2. Methods
This study aims to provide a comprehensive overview of AI tools used in the detection and diagnosis of NPH. The objective is to present a detailed analysis of relevant studies that utilize a wide range of methods, specifically focusing on segmentation, treatment monitoring, confirmation, and disease progression. We sought to analyze the relevance and impact of each work, presenting a comprehensive summary of its contribution to the field.
Figure 1 summarizes the procedure used in this study. This review follows the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines.
2.1. Literature Search
To gather reliable information on studies related to the diagnosis, detection, prediction, or evolution of NPH, we conducted an extensive search of high-quality databases. We utilized PubMed, Scopus, ACM Digital Library, and IEEEXplore, which are known for their comprehensive coverage of biomedical, life sciences, engineering, and technology research. The databases were searched from inception to 20 February 2025.
2.2. Search Query
We employed a carefully crafted search query consisting of fifteen specific keywords essential for understanding NPH and its diagnosis: “Normal pressure hydrocephalus”, “NPH”, “diagnosis”, “detection”, “Machine learning”, “ML”, “Deep Learning”, “DL”, “Feature Generation”, “segmentation”, “Magnetic Resonance Imaging”, “MRI”, “Computed Tomography Scan”, “CT scan”, and “image analysis”. To ensure a comprehensive search, we considered the full article, including the title, abstract, and body. We used the logical AND, OR function to account for variations in synonyms and abbreviations, without relying on quotation marks, as follows:
(“Normal Pressure Hydrocephalus” OR “NPH”) AND (“Computed Tomography” OR “CT Scan” OR “Magnetic Resonance Imaging” OR “MRI”) AND (“Machine Learning” OR “ML” OR “deep learning” OR “DL” OR “Feature generation” OR “risk assessment” OR “safety-critical systems”) AND (“Diagnosis” OR “Detection” OR “Segmentation” OR “clinical validation”) AND Date: (2010/1/1:2025/02/20)
2.3. Filtering and Selection
Data extraction was performed independently by two reviewers (L.R.M.-D. and G.X.G.) using a standardized data extraction form developed a priori. The form was pilot-tested on three randomly selected included studies and refined accordingly. Extracted data were cross-checked for accuracy, with any discrepancies resolved through discussion. When necessary, information was unclear or missing from published reports; we attempted to contact study authors via email with a maximum of three attempts over a four-week period.
After obtaining the initial results, we meticulously reviewed each article, discarding those that did not meet the predefined criteria or were unrelated to the specific analysis methods or application areas of interest. The selection criteria used for article filtering included the following:
The article must have been published in a scientific database.
The article must have been an empirical study, research article, or case study.
The article must have focused on AI techniques for NPH detection and diagnosis using MRI or CT scans.
The article must have included sufficient methodological details.
Articles that were solely bibliographic reviews or that did not meet all the above criteria were excluded from the study. The purpose of these criteria was to ensure that the articles included in the study were high-quality, empirical studies that could provide valuable insights into the use of AI techniques for NPH detection and diagnosis. Excluding articles that were not empirical studies, bibliographic reviews, or case studies, the authors were able to ensure that the articles included in the study were based on real-world data and that they provided concrete examples of how AI techniques could be used to improve NPH detection and diagnosis. We also excluded articles that did not include sufficient methodological details. Methodological details allow other researchers to replicate the results of a given study.
Two independent reviewers (L.R.M.-D. and H.F.P.-Q.) screened all titles and abstracts identified from the database searches against the predefined eligibility criteria. Full-text articles of potentially relevant studies were then retrieved and independently assessed by the same reviewers. Any discrepancies at either stage were resolved through discussion, with a third reviewer (N.P.) consulted when consensus could not be reached. The inter-rater agreement was calculated using Cohen’s kappa coefficient, yielding substantial agreement (κ = 0.82).
Due to the anticipated heterogeneity in AI methodologies, imaging modalities, outcome measures, and study populations, we planned a priori to explore potential sources of heterogeneity through narrative analysis rather than statistical methods. We specifically examined variability across the following domains: (1) AI approach (ML vs. DL vs. hybrid); (2) imaging modality (MRI vs. CT); (3) control group composition (healthy controls vs. disease-specific controls); (4) key radiological features utilized; and (5) sample size and population characteristics. These factors were systematically compared across studies to identify patterns that might explain differences in diagnostic performance.
Given the heterogeneity of the included studies and our narrative synthesis approach, formal sensitivity analyses were not performed. However, we conducted a qualitative assessment of the robustness of findings by examining how results varied across studies with different methodological quality characteristics, particularly focusing on validation methods (e.g., studies using external validation vs. cross-validation only) and sample size (larger vs. smaller studies).
