods can be broadly categorized into four main categories [24]:
- Bagging: involves training multiple models independently on different subsets of the training data and then averaging their predictions, to reduce variance (e.g., random forest classifier).
- Boosting: sequentially trains models, where each new model focuses on correcting the errors made by the previous ones, thereby improving accuracy (e.g., XGBoost classifier).
- Stacking: combines multiple models by training a meta-learner to optimally weigh their predictions.
- Voting: aggregates the predictions of multiple base models (that can be ensemble models already) to make a final decision. Voting classifiers can be implemented in two main ways:
- −
- Majority Voting: predicts the class with the majority vote from the base models. This approach is simple and works well when individual models are diverse and have similar levels of accuracy.
- −
- Weighted Voting: averages the predicted probabilities from each base model and selects the class with the highest average probability, which is particularly effective when base models output reliable probabilities.
1.4. Related Work
In recent years, research on applying machine learning to adrenal imaging has been increasing, with the development of many promising preliminary studies that leverage ML capabilities to differentiate benign from malignant lesions and improve lesion characterization in abdominal medical images, mainly from CT and MRI [5,9]. Zhang et al. identified three main shortcomings in the reviewed radiomics studies: the lack of prospective studies, the lack of external multicenter validation, and the lack of comparison of diagnostic performance between the radiologist and the ML tools [9]. In addition, Barat et al. pointed out that the incoherence of the recommended practices hinders the clinical application of the studies. Some studies suggest that ML algorithms should use biological and imaging data for better accuracy, while others ignore that fact [5]. All of the studies reviewed in this section analyze axial slices, reflecting their standard use for the evaluation of adrenal lesions. The following subsections analyze the state of the art, categorizing the studies into three groups based on the type of machine learning pipeline employed and their specific classification objectives.
1.4.1. Radiomics-Based ML Pipelines: Adenomas vs. Other Adrenal Lesions
Most papers in this area have developed radiomics-based ML pipelines to differentiate between adrenal adenomas and other adrenal lesions. Ho et al.’s was the only study to compare MRI and CT datasets when applying a logistic regression model using shape-based, first- and second-order radiomic features to classify lipid-poor adenomas and carcinomas [25]. Other studies utilized a logistic regression model to differentiate metastases from adenomas using only first-order [26] and metastases from lipid-poor adenomas using shaped-based and first-order radiomic features [27], in this case lipid-poor adenomas. Liu et al. also used first-order radiomic features to fit a dataset of adenomas and pheochromocytomas with a support vector machine (SVM) model [28]. Romeo et al. attempted a multiclass scenario to differentiate between lipid-poor, lipid-rich, and non-adenoma lesions. They extracted first-and second-order features and classified them with a decision tree [29]. Despite their varied approaches, these studies share limitations, including small (fewer than 70 lesions) and imbalanced MRI datasets, specifically of T1 Weighted Chemical Shift (CS)—In Phase (IP) and Out of Phase (OOP)—and T2 Weighted (T2W) images.
Other studies followed a similar approach using CT images, primarily analyzing multiphase CT to differentiate lipid-poor adrenal adenomas from pheochromocytomas [7,30,31] and adenomas from carcinomas [32,33]. Torresan et al. was the only study that applied an unsupervised ML model—KMeans—to classify first- and second-order radiomic features, though their dataset contained only 19 lesions [32]. Elmohr et al. extracted radiomic features across all categories and tested two different ML models, achieving the performance with Boruta Random Forest [33]. Xiao et al. also experimented with multiple models, and their multi-layer perceptron achieving the best results using first-order, shape-based, and second-order features [30]. Liu et al. innovated by incorporating clinical data (eg., necrosis changes) alongside first-order radiomic features. The authors also tested multiple ML models, and the best was the logistic regression [7]. Yi et al. also applied logistic regression to classify first-, second-, and higher-order radiomic features [31]. Unenhanced CT images were used to distinguish between adenomas and metastases [34], lipid-poor adenomas and pheochromocytomas [35], and adenomas and other unspecified lesions [36]. In addition, Altay et al. trained a logistic regression with second-order features for multiclass classification with a dataset with lipid-rich and lipid-poor adenomas, metastases, and pheochromocytomas [37]. Tu et al. also trained a logistic regression but only with first-order features [34]. Yi et al. followed a similar approach in their studies [31,35]. Feliciani et al. focused their models on analyzing higher-order radiomic features and used an LASSO regression model for feature selection [36].
Studies leveraging MRI and CT data have extensively explored radiomics-based ML pipelines to differentiate adenomas from other lesions. MRI-focused studies have primarily targeted lipid-poor and lipid-rich adenomas and pheochromocytomas, employing logistic regression, SVM, and decision trees to classify first- and second-order features. Similarly, CT-based studies have demonstrated the effectiveness of radiomics for differentiating adenomas from carcinomas, metastases, and pheochromocytomas, with logistic regression, random forest, and MLP models frequently achieving high accuracy. Despite their potential, these studies share common limitations, including small and imbalanced datasets, reliance on manual segmentation, and variability in feature extraction techniques.
1.4.2. Radiomics-Based ML Pipelines: Benign vs. Malignant Adrenal Lesions
The studies in this group focus on the differentiation between benign and malignant adrenal lesions. Two studies used T1 IP/OOP and T2W MRI data for radiomic feature extraction [38,39]. Barstugan et al. extracted second- and higher-order radiomic features and classified them using an SVM. Their dataset had less than 10% malignant lesions [38]. Stanzione et al. extracted features across all categories and used an extra trees model to classify them [39]. In both studies, medical experts performed manual segmentation of lesions.
CT images were also used, specifically multiphase CT scans. Moawad et al. focused their analysis on small adrenal lesions and employed a logistic regression model to classify shape-based, and first- and second-order features [40]. Koyuncu et al. extracted features from all categories, including higher-order wavelet features, and utilized a particle swarm optimization–neural network classifier [41]. In both studies, the lesions were manually segmented by medical experts. In contrast, Andersen et al. used semi-automatic methods for lesion segmentation. Their study targeted adrenal metastases, using a random forest for classification [42].
1.4.3. Deep Learning for Adrenal Lesion Analysis
The application of deep learning for adrenal lesion analysis is still in its infancy, primarily due to the lack of high-quality datasets in this area. Despite that, several studies have successfully utilized DL for various tasks, including distinguishing large lipid-poor adenomas from carcinomas [43], adenomas from malignant non-adenomas [44], classifying multiple types of adrenal lesions in a multiclass scenario [45], and segmenting and classifying adrenal glands [46] and functional and non-functional adrenal tumors [47]. All these studies have analyzed multiphase CT images.
These studies offer various approaches to address specific challenges in this field. Singh et al. utilized a 3D-DenseNet-121 architecture to classify 3D lesion patches manually segmented by radiologists, leveraging traditional augmentations for enhanced generalization [43]. Kusunoki et al. designed a custom DCNN to classify lesions from manually cropped regions of interest, comparing performance across multiple CT phase combinations [44]. Bi et al. introduced a novel Deep Multi-Scale Resemblance Network (DMRN), which incorporated similarity feature learning to classify lesions into five distinct categories [45]. Due to their relatively small datasets, Singh, Kusunoki, and Bi implemented cross-validation to maximize training efficiency and ensure robust model evaluation. Robinson-Weiss et al. integrated U-Net for gland segmentation, followed by DenseNet-based patch classification to distinguish between normal and abnormal glands [46]. Alimu et al. developed a three-stage pipeline combining U-Net for kidney, adrenal, and tumor segmentation with a variational autoencoder (VAE) for refined lesion classification [47].
1.5. Goals and Contributions
The main goal of this study is to develop an end-to-end machine learning pipeline for the automated detection and classification of adrenal lesions in MRI images. Specifically, the pipeline focuses on identifying adrenal adenomas and distinguishing them from other lesion types or the absence of lesions within the dataset.
During the course of this investigation, multiple approaches were explored, including an end-to-end deep learning model for both detection and classification in a single stage. However, this approach yielded lower-than-expected validation performance, likely due to the limited dataset size and class imbalance, which posed challenges for deep learning optimization. Given these findings, we concluded that the most effective strategy is the two-stage pipeline presented here.
This framework integrates a DL model for region-of-interest (ROI) detection on MRI slices and a traditional machine learning (ML) model to classify the detected regions in a binary setting—adenoma versus non-adenoma. By combining the strengths of DL for feature extraction and traditional ML for robust classification, this study aims to address key challenges in adrenal lesion analysis, such as overlapping imaging features, imbalanced datasets, and the need for automation in diagnostic workflows.
By employing our pipeline, manual lesion segmentation during inference is no longer required, significantly streamlining the diagnostic process. Additionally, the pipeline filters out non-lesion slices, which are the vast majority in routine medical imaging, allowing for focused lesion analysis. Furthermore, training on multiple MRI sequences enhances robustness to variations in imaging protocols, making the method more adaptable to real-world clinical settings. This automated approach reduces the reliance on expert annotations, minimizes human error, and accelerates the analysis, making it a practical and efficient solution for clinical applications. Moreover, the usage of a second stage with a traditional ML model helps mitigate the challenges posed by small and imbalanced datasets, where deep learning alone struggles. Additionally, it allows for greater interpretability, as feature-based models provide insights into the classification decision-making process, which could be valuable for future clinical applications.
The contributions of our developed ML pipeline are as follows:
- The pipeline filters out non-lesion slices, which are the majority in medical imaging, allowing for a more efficient and targeted lesion analysis.
- The pipeline was trained using multiple MRI sequences, enhancing robustness against variations in imaging protocols.
- The two-stage approach mitigates deep learning challenges by using traditional ML for classification, which is more effective on small and imbalanced datasets.
- Unlike end-to-end deep learning, using an ensemble ML classifier allows for greater interpretability, making it easier to understand decision-making processes.
- The pipeline eliminates the need for manual segmentation, which is common in radiomics-based studies, improving clinical usability and efficiency.
2. Materials and Methods
This study involved three main steps: dataset creation and preparation, model training and selection for lesion detection and classification, and end-to-end pipeline evaluation. These steps are briefly described in Figure 1 and will be detailed in the following subsections. The code developed to train, validate both detection and classification models, and test the end-to-end pipeline is available at https://github.com/bbg-fct/Adrenal-Adenoma-Detection-ML-Pipeline/tree/main (accessed 1 March 2025).
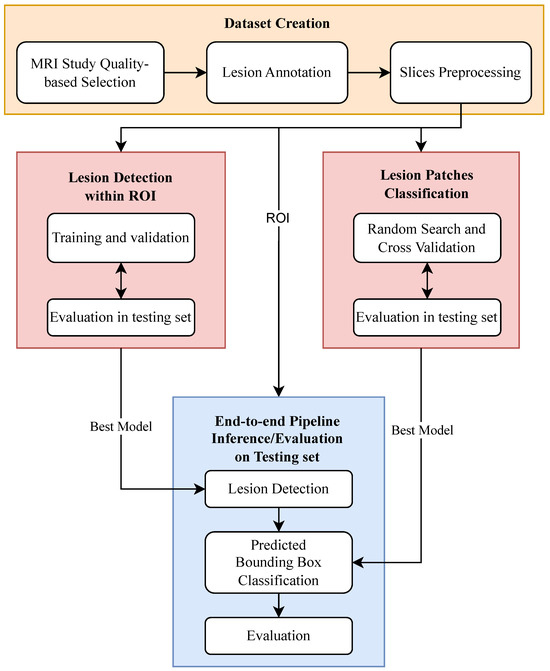
Figure 1.
Overview of the methodology applied in the study, illustrating the machine learning pipeline for adrenal lesion detection and classification, including MRI dataset preparation, lesion detection, and binary classification of adenomas versus non-adenomas.
2.1. Dataset Creation and Preparation
This retrospective study was approved by the Ethics Committee of Hospital Garcia de Orta, and the requirement for informed consent was waived as all data included in this study were anonymized beforehand. All the MRI studies, acquired in that institution between 2008 and 2019, were pre-analyzed by radiologists during routine clinical practice, with medical reports already available for each study. For this research, a medical expert reviewed the MRI studies and annotated the adrenal lesions by adding bounding boxes to the images. Only one study was selected for each patient based on its overall quality. All the selected studies had at least axial and coronal T2W, T1W CS, and T1W contrast-enhanced (CE) sequences and the presence of at least one adrenal lesion.
Table 1 provides an overview of the dataset. The original dataset was balanced in terms of the patient’s gender but showed an imbalance in the types of adrenal lesions, with adenomas being much more prevalent than other lesions. The lesion dimensions also had limited variability.

Table 1.
Dataset description. Summary of the original and final dataset characteristics.
Additional processing was required to create the final dataset that was used to train and test the ML models. Each DICOM study was exported from the hospital’s system and converted to an image dataset, with the labels stored using Fiftyone [48] toolkit and CVAT (https://www.cvat.ai/ (accessed in 26 January 2025)) annotation tool. The PyDicom package (https://github.com/pydicom/pydicom (accessed in 29 January 2025)) was employed to extract relevant metadata from the DICOM studies and to convert the DICOM files into 512 × 512 PNG images, which was the default size of the slices. Similarly to most studies on adrenal lesions, our final dataset consisted exclusively of axial slices, which is the standard imaging orientation for evaluating these lesions. To create a dataset to train a lesion detection model, 12,593 slices were selected using an automated “window-based selection” approach, to address the imbalance between the lesion and non-lesion slices. For each group of lesion slices in the MRI studies (most lesions spanned multiple consecutive slices), we included three slices before and three slices after the lesion group, with two slices of interval. We used this interval to avoid slices with small portions of lesion—that were not labeled as lesion slices due to potential annotation inaccuracies—to be considered as non-lesion slices. Table 2 shows a graphical explanation of the slice inclusion algorithm.

Table 2.