To assess the certainty of evidence for key outcomes, we adapted the GRADE (Grading of Recommendations, Assessment, Development, and Evaluations) approach for diagnostic accuracy studies to the AI context. For each main comparison (e.g., ML vs. DL approaches, MRI vs. CT-based methods), we considered the following domains: risk of bias (using our modified QUADAS-2 assessment), inconsistency (unexplained heterogeneity in results), indirectness (applicability of findings to the review question), imprecision (sample size and confidence intervals when reported), and publication bias. The certainty of evidence was categorized as high, moderate, low, or very low, reflecting our confidence that the true effect lies close to the estimated effect.
Our review has several methodological limitations. First, despite our comprehensive search strategy, we may have missed relevant studies, particularly those published in non-indexed journals or in languages other than English. Second, our assessment of the risk of bias was hampered by incomplete reporting in many of the included studies, particularly regarding patient selection and reference standards. Third, the rapid evolution of AI techniques means that newer approaches may not be adequately represented in our review. Finally, our inability to conduct a meta-analysis due to heterogeneity limits our ability to provide precise estimates of diagnostic accuracy for different AI approaches.
2.4. Article Selection
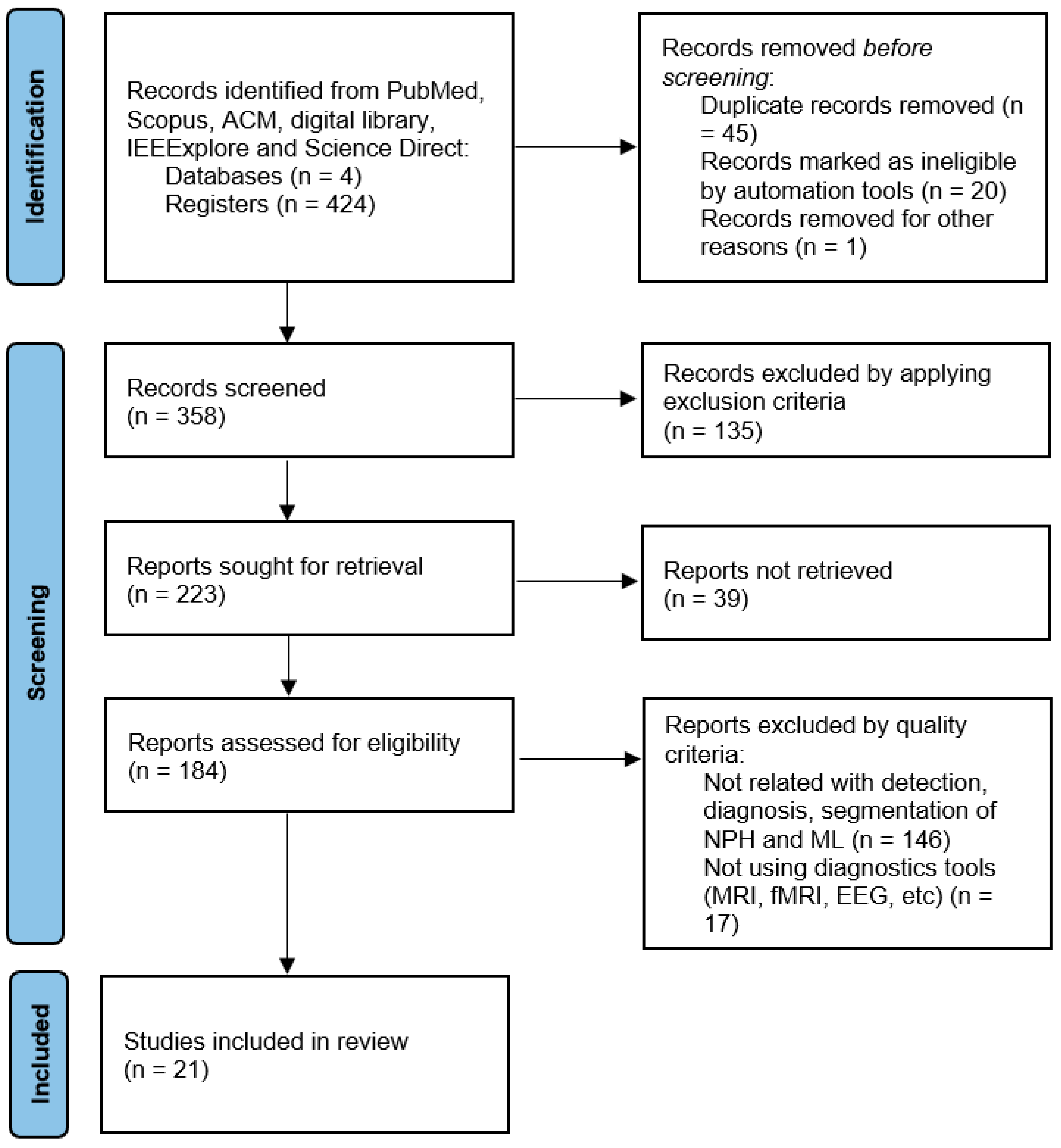
The search results from different databases yielded varying numbers of articles. Scopus returned a total of 189 articles, and after removing the review articles, 102 articles remained. After filtering out articles that did not meet the defined keywords, the number was reduced to ten articles. In the case of ScienceDirect, forty articles were obtained, and after removing the reviews, two articles remained. PubMed initially had eleven articles that met the criteria, but upon excluding the review articles, there were 6 articles remaining. ACM returned 148 articles, and after removing the review articles, 104 articles remained. Further examination of these articles led to the identification of 2 articles that contained relevant information associated with the search criteria. Similarly, IEEExplore returned 36 articles, and after the review filter, 17 articles were left, and ultimately, only 1 article met all the inclusion criteria.
Figure 2 illustrates the entire search approach followed in this study, how we located 21 articles to review, and the trend analysis [
31].
2.5. Risk of Bias
To assess the risk of bias in the included studies, we employed a multi-faceted evaluation framework tailored to AI applications in medical imaging. Each study was systematically assessed for methodological quality and potential sources of bias using criteria adapted from the QUADAS-2 (Quality Assessment of Diagnostic Accuracy Studies) tool, modified to address the specific requirements of AI-based diagnostic studies. Our assessment focused on four key domains: patient selection, index test (AI algorithm), reference standard, and flow and timing.
We evaluated each study for potential selection bias by examining inclusion/exclusion criteria, patient recruitment methods, and demographic representation. Technical bias was assessed by examining model architecture description completeness, input data preprocessing, feature selection justification, and hyperparameter optimization approaches. For validation rigor, we examined whether studies utilized appropriate cross-validation techniques, independent test sets, or external validation cohorts. Additionally, we assessed reporting transparency, including whether studies provided a comprehensive performance metrics, confidence intervals, and limitations of the AI algorithms. Studies with insufficient methodological details, inadequate validation protocols, or potential confounding factors were noted as having a higher risk of bias.
2.6. Synthesis of Results
We employed a narrative synthesis approach to analyze and present the findings from the included studies, such as the heterogeneity in AI methodologies, imaging modalities, and outcome measures precluded a formal meta-analysis. The results were synthesized and organized into categories based on the AI methodology employed (ML vs. DL), diagnostic tasks performed (classification, segmentation, and feature extraction), and imaging modalities utilized (MRI, CT).
For each category, we systematically extracted and tabulated key information, including study design, sample size, participant characteristics, AI algorithm specifications, radiological features utilized, performance metrics, and limitations. The performance results were presented using standardized metrics where possible (accuracy, sensitivity, specificity, ROC-AUC, and Dice coefficients) to facilitate cross-study comparisons. To enhance interpretability and highlight patterns across studies, we developed comparative tables detailing the control groups used, specific radiological features examined, and feature/radiomics approaches employed. This structured synthesis enabled comprehensive evaluation of the current state of AI applications for NPH diagnosis while identifying methodological trends, knowledge gaps, and promising directions for future research.
The methodology was established a priori by the review team based on the PRISMA 2020 guidelines and standard systematic review methodology for diagnostic test accuracy studies. The data extraction template used in this review and the complete dataset of extracted information from included studies are available upon reasonable request from the corresponding author. No specialized analytic code was required for this narrative synthesis.
3. Results
We identified, reviewed, and analyzed a total of twenty-one relevant articles for this study. The articles were selected based on our search methodology and contributed significantly to understanding NPH diagnosis using AI techniques.
Table 1 presents a comprehensive summary of all included studies, their methodologies, and key performance metrics.
Our analysis revealed distinct methodological distributions among the studies: eleven articles (52.4%) employed machine learning (ML) methods, including LogReg [
32,
33,
34,
35,
36], SVM [
37,
38], random forest (RF {XE “RF” \t “
Random Forest”}) [
30,
37,
39,
40,
41,
42], XGBoost (XGB { XE “XGB” \t “
xgBoost”}) [
37,
42], multilayer perceptron (MLP {XE “MLP” \t “
multilayer perceptron”}) [
42], gaussian naive bayes (GaussNB { XE “GaussNB” \t “
Gaussian Naive Bayes”}) [
42], adaptive boosting (AdaBoost { XE “AdaBoost” \t “
Adaptive Boosting”}) [
42] and other ML methods. Seven articles used DL methods, with deep convolutional network (DCNNs {XE “DCNN” \t “
Deep Convolutional Network”}) [
27,
28,
30,
39,
43,
44,
45] being the most common (used in six studies or 28.6% of the total). Notably, three studies (14.3%) implemented hybrid methodologies combining both ML and DL techniques, indicating an emerging trend toward integrated approaches.