Axial slices inclusion automated procedure (direction: caudal-to-cranial).
Following the slice selection procedure, a region of interest (ROI) measuring 150 × 150 pixels was extracted from each image, with the adrenal lesions positioned at the center. For the lesion slices, this extraction was performed deterministically using bounding boxes provided by medical expert annotations. The coordinates of the extracted ROI were also used to crop the non-lesion slices. This process could be automated using adrenal or kidney detection models; such automation falls outside the scope of this work.
To ensure effective machine learning training and evaluation, the final dataset was divided into three subsets: train, validation, and test. In this split, it was ensured that images from each patient were included in only one dataset to prevent data leakage and that an even distribution of lesion sizes across all datasets was maintained. These datasets with the ROIs were used to train and evaluate the lesion detection DL model. For the classification model, we used the same dataset split, but focused solely on lesion patches (cropped bounding boxes from the medical expert annotation) excluding all non-lesion slices, as our primary interest was the classification of the lesions. Both the ROIs used for detection and the lesion patches used for classification were normalized using the min-max algorithm.
2.2. Lesion Detection
To deploy our DL model, we utilized the Detectron2 Framework [49], monitoring the performance metrics during training with Weights & Biases (https://docs.wandb.ai/guides/ (accessed on 29 January 2025)). After experimenting with several CNN-based models on our MRI dataset, we empirically selected the Fully Convolutional One-Stage Object Detection (FCOS) model [50], as it achieved the best results during validation and testing in terms of detection performance. Nevertheless, the FCOS model has demonstrated success in medical imaging tasks [51,52], further supporting its suitability for this study. Additionally, FCOS has important features for our use case:
- Anchor-Free Design: particularly suited for datasets with diverse object sizes, such as lesions of varying dimensions across slices, as it avoids the constraints of fixed anchors and generalizes better to irregularly sized objects.
- One-stage Architecture: offers greater computational efficiency during training and inference, making it ideal for applications with limited computational resources and real-time applications.
- Focal Loss: crucial for addressing the high class imbalance inherent in dense sampling. Dense sampling generates many predictions, most of which correspond to the background class. Focal loss mitigates this by reducing the weight of easy background samples and focusing on harder-to-classify examples, such as lesion regions.
- Feature Pyramid Network (FPN): essential for detecting lesions of various sizes, as the FPN enables multi-scale detection by learning features at different resolutions.
During the training of our DL model, we monitored standard evaluation metrics commonly used for COCO datasets (https://cocodataset.org/#detection-eval (accessed on 1 February 2025)). The primary metric, Average Precision (AP) or mean Average Precision (mAP), combines precision and recall, calculated as the area under the precision–recall curve over a range of Intersection Over Union (IoU) thresholds. This metric is highly informative but also among the most challenging to achieve. Another relevant metric is AP50, which considers a prediction correct if the IoU with the ground truth exceeds 50%. These metrics were tracked during training to ensure the model was learning effectively and to identify and rectify any erroneous configurations. At the testing phase, in addition to the standard detection metrics, we calculated a detection confusion matrix and its associated metrics (detailed in Section 2.3), considering two classes: lesion and non-lesion. This required specifying a predefined IoU value, which served as the threshold for determining whether predictions were accurate.
2.3. Lesion Classification
This stage of our pipeline involved three main steps: feature extraction, feature selection, and model fitting. Feature extraction was performed using SimpleITK [53] and PyRadiomics [54], while machine learning models were implemented through the scikit-learn package [55]. To optimize the performance of our ML model, we conducted a grid search to explore hyperparameter combinations and employed LASSO regression and Principal Component Analysis (PCA) for feature selection and reduction. Additionally, a random search of the internal parameters of each model paired with cross-validation was used for model fitting. The grid search evaluated variations in the number of selected features, the proportion of excluded lesion slices (avoiding slices closer to the lesion center), and the number of cross-validation folds. LASSO regression was also applied with cross-validation. The final classification model was a voting classifier comprising three base learners: random forest, logistic regression, and XGBoost, with weighted voting.
Given that we used cross-validation to select our best-performing classifier, we merged the training and validation datasets into a single training set. This approach allowed us to maximize the available data for model training. During cross-validation, the model’s performance was evaluated using the recall score, ensuring a robust assessment of its ability to identify positive cases correctly. At the testing phase, we computed a traditional binary—positive class is adenoma, negative class is non-adenoma—confusion matrix and its related metrics:
- Accuracy: The proportion of correctly classified instances out of the total number of instances.
- Precision: The ratio of true positives to the total predicted positives:
- Recall (Sensitivity): The ratio of true positives to the actual positives:
- Specificity (%): The ratio of true negatives to the actual negatives:
- F1-Score: The harmonic mean of precision and recall:
2.4. End-to-End Evaluation
After individually evaluating and selecting the best models for lesion detection and classification, the models were combined sequentially to develop the complete pipeline. In this setup, all the predicted bounding boxes generated by the detection model were passed directly to the classifier without applying additional filters or thresholds. This evaluation was conducted on the entire test dataset to assess the pipeline’s overall performance using end-to-end metrics, capturing its ability to detect and classify adrenal lesions in a fully automated workflow.
To ensure a comprehensive evaluation of the complete ML pipeline, we created a multiclass confusion matrix (described in Table 3). The first column represents all samples analyzed by both stages of the pipeline that resulted in adenoma predictions: (1) correct classifications, (2) incorrect classifications as non-adenoma lesions, or (3) incorrect predictions due to the absence of lesions. The second column follows the same logic for non-adenoma predictions. The third column includes samples that only passed through the detection stage, because no lesions were detected by the DL model. To populate this matrix, a predefined IoU threshold was used to determine whether lesions were correctly localized by the detection model. A higher IoU threshold enforces stricter detection criteria.

Table 3.
Descriptive confusion matrix for end-to-end evaluation.
The computed end-to-end metrics are derived from this multiclass confusion matrix. We calculated macro, weighted, and class-specific metrics, for a holistic evaluation of our pipeline. Macro metrics provided an unweighted average across all classes, treating each class equally, regardless of its frequency. Weighted metrics, on the other hand, accounted for the imbalance in class distribution, offering insights into the pipeline’s performance in proportion to the occurrence of each class. Finally, class-specific metrics allowed us to evaluate the model’s performance in individual classes.
All the experiments were made in a Docker environment using a container based on the pytorch/pytorch:2.5.0-cuda12.4-cudnn9-runtime image. The system was configured with Python 3, essential development tools, and the required Python libraries, including Detectron2, PyRadiomics, and scikit-learn. The host computer had a 12th Gen Intel(R) Core(TM) i7-12700 CPU, 32 GB DDR4 RAM, and an NVIDIA GeForce RTX 3060 GPU with CUDA 12.4 and cuDNN 9 for efficient training. This environment ensured reproducibility and consistency across all experiments.
3. Results
This section presents the experimental results of our study, detailing the performance of the lesion detection and classification models, as well as the overall evaluation of the end-to-end pipeline.
3.1. Lesion Detection
Multiple experiments were conducted to select the best model, exploring different hyperparameters, augmentations, and MRI slice types. The best-performing model was trained using all available MRI sequences over a total of 8 h and 43 min. Table 4 presents the augmentations applied during training. These included common transformations such as image flipping, rotation, and resizing, along with contrast and brightness adjustments. The latter were particularly critical for ensuring robustness across the diverse MRI sequences (T1W, T2W, and T1W CE) included in the dataset. Additionally, moderate elastic transformations were applied to improve the model’s ability to handle organ deformations visible in MRI studies.

Table 4.
Best data augmentations for training the lesion detection DL model.
The best hyperparameters for the FCOS detection model are presented in Table 5. A small learning rate (0.0001) was chosen to ensure stable training, which is particularly important given the relatively modest size of our dataset, for deep learning standards. The backbone architecture used in the FCOS detection model was ResNet-50, which was pre-trained on ImageNet. To improve training efficiency and mitigate overfitting, the first two stages of the ResNet backbone were frozen, allowing the higher-level layers to adapt to the specific features of MRI images. The focal loss parameters were left at their defaults. A detection score threshold of 0.5 was applied, balancing the trade-off between sensitivity and precision. Additionally, a slightly higher non-maximum suppression (NMS) threshold of 0.6 was used to retain more overlapping bounding boxes, which helped detect small or irregularly shaped lesions. This deliberate configuration reflects the need to prioritize lesion sensitivity while maintaining reliable performance across varying lesion characteristics.

Table 5.
Best model hyparameters for lesion detection.
The model’s performance metrics are detailed in Table 6. The AP of 42.68% demonstrates the model’s capability to handle challenging detection scenarios, while an AP50 of 85.80% confirms its strong performance under less strict IoU thresholds. Additionally, traditional classification metrics, computed for an IoU threshold of 50%, showcase the model’s high recall (89.47%) and F1-score (87.62%) while detecting adrenal lesions.

Table 6.
Evaluation metrics for the best lesion detection model on the testing set.
3.2. Lesion Classification
The lesion classification model was trained using all available MRI sequences. This decision aligns with the best training approach for the detection model, which also utilized the same MRI sequences. To select the best adrenal lesion classification model, we conducted an extensive search for global parameters using a grid search and a random search for internal parameters of the individual base learners. Table 7 summarizes the hyperparameter results from both search strategies. As a result, the optimal model incorporated 15 radiomic features selected through LASSO regression and PCA, comprising first-order features (Energy, Kurtosis, Skewness), second-order features (GLRLM, GLSZM), and higher-order features (GLDM). The training set consisted of the most representative 20% of lesion slices, emphasizing slices that best captured the characteristics of adrenal lesions, i.e., closer to the center of the lesion. Cross-validation with 10 folds was employed, using recall as the evaluation metric to prioritize sensitivity in the model’s predictions.

Table 7.
Best parameters for the classification model, results from random search and grid search.
This optimization process culminated in a robust model configuration, whose performance is detailed in the subsequent results, detailed in Table 8. These results were obtained in the complete testing set without any slice exclusion.

Table 8.
Evaluation metrics for the classification model on the testing set—non-adenoma vs. adenoma.
The classification model achieved strong performance in distinguishing adrenal adenomas from non-adenoma lesions, as indicated by its evaluation metrics. With an accuracy of 88.97%, the model demonstrated high overall correctness in its predictions. The precision of 90.31% reflects a low false adenoma prediction rate. The recall (sensitivity) of 94.20% highlights the model’s ability to detect adenomas effectively, minimizing the number of missed adenomas. However, the specificity of 77.13% indicates a relatively higher false positive rate when identifying non-adenomas, suggesting that some non-adenoma lesions were misclassified as adenomas. These results suggest that the classification model is highly effective in identifying adrenal adenomas, although improvements could be made to enhance the classification of non-adenomas.
3.3. End-to-End Pipeline
The results presented in this section originate from the sequential pipeline, which integrates the best-performing detection and classification models, following the methodology detailed in Section 2.4. Figure 2a is populated version of the descriptive matrix provided in Table 3. This matrix gives a complete overview of the results of our pipeline in the testing dataset. Additionally, Figure 2b focuses specifically on the detection and classification outcomes for adrenal adenomas, which represent the primary focus of this study.

Figure 2.
Confusion matrices. (a) Overall end-to-end confusion matrix. (b) Adenoma-specific end-to-end confusion matrix.
These matrices lay the foundation for calculating the metrics necessary to evaluate the overall quality of the machine learning pipeline. Table 9 presents the computed metrics derived from both the overall confusion matrix (Figure 2a) and the adenoma-specific confusion matrix (Figure 2b).

Table 9.
Overall and adenoma-specific metrics.
The pipeline achieved an overall accuracy of 85.06%, indicating a high proportion of correct predictions across all classes. The macro metrics, which treat each class equally, further highlight the robustness of the pipeline, with a macro precision of 86.27%, a macro recall of 97.62%, and a macro F1-score of 90.13%. However, the weighted metrics, which account for class imbalances, showcased significantly higher values for precision (98.14%) and F1-score (97.21%). This gap between macro and weighted metrics suggests that the model performs exceptionally well on the adenoma and non-lesion classes (the majority classes) while facing challenges with the less frequent non-adenoma class. Specifically, the increased weighted precision and F1-score underscore difficulties in accurately classifying non-adenoma lesions, likely due to their lower representation in the dataset and overlapping imaging characteristics with adenomas.
For adenoma-specific results, the pipeline demonstrated an accuracy of 87.45%, reflecting its effectiveness in handling this class. The precision of 82.22% and recall of 87.63% indicate that the model maintains a good balance between minimizing false positives and correctly identifying adenomas. The F1-score of 84.84% highlights the overall balance between precision and recall. Additionally, the specificity of 87.33% for adenomas indicates that the pipeline correctly identifies non-adenomas as not adenomas in 87.33% of cases. These results underline the pipeline’s capability to classify adenomas with high reliability, which is the primary focus of this study.
In addition to presenting the quantitative results of our pipeline, it is essential to highlight qualitative examples of its performance. Figure 3 showcases correct detections and classifications of adenomas across different patients, highlighting the diversity in adenoma sizes, shapes, and imaging characteristics. The first row of the figure shows T1-W CS OOP slices and the second row presents T1-W CE slices. The green bounding boxes represent the medical annotations and the red boxes represent the model’s predictions.

Figure 3.
Examples of true positive detections for adenomas across different MRI sequences and different patients. Each row corresponds to a different MRI sequence: the first row shows T1-Weighted Chemical Shift (Out-of-Phase) images, and the second row shows T1-Weighted Contrast-Enhanced images. Green bounding boxes are the ground truth; red is the high-score prediction. Subfigures are labeled (a–f) for reference.