The distribution of AI methods reflects the evolving landscape of computational approaches for NPH diagnosis—from traditional ML algorithms that rely on predefined features to more advanced DL methods capable of automatic feature extraction. This methodological diversity underscores the complexity of NPH diagnoses and the varied approaches researchers are employing to address this challenge.
Table 1.
Summary of AI methods and performance for NPH detection.
Table 1.
Summary of AI methods and performance for NPH detection.
| Study | AI Method | Task | Dataset | Performance Metrics | Key Features |
|---|
| Traditional ML Methods |
| Rau et al. (2021) [39] | SVM | Classification (NPH vs. HC) | MRI scans | Accuracy: 0.93, ROC-AUC: 0.99 | Periventricular regions, lateral ventricles, Sylvian fissures |
| Xu et al. (2022) [38] | ANN, RF, SVM, XGB | Classification (NPH vs. HC) | MRI scans | ROC-AUC: 0.96 ± 0.05 (ANN), 0.96 ± 0.06 (RF), 0.94 ± 0.05 (SVM), 0.94 ± 0.07 (XGB) | Evans ratio, frontal horns’ length |
| Bianco et al. (2022) [41] | LogReg | Classification (NPH vs. AD) | MRI | Accuracy: 89.6–94.3%, ROC-AUC: 0.96–0.99 | Evans index, callosal angle, disproportionate sulci, volumetrics |
| Vlasak et al. (2022) [43] | RF, RF + 3D MCV, RF + MGAC | Brain segmentation | CT scans | Ventricles: 84%, Gray-white matter: 87%, Subarachnoid space: 35% | MRI phase contrast features |
| Iida et al. (2021) [34] | LogReg | Analysis of NPH | MRI | Accuracy not reported | Parkinsonism subtype, midbrain dimensions |
| Zhang et al. (2022) [40] | RF, RF + 3D MCV, RF + MGAC | Brain segmentation | CT scans | Ventricles: 84%, Gray-white matter: 87%, Subarachnoid space: 35% | Ventricles, Gray-white matter, Subarachnoid space |
| Galeano et al. (2011) [46] | Correlation-based feature selection | Classification | ICP signals, CT | Accuracy: 85.7% | Skewness of Single-Wave Amplitude, P1 subpeak amplitude, Leading Edge Slope |
| Griffa et al. (2022) [36] | Discriminant analysis | Classification | Phase-contrast MRI | Accuracy: 58–77% (controls), 16–84% (CVD), 11–75% (NPH) | CSF flow/velocity at aqueduct of Sylvius |
| Deep Learning Methods |
| Irie et al. (2020) [29] | DCNN | NPH classification | MRI | Accuracy: 99.1%, Sensitivity: 98.5%, Precision: 98.2% | Color-based transformation features |
| Mao et al. (2022) [45] | DCNN | Hydrocephalus classification | MRI | Accuracy: 99.1%, Sensitivity: 98.5%, Precision: 98.2% | Color-based transformation features |
| Tsou et al. (2021) [44] | MultiResUNet, UNet | ROI segmentation | Phase-contrast MRI | DSC: 0.95–0.96, ICC: 0.99, Pearson: 0.99 | Ventricular volume, intracranial volume |
| Haber et al. (2022) [28] | DCNN | NPH detection | CT | Sensitivity: 100%, Specificity: 89%, ROC-AUC: 0.96 | CT image features |
| Rudhra et al. (2021) [31] | DCNN, MultiResUNet, UNet | ROI segmentation | MRI | DSC: 0.933, ICC: 0.95, Pearson: 0.95 | Watershed segmentation features |
| Hybrid Methods |
| Rudhra et al. (2021) [31] | DCNN + traditional ML segmentation | Hydrocephalus classification | MRI | DSC: 0.87–0.93 | Feature maps from ML segmentation |
| Zhang et al. (2022) [40] | RF + MGAC | Segmentation | CT | Ventricles: 84%, Gray-white matter: 87%, Subarachnoid space: 35% | Ventricles, gray-white matter, subarachnoid space |
3.1. ML Methods
Traditional ML methods for NPH detection can be categorized into three main functional areas: classification approaches for differential diagnosis, segmentation and feature analysis techniques, and key feature identification. This organizational structure reflects the progressive stages of ML application in NPH diagnosis: first establishing accurate classification between NPH and other conditions, then developing methods to segment and analyze relevant brain structures and finally identifying the most discriminative features for diagnosis.