Notably, the model successfully identifies very small adenomas (e.g., Figure 3a,d) with more elongated, horizontal shapes, and larger adenomas with more square-like shapes (e.g., Figure 3c,f). Furthermore, the pipeline’s robustness in handling variations in image intensity is evident, highlighting its capacity to adapt to diverse imaging conditions, including contrast-enhanced and non-contrast-enhanced sequences.
While the pipeline demonstrates strong performance in detecting and classifying adrenal lesions, examining its limitations through examples of incorrect detections and classifications is crucial. Figure 4 presents six subfigures showcasing these errors, organized by the ground truth labels: non-lesion, non-adenoma, and adenoma. For each ground truth, a pair of images illustrates specific challenges: misclassifications, false detections, and missed lesions. All subfigures show the same MRI sequence—T1W CS OOP.

Figure 4.
Examples of incorrect detection and/or classifications for different patients. All presented images are T1-Weighted Chemical Shift (Out-of-Phase). GT: Ground truth; Pred: Predicted. Green bounding boxes are the ground truth; red is the high-score prediction. Subfigures are labeled (a–f) for reference. (a) GT: Non-lesion; Pred: Non-adenoma; (b) GT: Non-lesion; Pred: Adenoma; (c) GT: Non-adenoma; Pred: No lesion; (d) GT: Non-adenoma; Pred: Adenoma; (e) GT: Adenoma; Pred: Non-lesion; (f) GT: Adenoma; Pred: Non-adenoma.
4. Discussion
In this study, we developed a two-stage ML pipeline for automated adrenal lesion detection and binary classification (non-adenoma vs. adenoma) in MRI images across multiple sequences. Each stage was trained separately using the same inter-patient split and the same training, validation, and testing patients to avoid data leakage within and between stages. After independently validating each stage, we evaluated the complete pipeline on the full test set in a real-world scenario, where all detected lesions were passed to the classifier without additional filtering.
To provide a contextual benchmark for our study, we selected similar works from the literature to compare methodologies and results. From the studies that use radiomics-based feature extraction and classification with traditional ML, we selected two that focus on differentiating adenomas from other adrenal lesions (1) and analyzed MRI datasets (2). Studies that exclusively analyzed lipid-poor adenomas were excluded, as our dataset did not contain a significant number of such cases. For deep learning-based studies, since no prior work specifically analyzed MRI images, we selected one study based on its similar objective to ours but that analyzes CT images. Table 10 summarizes the imaging modality, dataset characteristics, machine learning models, and key evaluation metrics (accuracy, specificity, and recall) of these studies, highlighting methodological differences and performance variations.

Table 10.
Comparison of similar studies on adrenal lesion classification, detailing the dataset composition (imaging modality, and number and type of lesions), machine learning approach, and key evaluation metrics (accuracy, specificity, and recall). This table highlights methodological differences and performance variations across studies, providing a contextual benchmark for this study’s proposed pipeline.
Compared to the similar studies in Table 10, our proposed pipeline demonstrates competitive performance while addressing key limitations of prior research. Our dataset contains 206 adenomas and 45 non-adenomas, making it significantly larger than those used in Schieda et al. [26] (29 adenomas, 15 metastases) and Liu et al. [28] (38 adenomas, 20 pheochromocytomas), both of which suffered from small and highly imbalanced datasets. These studies employed logistic regression and SVM, respectively, on manually extracted radiomic features, whereas our pipeline integrates deep learning-based lesion detection (FCOS) with an ensemble classifier, enabling fully automated processing without manual segmentation. Kusunoki et al. [44] achieved higher accuracy (94%), but their dataset was limited to 83 adenomas and 24 non-adenomas, and they used multiphase CT. Our pipeline achieves a high specificity (87.33%), indicating strong performance in correctly identifying non-adenomas (i.e., non-adenoma lesions + non-lesion images), which was a key challenge in previous ML-based studies.
While our proposed pipeline demonstrates strong performance in automated adrenal lesion classification, several limitations should be stated. First, our dataset, although larger than previous MRI-based studies, remains relatively small for deep learning standards, which affects detection performance in the first stage of our pipeline. Additionally, it is imbalanced in terms of lesion types, which impacts classification performance in the second stage.
A closer examination of the overall end-to-end confusion matrix (see Figure 2a) reveals that the recall (true positive rate) for non-adenomas was only 29.41%, indicating a substantial number of false negatives. Specifically, 103 non-adenomas slices were misclassified as adenomas and 53 were incorrectly classified as no lesion, suggesting that the model is biased towards adenoma detection. While this aligns with our primary focus, it remains a limitation that must be addressed in future work. This poor performance in non-adenoma classification can be attributed to two main factors. One contributing factor is dataset imbalance—non-adenomas constitute only 17.2% of the total lesions—limiting the model’s exposure to this class during training. Another challenge is the significant feature overlap between adenomas and certain non-adenoma lesions (e.g., pheochromocytomas and metastases), as both can exhibit similar imaging characteristics, thereby complicating differentiation.
To address these issues, future work should focus on increasing the representation of non-adenoma lesions through dataset expansion or data augmentation techniques. Additionally, incorporating external validation datasets, exploring alternative classifiers, and integrating supplementary imaging features or clinical data may enhance the model’s ability to distinguish between adenomas and non-adenomas.
Second, our pipeline does not include normal patients (i.e., patients without any adrenal lesions). This omission limits our ability to assess the false positive rate (FPR) for non-lesion slices in a real clinical setting, where normal scans are common. Based on our confusion matrix (Figure 2a), only 8.76% of non-lesion slices were incorrectly classified as adenomas or non-adenomas. However, these non-lesion slices were extracted from patients with adrenal lesions, meaning they do not necessarily represent true normal adrenal anatomy, as the slices may not even include the adrenal glands. As a result, the FPR for non-lesion slices in a real clinical setting could be higher than our reported value. Future studies should include normal patients to provide a more comprehensive evaluation of the pipeline’s performance in clinical practice.
Third, although our method eliminates the need for manual lesion segmentation, enhancing clinical applicability, it also means that lesion localization is less precise than segmentation-based approaches. This trade-off should be considered when comparing our pipeline to radiomics-based methods that rely on manually segmented regions of interest. The pipeline’s performance could be further improved by incorporating a lesion segmentation step, which would provide more precise lesion localization and potentially enhance classification accuracy. However, this was not possible due to the lack of lesion segmentation annotations in our dataset.
Lastly, the impact of MRI sequence variations on model generalizability was not explicitly tested. Different institutions may use different MRI acquisition protocols, which can affect image contrast, spatial resolution, and signal intensity, potentially influencing model performance on unseen data. However, our pipeline was trained using multiple MRI sequences from the same institution and included augmentations such as brightness and contrast variations, which can help improve robustness to different acquisition protocols. Additionally, all images were preprocessed using min-max normalization, ensuring a consistent intensity range across MRI scans, which can further aid in reducing variations between acquisition settings. A possible approach to further address this challenge is the use of domain adaptation techniques, such as feature normalization or style transfer methods, to align images from different protocols. Additionally, data standardization techniques, including intensity normalization and histogram matching, could help reduce variability. Future work should include validation across datasets from multiple institutions to assess generalizability and determine the most effective adaptation strategies for clinical deployment.
Future research should aim to expand the dataset, particularly by increasing the representation of non-adenoma lesions and including normal patients to better assess real-world performance. Additionally, external validation on an independent dataset would be valuable in confirming the pipeline’s robustness across different imaging protocols and scanner manufacturers. Another important extension would be the inclusion of lipid-poor adenomas, which are more challenging to differentiate from malignant lesions and were underrepresented in our dataset. Future studies could explore integrating clinical and biochemical data alongside MRI features to enhance model interpretability and improve classification performance. Given that CT is the most widely used modality in adrenal lesion assessment, adapting this pipeline for multiphase CT images could also extend its clinical applicability. Exploring alternative deep learning architectures or fine-tuning hyperparameters through additional grid/random search comparisons could further improve classification and detection accuracy, particularly for non-adenoma lesions. Finally, future studies should include a comparative performance analysis between the pipeline and expert radiologists to further validate clinical applicability. This would involve reader studies, where radiologists classify adrenal lesions using the same dataset, allowing for a direct assessment of the model’s strengths and limitations in comparison to human expertise. Such studies would help determine whether the pipeline can be a decision-support tool to enhance radiologists’ diagnostic accuracy and efficiency.
5. Conclusions
This study presents a fully automated machine learning pipeline for adrenal lesion detection and classification in MRI images, combining deep learning-based lesion detection with an ensemble classifier. The developed pipeline is capable of filtering out non-lesion slices that are the vast majority in common medical imaging studies and analyze the lesion slices differentiating adenomas from non-adenoma lesions, which is important to avoid unnecessary follow-up exams and surgeries. In addition, the pipeline was trained on multiple MRI sequences, enhancing its robustness to variations in imaging protocols. By integrating multi-sequence MRI analysis and filtering out non-lesion slices, our approach enhances adaptability to real-world clinical workflows while eliminating the need for manual segmentation.
Author Contributions
Conceptualization, B.G., P.V., M.R. and L.V.; methodology, B.G., P.V. and L.V.; software, B.G.; validation, B.G. and P.V.; investigation, B.G.; resources, P.V., G.S. and M.R.; data curation, G.S. and B.G.; writing—original draft preparation, B.G.; writing—review and editing, B.G., L.V., P.V., M.R. and G.S.; visualization, B.G.; supervision, P.V. and L.V.; project administration, B.G.; funding acquisition, P.V. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by the FCT—Portuguese Foundation for Science and Technology and Bee2Fire Lda under a PhD grant with reference PD/BDE/150624/2020.
Institutional Review Board Statement
This study was conducted in accordance with the Declaration of Helsinki and approved by the Ethics Committee from Hospital Garcia de Orta (Almada–Portugal), accepted on 31 August 2020. The committee does not issue approval reference numbers.
Informed Consent Statement
Patient consent was waived as all data were fully anonymized prior to inclusion in this study.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Acknowledgments
We would like to express our gratitude to Júlio Pires from Garcia de Orta Hospital, for facilitating the initial introduction between Bernardo Gonçalves and his advisers with Miguel Ramalho from Garcia de Orta Hospital. This pivotal connection led to the development and establishment of this study.
Conflicts of Interest
Author Bernardo Gonçalves was employed by the company Bee2Fire Lda. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Dhamija, E.; Panda, A.; Das, C.J.; Gupta, A.K. Adrenal imaging (Part 2): Medullary and secondary adrenal lesions. Indian J. Endocrinol. Metab. 2015, 19, 16–24. [Google Scholar] [CrossRef]
- Panda, A.; Das, C.J.; Dhamija, E.; Kumar, R.; Gupta, A.K. Adrenal imaging (Part 1): Imaging techniques and primary cortical lesions. Indian J. Endocrinol. Metab. 2015, 19, 8–15. [Google Scholar] [CrossRef]
- Bracci, B.; Santis, D.D.; Gaudio, A.D.; Faugno, M.C.; Romano, A.; Tarallo, M.; Zerunian, M.; Guido, G.; Polici, M.; Polidori, T.; et al. Adrenal Lesions: A Review of Imaging. Diagnostics 2022, 12, 2171. [Google Scholar] [CrossRef]
- Platzek, I.; Sieron, D.; Plodeck, V.; Borkowetz, A.; Laniado, M.; Hoffmann, R.T. Chemical shift imaging for evaluation of adrenal masses: A systematic review and meta-analysis. Eur. Radiol. 2019, 29, 806–817. [Google Scholar] [CrossRef]
- Barat, M.; Gaillard, M.; Cottereau, A.S.; Fishman, E.K.; Assié, G.; Jouinot, A.; Hoeffel, C.; Soyer, P.; Dohan, A. Artificial intelligence in adrenal imaging: A critical review of current applications. Diagn. Interv. Imaging 2023, 104, 37–42. [Google Scholar] [CrossRef]
- Zheng, Y.; Liu, X.; Zhong, Y.; Lv, F.; Yang, H. A Preliminary Study for Distinguish Hormone-Secreting Functional Adrenocortical Adenoma Subtypes Using Multiparametric CT Radiomics-Based Machine Learning Model and Nomogram. Front. Oncol. 2020, 10, 570502. [Google Scholar] [CrossRef]
- Liu, H.; Guan, X.; Xu, B.; Zeng, F.; Chen, C.; Yin, H.L.; Yi, X.; Peng, Y.; Chen, B.T. Computed Tomography-Based Machine Learning Differentiates Adrenal Pheochromocytoma From Lipid-Poor Adenoma. Front. Endocrinol. 2022, 13, 833413. [Google Scholar] [CrossRef]