The classification approaches (
Section 3.1.1) address the clinical challenge of differentiating NPH from conditions with similar presentations, such as Alzheimer’s disease and Parkinson’s disease, as well as from healthy controls. These methods employ supervised learning algorithms trained on labeled datasets to establish decision boundaries between diagnostic categories. We examine these approaches based on their comparative diagnostic targets (healthy controls, AD, PSP, or multiple conditions), which represent different clinical scenarios with varying levels of diagnostic complexity.
The segmentation and feature analysis techniques (
Section 3.1.2) focus on isolating and quantifying relevant brain structures and CSF dynamics. These methods address the technical challenge of accurately delineating ventricles, brain parenchyma, and subarachnoid spaces—crucial regions for NPH diagnosis—and extracting meaningful measurements from these segmentations. We analyze these approaches based on their anatomical targets and technical methodologies. The key features section (
Section 3.1.3) synthesizes findings across studies to identify the most diagnostically valuable measurements and biomarkers. This analysis helps establish which parameters should be prioritized in future ML model development for NPH diagnosis and provides insight into the underlying pathophysiological mechanisms of the condition. Together, these three areas provide a comprehensive view of traditional ML applications in NPH detection, from the clinical challenge of differential diagnosis to the technical challenges of image analysis and feature extraction.
3.1.1. Purpose-Based Classification
NPH vs. Healthy Controls
In a study by employed an SVM to detect NPH patterns in MRI scans, achieving an impressive accuracy of 0.93 and an ROC-AUC of 0.99 [
38]. These metrics represent exceptional discriminative capability that approaches the performance of specialized neuroradiologists, highlighting the potential of ML algorithms to serve as reliable diagnostic support tools for NPH detection. The algorithm demonstrated the highest discriminative power in specific brain regions, including periventricular regions, lateral ventricles, and Sylvian fissures—areas known to be crucial for NPH diagnosis in clinical practice. This alignment between the algorithm’s focus and established radiological markers validates the clinical relevance of the approach.
Their results suggest that MRI morphometric and advanced image processing may effectively capture hallmarks of NPH useful for developing AI systems, especially when integrated into a multiparametric approach. However, the complexity of NPH means that simply using common measures and segmentation algorithms may be insufficient; determining optimal diagnostic signatures will require systematically investigating which feature combinations provide the greatest discriminatory power.
In the same study the results suggest MRI morphometric and advanced image processing may capture hallmarks of NPH useful for developing AI, especially when fused into a multiparametric approach [
38]. But the complexity of NPH means simply using common measures and segmentation algorithms may be insufficient; determining optimal signatures for diagnosis and monitoring will require systematically investigating which features, in which combinations, provide the greatest discriminatory power.
In another study developed multiple ML models to explore the effectiveness of commonly used morphological parameters for NPH diagnosis [
37]. Their feature importance analysis revealed the most weighted diagnostic features were Evans ratio (24.79%), frontal horns’ length (10.64%), and disproportionately enlarged subarachnoid space hydrocephalus—quantifying for the first time the relative importance of these clinical markers. When comparing model performance, the artificial neural network achieved the highest ROC-AUC of 0.96 ± 0.05, marginally outperforming random forest (0.96 ± 0.06), SVM (0.94 ± 0.05), and XGBoost (0.94 ± 0.07). The minimal performance differences between these diverse algorithms (all within 2 percentage points) suggests that feature selection may be more critical than algorithm choice for accurate NPH diagnosis.
Another team implemented a multinomial LogReg algorithm to predict probabilities across three different diagnoses: healthy elderly controls, NPH, and Alzheimer’s disease (AD) [
32]. This approach represents an important advance over binary classifiers, addressing the clinical reality where differential diagnosis across multiple conditions is often required. Their model demonstrated exceptional classification accuracy of 96.3%, with only two misclassifications out of 15 NPH cases. This impressive performance indicates that even relatively simple ML algorithms, when provided with appropriate features, can effectively differentiate between clinically similar neurodegenerative conditions. The model’s ability to provide confidence values for each case (averaging 97% for NPH diagnoses) offers an additional dimension valuable for clinical decision support. The high sensitivity (100%) and specificity (87%) in the binary LogReg model further confirm the robustness of this approach for NPH-AD differentiation.