- Anagnostis, P.; Karagiannis, A.; Tziomalos, K.; Kakafika, A.; Athyros, V.; Mikhailidis, D. Adrenal incidentaloma: A diagnostic challenge. Hormones 2009, 8, 163–184. [Google Scholar] [CrossRef]
- Zhang, H.; Lei, H.; Pang, J. Diagnostic performance of radiomics in adrenal masses: A systematic review and meta-analysis. Front. Oncol. 2022, 12, 975183. [Google Scholar] [CrossRef]
- Elsayes, K.M.; Elmohr, M.M.; Javadi, S.; Menias, C.O.; Remer, E.M.; Morani, A.C.; Shaaban, A.M. Mimics, pitfalls, and misdiagnoses of adrenal masses on CT and MRI. Abdom. Radiol. 2020, 45, 982–1000. [Google Scholar] [CrossRef]
- Matos, A.P.; Semelka, R.C.; Herédia, V.; AlObaidiy, M.; Gomes, F.V.; Ramalho, M. Modified approach to the characterization of adrenal nodules using a standard abdominal magnetic resonance imaging protocol. Radiol. Bras. 2017, 50, 19–25. [Google Scholar] [CrossRef][Green Version]
- Alwiyah, A.; Setyowati, W. A comprehensive survey of machine learning applications in medical image analysis for artificial vision. Int. Trans. Artif. Intell. (ITALIC) 2023, 2, 90–98. [Google Scholar] [CrossRef]
- Arasi, M.A.; Babu, S. Survey of machine learning techniques in medical imaging. Int. J. Adv. Trends Comput. Sci. Eng. 2019, 8, 2107–2116. [Google Scholar] [CrossRef]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; van der Laak, J.A.W.M.; van Ginneken, B.; Sánchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef]
- Shen, D.; Wu, G.; Suk, H.I. Deep learning in medical image analysis. Annu. Rev. Biomed. Eng. 2017, 19, 221–248. [Google Scholar] [CrossRef]
- Hosny, A.; Parmar, C.; Quackenbush, J.; Schwartz, L.H.; Aerts, H.J.W.L. Artificial intelligence in radiology. Nat. Rev. Cancer 2018, 18, 500–510. [Google Scholar] [CrossRef]
- Anaya-Isaza, A.; Mera-Jiménez, L.; Zequera-Diaz, M. An overview of deep learning in medical imaging. Inform. Med. Unlocked 2021, 26, 100723. [Google Scholar] [CrossRef]
- Elyan, E.; Vuttipittayamongkol, P.; Johnston, P.; Martin, K.; Sarker, M.M.K. Computer vision and machine learning for medical image analysis: Recent advances, challenges, and way forward. Artif. Intell. Surg. 2022, 2, 24–45. [Google Scholar] [CrossRef]
- Aggarwal, R.; Sounderajah, V.; Martin, G.; Ting, D.S.W.; Karthikesalingam, A.; King, D.; Ashrafian, H.; Darzi, A. Diagnostic accuracy of deep learning in medical imaging: A systematic review and meta-analysis. NPJ Digit. Med. 2021, 4, 65. [Google Scholar] [CrossRef]
- Mall, P.K.; Singh, P.K.; Srivastav, S.; Narayan, V.; Paprzycki, M.; Jaworska, T.; Ganzha, M. A comprehensive review of deep neural networks for medical image processing: Recent developments and future opportunities. Healthc. Anal. 2023, 4, 100216. [Google Scholar] [CrossRef]
- Zaidi, S.S.A.; Ansari, M.S.; Aslam, A.; Kanwal, N.; Asghar, M.; Lee, B. A survey of modern deep learning based object detection models. Digit. Signal Process. 2022, 126, 103514. [Google Scholar] [CrossRef]
- Incoronato, M.; Aiello, M.; Infante, T.; Cavaliere, C.; Grimaldi, A.M.; Mirabelli, P.; Monti, S.; Salvatore, M. Radiogenomic analysis of oncological data: A technical survey. Int. J. Mol. Sci. 2017, 18, 805. [Google Scholar] [CrossRef]
- Wagner, M.W.; Namdar, K.; Biswas, A.; Monah, S.; Khalvati, F.; Ertl-Wagner, B.B. Radiomics, machine learning, and artificial intelligence—what the neuroradiologist needs to know. Neuroradiology 2021, 63, 1957–1967. [Google Scholar] [CrossRef]
- Mienye, I.D.; Sun, Y. A survey of ensemble learning: Concepts, algorithms, applications, and prospects. IEEE Access 2022, 10, 99129–99149. [Google Scholar] [CrossRef]
- Ho, L.M.; Samei, E.; Mazurowski, M.A.; Zheng, Y.; Allen, B.C.; Nelson, R.C.; Marin, D. Can Texture Analysis Be Used to Distinguish Benign From Malignant Adrenal Nodules on Unenhanced CT, Contrast-Enhanced CT, or In-Phase and Opposed-Phase MRI? Am. J. Roentgenol. 2019, 212, 554–561. [Google Scholar] [CrossRef]
- Schieda, N.; Krishna, S.; McInnes, M.D.F.; Moosavi, B.; Alrashed, A.; Moreland, R.; Siegelman, E.S. Utility of MRI to Differentiate Clear Cell Renal Cell Carcinoma Adrenal Metastases From Adrenal Adenomas. AJR Am. J. Roentgenol. 2017, 209, W152–W159. [Google Scholar] [CrossRef]
- Tu, W.; Abreu-Gomez, J.; Udare, A.; Alrashed, A.; Schieda, N. Utility of t2-weighted mri to differentiate adrenal metastases from lipid-poor adrenal adenomas. Radiol. Imaging Cancer 2020, 2, e200011. [Google Scholar] [CrossRef]
- Liu, J.; Xue, K.; Li, S.; Zhang, Y.; Cheng, J. Combined Diagnosis of Whole-Lesion Histogram Analysis of T1- and T2-Weighted Imaging for Differentiating Adrenal Adenoma and Pheochromocytoma: A Support Vector Machine-Based Study. Can. Assoc. Radiol. J. 2021, 72, 452–459. [Google Scholar] [CrossRef]
- Romeo, V.; Maurea, S.; Cuocolo, R.; Petretta, M.; Mainenti, P.P.; Verde, F.; Coppola, M.; Dell’Aversana, S.; Brunetti, A. Characterization of Adrenal Lesions on Unenhanced MRI Using Texture Analysis: A Machine-Learning Approach. J. Magn. Reson. Imaging 2018, 48, 198–204. [Google Scholar] [CrossRef]
- Xiao, D.X.; Zhong, J.P.; Peng, J.D.; Fan, C.G.; Wang, X.C.; Wen, X.L.; Liao, W.W.; Wang, J.; Yin, X.F. Machine learning for differentiation of lipid-poor adrenal adenoma and subclinical pheochromocytoma based on multiphase CT imaging radiomics. BMC Med. Imaging 2023, 23, 159. [Google Scholar] [CrossRef]
- Yi, X.; Guan, X.; Zhang, Y.; Liu, L.; Long, X.; Yin, H.; Wang, Z.; Li, X.; Liao, W.; Chen, B.T.; et al. Radiomics improves efficiency for differentiating subclinical pheochromocytoma from lipid-poor adenoma: A predictive, preventive and personalized medical approach in adrenal incidentalomas. EPMA J. 2018, 9, 421–429. [Google Scholar] [CrossRef]
- Torresan, F.; Crimì, F.; Ceccato, F.; Zavan, F.; Barbot, M.; Lacognata, C.; Motta, R.; Armellin, C.; Scaroni, C.; Quaia, E.; et al. Radiomics: A new tool to differentiate adrenocortical adenoma from carcinoma. BJS Open 2021, 5, zraa061. [Google Scholar] [CrossRef]