NPH vs. AD
Another study proposed a comprehensive approach using FreeSurfer for brain segmentation followed by logistic regression trained on multiple MRI markers [
36]. Their systematic integration of multiple imaging features—including Evans index, callosal angle, disproportionate sulci, and volumetrics—yielded models with high accuracy (89.6–94.3%) and ROC-AUC values (0.96–0.99). The remarkably high-performance metrics suggest that integrating multiple complementary features can substantially improve diagnostic accuracy compared to single-feature approaches. However, as the authors acknowledge, the limited sample size and exploratory nature of the study necessitate additional validation before clinical translation. This limitation is common across many studies in our review and highlights the importance of larger multicenter validation studies before implementing these promising AI approaches in clinical practice.
NPH vs. Progressive Supranuclear Palsy
In a recent study utilized ML algorithms to differentiate between NPH and progressive supranuclear palsy based on cortical thickness and volumetric data [
40]. Using RF, they achieved a ROC-AUC of 0.96 for distinguishing NPH from healthy controls, indicating excellent discriminative performance. This high performance is particularly valuable considering the clinical challenge of differentiating these conditions, which can present with similar movement disorders. Their finding of more severe and widespread cortical involvement in NPH compared to PSP could be attributed to the marked lateral ventricular enlargement characteristic of NPH—providing both a diagnostic marker and insight into disease mechanisms. This study effectively demonstrates how ML can not only improve diagnosis but also advance understanding of disease pathophysiology.
NPH vs. Multiple Conditions (AD vs. Healthy Controls)
This team conducted a comparative analysis of eight state-of-the-art ML algorithms applied to MRI features from 30 NPH patients and 15 healthy controls [
40]. Their systematic performance comparison revealed a clear hierarchy among algorithms: AdaBoost and XGBoost demonstrated the highest accuracy (80.4% and 79.6%, respectively), while MLP showed the lowest (72.0%). This 8.4 percentage point performance gap between algorithms, despite using identical features and validation methodology, highlights the importance of algorithm selection for NPH classification. Notably, ensemble methods (AdaBoost, XGBoost, Random Forest) consistently outperformed single-model approaches, suggesting that combining multiple predictive models may better capture the complex patterns associated with NPH. The relatively modest accuracy across all methods (below 85%) also reflects the inherent difficulty of NPH diagnosis, even with advanced ML techniques.
In a study investigated ventriculomegaly and NPH-like features in patients with myotonic dystrophy type 1 [
33]. Their binomial LogReg analysis revealed that while age, gender, and CTG repeat numbers did not significantly affect the z-Evans Index, callosal angle and pathological brain atrophy showed significant associations (Adjusted Odds Ratio of 1.0, 95% CI,
p < 0.01). These findings indicate that specific morphological features, rather than demographic or genetic factors, are most closely associated with ventriculomegaly in this patient population. The lack of association between enlarged perivascular spaces and the z-Evans Index suggests that not all radiological findings common to NPH have equal diagnostic value across different patient populations—an important consideration for developing robust AI diagnostic tools.
Other group measured various morphometric parameters and examined their association with neuropsychological tests in NPH patients [
34]. Their LogReg analysis identified significant discriminative factors including Parkinsonism subtype, midbrain anteroposterior diameter, and midbrain tegmentum diameter—highlighting the importance of considering both morphological and clinical characteristics. Their finding that frontal horn length alone achieved 95.0% accuracy for predicting NPH, while combining frontal horn length and callosal angle improved accuracy to 96.3%, demonstrates the potential benefits of feature combinatorics for NPH diagnosis. Interestingly, adding additional parameters (midbrain longitudinal length, Evans ratio, frontal horn ratio, and biparietal diameter) did not substantially improve accuracy, suggesting that feature selection should prioritize quality over quantity. This “less is more” principle could guide more efficient and generalizable AI model development for NPH.
3.1.2. Segmentation and Feature Analysis
Brain Region Segmentation
In the study, developed an automated segmentation and connectivity analysis method for identifying brain regions relevant to NPH diagnosis [
39]. Their comparative analysis of multiple algorithms revealed substantial performance differences: their proposed method achieved segmentation accuracies of 84% for ventricles, 87% for gray-white matter, and 35% for subarachnoid space, significantly outperforming traditional techniques. The notably poor performance for subarachnoid space segmentation (35% accuracy) across all methods highlights a persistent technical challenge in NPH imaging analysis, as this region is crucial for diagnosing disproportionately enlarged subarachnoid space hydrocephalus (DESH). The substantial performance gap between ventricle/gray-white matter segmentation (84–87%) and subarachnoid space segmentation (35%) indicates that future methodological development should focus specifically on improving this challenging aspect of NPH-related image analysis.
Morphometric Parameter Evaluation
In another study employed a RF algorithm to predict NPH and evaluate six commonly used morphometric parameters [
41]. Their feature importance analysis revealed that frontal horn length alone achieved 95.0% accuracy, the highest among all individual parameters. Surprisingly, combining all six parameters decreased accuracy to 89.3%, demonstrating that more parameters do not necessarily improve performance. This counterintuitive finding challenges the common assumption that incorporating more features improves ML performance and highlights the risk of overfitting when using redundant or irrelevant parameters. Their results suggest that ML algorithms for NPH diagnosis should be trained on carefully selected informative features rather than broadly incorporating all available measurements. This principle of parsimonious feature selection could improve both model accuracy and generalizability.
CSF Dynamics Analysis
In recent study conducted an intensive investigation on intracranial pressure signals and CT scans using correlation-based feature selection [
47]. Their method achieved 85.7% classification accuracy (12 of 14 patients correctly classified) using just three features: Skewness of Single-Wave Amplitude, Skewness of P1 subpeak absolute amplitude, and Skewness Leading Edge Slope. The high accuracy achieved with this minimal feature set demonstrates the diagnostic power of CSF pressure dynamics for NPH classification. The method showed higher specificity (88.89% for non-NPH patients) than sensitivity (80% for NPH patients), indicating it may be more reliable for ruling out than confirming NPH. While limited by small sample size (14 patients), these promising results suggest that incorporating CSF pressure dynamics into AI diagnostic systems could complement traditional imaging-based approaches.
Griffa et al. assessed the cerebrospinal fluid tap test as a prognostic tool for shunt surgery in NPH patients [
36]. Their comparison of different predictive models revealed a clear advantage for multimodal approaches: the combined clinical + imaging model substantially outperformed both clinical-only and imaging-only models (out-of-sample accuracy: 0.70 vs. 0.57/0.63, ROC-AUC: 0.83 vs. 0.54/0.59). The statistically significant improvement (
p = 0.028) achieved by combining modalities demonstrates the value of integrating diverse data types for NPH prediction. This finding has important implications for AI development, suggesting that models incorporating both imaging and clinical data will likely achieve higher diagnostic performance than those limited to a single data type. The impressive performance metrics of their integrated model (ROC-AUC of 1.0 [0.99–1.0] on the whole dataset) further supports the potential of sophisticated multimodal ML approaches for NPH management.
Phase-Contrast MRI Analysis
In this work they used phase-contrast MRI to diagnose NPH and differentiate it from similar disorders [
46]. Their discriminant analyses of CSF flow parameters showed substantial variability in classification accuracy: controls were classified with moderate accuracy (58–77%), while cerebrovascular disease and NPH patients showed lower accuracy (16–84% and 11–75%, respectively). The wide performance ranges indicate that some parameters are substantially more informative than others for diagnosis. The relatively poor performance for NPH classification (as low as 11% for some parameters) highlights the challenge of relying solely on phase-contrast MRI for definitive diagnosis. Their findings suggest that while phase-contrast MRI provides valuable information, it should be integrated with other imaging and clinical parameters rather than used in isolation for NPH diagnosis.
3.1.3. Key Features in ML Methods
Morphological parameters have proven to be robust indicators for NPH diagnosis across multiple studies. The callosal angle has emerged as a particularly reliable marker, with [
33] demonstrating its significant association with the z-Evans Index in their analysis. Frontal horn length has shown remarkable diagnostic accuracy, with [
37] reporting 95.0% accuracy using this parameter alone [
41]. Midbrain dimensions, including anteroposterior and tegmentum diameter, have been identified as significant differentiating factors by [
34]. The presence of disproportionately enlarged subarachnoid space hydrocephalus (DESH) has also been recognized as a key diagnostic feature, ranking among the top three most weighted diagnostic features in recent studies.
Clinical parameters have provided valuable complementary information to imaging features in NPH diagnosis. CSF pressure measurements, analyzed through techniques such as infusion tests, have shown significant diagnostic potential, with [
47] achieving 85.7% classification accuracy using pressure-derived features. Neuropsychological tests have been effectively incorporated into diagnostic models, with [
35] demonstrating improved accuracy when combining these assessments with imaging features. Their combined clinical and imaging model achieved an impressive ROC-AUC of 0.83, significantly outperforming models using either type of feature alone. Gait assessment, while less commonly incorporated into ML models, has shown promise as an additional diagnostic parameter, particularly when combined with other clinical and imaging features [
34,
35].