- Elmohr, M.M.; Fuentes, D.; Habra, M.A.; Bhosale, P.R.; Qayyum, A.A.; Gates, E.; Morshid, A.I.; Hazle, J.D.; Elsayes, K.M.; I, A.M.; et al. Machine learning-based texture analysis for differentiation of large adrenal cortical tumours on CT. Clin. Radiol. 2019, 74, 818.e1–818.e7. [Google Scholar] [CrossRef]
- Tu, W.; Verma, R.; Krishna, S.; McInnes, M.D.F.; Flood, T.A.; Schieda, N. Can Adrenal Adenomas Be Differentiated From Adrenal Metastases at Single-Phase Contrast-Enhanced CT? Am. J. Roentgenol. 2018, 211, 1044–1050. [Google Scholar] [CrossRef]
- Yi, X.; Guan, X.; Chen, C.; Zhang, Y.; Zhang, Z.; Li, M.; Liu, P.; Yu, A.; Long, X.; Liu, L.; et al. Adrenal incidentaloma: Machine learning-based quantitative texture analysis of unenhanced CT can effectively differentiate sPHEO from lipid-poor adrenal adenoma. J. Cancer 2018, 9, 3577–3582. [Google Scholar] [CrossRef]
- Feliciani, G.; Serra, F.; Menghi, E.; Ferroni, F.; Sarnelli, A.; Feo, C.; Zatelli, M.C.; Ambrosio, M.R.; Giganti, M.; Carnevale, A. Radiomics in the characterization of lipid-poor adrenal adenomas at unenhanced CT: Time to look beyond usual density metrics. Eur. Radiol. 2024, 34, 422–432. [Google Scholar] [CrossRef]
- Altay, C.; Başara Akın, I.; Özgül, A.H.; Adıyaman, S.C.; Yener, A.S.; Seçil, M. Machine learning analysis of adrenal lesions: Preliminary study evaluating texture analysis in the differentiation of adrenal lesions. Diagn. Interv. Radiol. 2023, 29, 234–243. [Google Scholar] [CrossRef]
- Barstugan, M.; Ceylan, R.; Asoglu, S.S.S.; Cebeci, H.; Koplay, M. Adrenal tumor characterization on magnetic resonance images. Int. J. Imaging Syst. Technol. 2020, 30, 252–265. [Google Scholar] [CrossRef]
- Stanzione, A.; Cuocolo, R.; Verde, F.; Galatola, R.; Romeo, V.; Mainenti, P.P.; Aprea, G.; Guadagno, E.; Caro, M.D.B.D.; Maurea, S. Handcrafted MRI radiomics and machine learning: Classification of indeterminate solid adrenal lesions. Magn. Reson. Imaging 2021, 79, 52–58. [Google Scholar] [CrossRef]
- Moawad, A.W.; Ahmed, A.; Fuentes, D.T.; Hazle, J.D.; Habra, M.A.; Elsayes, K.M. Machine learning-based texture analysis for differentiation of radiologically indeterminate small adrenal tumors on adrenal protocol CT scans. Abdom. Radiol. 2021, 46, 4853–4863. [Google Scholar] [CrossRef]
- Koyuncu, H.; Ceylan, R.; Asoglu, S.; Cebeci, H.; Koplay, M. An extensive study for binary characterisation of adrenal tumours. Med. Biol. Eng. Comput. 2019, 57, 849–862. [Google Scholar] [CrossRef]
- Andersen, M.B.; Bodtger, U.; Andersen, I.R.; Thorup, K.S.; Ganeshan, B.; Rasmussen, F. Metastases or benign adrenal lesions in patients with histopathological verification of lung cancer: Can CT texture analysis distinguish? Eur. J. Radiol. 2021, 138, 109664. [Google Scholar] [CrossRef]
- Singh, Y.; Kelm, Z.S.; Faghani, S.; Erickson, D.; Yalon, T.; Bancos, I.; Erickson, B.J. Deep learning approach for differentiating indeterminate adrenal masses using CT imaging. Abdom. Radiol. 2023, 48, 3189–3194. [Google Scholar] [CrossRef]
- Kusunoki, M.; Nakayama, T.; Nishie, A.; Yamashita, Y.; Kikuchi, K.; Eto, M.; Oda, Y.; Ishigami, K. A deep learning-based approach for the diagnosis of adrenal adenoma: A new trial using CT. Br. J. Radiol. 2022, 95, 20211066. [Google Scholar] [CrossRef]
- Bi, L.; Kim, J.; Su, T.; Fulham, M.; Feng, D.D.; Ning, G. Deep multi-scale resemblance network for the sub-class differentiation of adrenal masses on computed tomography images. Artif. Intell. Med. 2022, 132, 102374. [Google Scholar] [CrossRef]
- Robinson-Weiss, C.; Patel, J.; Bizzo, B.C.; Glazer, D.I.; Bridge, C.P.; Andriole, K.P.; Dabiri, B.; Chin, J.K.; Dreyer, K.; Kalpathy-Cramer, J.; et al. Machine Learning for Adrenal Gland Segmentation and Classification of Normal and Adrenal Masses at CT. Radiology 2023, 306, e220101. [Google Scholar] [CrossRef]
- Alimu, P.; Fang, C.; Han, Y.; Dai, J.; Xie, C.; Wang, J.; Mao, Y.; Chen, Y.; Yao, L.; Lv, C.; et al. Artificial intelligence with a deep learning network for the quantification and distinction of functional adrenal tumors based on contrast-enhanced CT images. Quant. Imaging Med. Surg. 2023, 13, 2675–2687. [Google Scholar] [CrossRef]
- Moore, B.E.; Corso, J.J. FiftyOne. GitHub. 2020. Available online: https://github.com/voxel51/fiftyone (accessed on 20 February 2025).
- Wu, Y.; Kirillov, A.; Massa, F.; Lo, W.Y.; Girshick, R. Detectron2. 2019. Available online: https://github.com/facebookresearch/detectron2 (accessed on 20 February 2025).
- Tian, Z.; Shen, C.; Chen, H.; He, T. FCOS: A simple and strong anchor-free object detector. IEEE Trans. Pattern Anal. Mach. Intell. 2020, 44, 69–76. [Google Scholar] [CrossRef]
- Guan, T.; Wang, Z.; Hu, W. Research on automatic detection algorithm for lumbar disc herniation based on FCOS. In Proceedings of the 2023 3rd International Conference on Computer Science, Electronic Information Engineering and Intelligent Control Technology (CEI), Wuhan, China, 15–17 November 2023; pp. 359–363. [Google Scholar] [CrossRef]
- Wang, Y.; Lin, X.; Zhang, X.; Ye, Q.; Zhou, H.; Zhang, R.; Ge, S.; Sun, D.; Yuan, K. Improved FCOS for detecting breast cancers. Curr. Med. Imaging Rev. 2022, 18, 1291–1301. [Google Scholar] [CrossRef]
- Lowekamp, B.C.; Chen, D.T.; Ibáñez, L.; Blezek, D. The design of SimpleITK. Front. Neuroinform. 2013, 7, 45. [Google Scholar] [CrossRef] [PubMed]
- van Griethuysen, J.J.M.; Fedorov, A.; Parmar, C.; Hosny, A.; Aucoin, N.; Narayan, V.; Beets-Tan, R.G.H.; Fillion-Robin, J.C.; Pieper, S.; Aerts, H.J.W.L. Computational radiomics system to decode the radiographic phenotype. Cancer Res. 2017, 77, e104–e107. [Google Scholar] [CrossRef]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).