3.2. DL Methods
Deep learning approaches for NPH detection generally operate without explicitly defined features, instead automatically extracting relevant patterns from imaging data. Irie et al. developed a fully automated 3D DCNN using whole-brain T1-weighted MRI from a balanced dataset of 23 NPH patients, 23 AD patients, and 23 controls [
29]. Their approach achieved high diagnostic accuracy of 0.90, successfully classifying 21/23 NPH cases, 19/23 AD cases, and 22/23 controls. The model’s sensitivity and specificity for NPH were both 0.91, demonstrating balanced performance across diagnostic categories. Their innovative use of Gradient-weighted Class Activation Mapping (Grad-CAM) revealed that the model focused on diagnostically relevant regions: the brain parenchyma surrounding the lateral ventricle for NPH cases and the medial temporal lobe for AD cases. This alignment between the model’s attention areas and known disease-specific regions provides interpretability and validates the biological relevance of the DL approach.
Architecture Types
While [
28] aimed to classify among three neurodegenerative disorders, [
45] focused more on obtaining or classifying distinct types of NPH, including NPH. They aimed to use DL algorithms to eliminate steps like pre-processing, segmentation, feature extraction, and classification. A color-based transformation technique is used for better processing of input tested images. Then, these pre-processed images are segmented by mean shift clustering, which is used to segment the image and provide a reliable and accurate estimated value. Then the features are extracted using Complete Local Binary Pattern. Finally, classification used DCNN with Emperor Penguin Optimization (EPO) for improving the system efficiency, with this optimization step being an additional step in the way of DL. The developed model achieves an accuracy of approximately 99.1%, sensitivity of approximately 98.5%, and precision value of approximately 98.2%, respectively. Additionally, the average training and validation accuracy of the system is 84.75% and 87.25%, to differentiate between the different types of hydrocephalus. Reference [
47] focused on developing an automated deep learning system to diagnose different hydrocephalus conditions. The study presented promising results in terms of classification performance and effectiveness.
We found that 3D CNN is commonly used in the field. The study by Mao et al. analyzed the relationship between cerebrospinal fluid variation and NPH in patients with different brain injuries. They used a 3D DCNN approach for pattern recognition and image classification. They collected MRIs from different brain damaged patients. A DL-based DCNN model was used to preprocess the image features, and the offline training and online reconstruction were conducted after the construction of the model. Four databases were selected for deep learning and analyzed using principal component analysis and 3D scale-invariant feature transformation. The weighted histogram of gradient orientation descriptor and ROC-AUC scores were the highest. Scale-invariant feature transformation, principal component analysis, and weighted histogram of gradient orientation had sensitivities of 87.5%, 88.2% and 90.1%, respectively, and specificities of 91.8%, 90.1%, and 94.2%, respectively. weighted histogram of gradient orientation also had the highest correct rate at 92.4%. The CSF volume in the subarachnoid space was 77.04% higher than that in the ventricles [
45].
3D CNNs offer significant advantages over 2D approaches for NPH diagnosis by capturing the full volumetric context of brain structures. Unlike 2D CNNs that process slices independently, 3D CNNs simultaneously model spatial relationships in all three dimensions (axial, sagittal, and coronal), preserving critical volumetric information about ventricular morphology and CSF distribution. This three-dimensional context is particularly important for NPH diagnosis, where the spatial relationship between ventricles, subarachnoid spaces, and surrounding brain tissue is diagnostically relevant. The 3D convolution operations extract features that represent spatial patterns across adjacent voxels in all directions, enabling more comprehensive analysis of subtle morphological changes that might be missed in slice-by-slice 2D analysis. This advantage was demonstrated in [
44] study, where 3D DCNN captured complex relationships between CSF volume in the subarachnoid space relative to ventricles (77.04% higher), a volumetric biomarker that would be difficult to quantify accurately with 2D approaches.
In another study they analyzed the relationship between CSF variation and hydrocephalus in patients with different cerebral injuries [
44]. They collected MRI scans from brain damaged patients and adopted a DCNN model to preprocess image features. They then conducted offline training and online reconstruction after constructing the model. Four databases were selected for DL analysis using principal component analysis and 3D scale-invariant feature transformation. The results showed that the weighted histogram of gradient orientation descriptor and ROC-AUC score were the highest. Scale-invariant feature transformation, principal component analysis and weighted histogram of gradient orientation achieved sensitivities of 87.5%, 88.2%, and 90.1%, respectively, while specificities were 91.8%, 90.1%, and 94.2%, respectively. The weighted histogram of gradient orientation method also had the highest correct rate at 92.4%. The study found that the CSF volume in the subarachnoid space was 77.04% higher than that in the ventricles. This study presented a DCNN approach to analyzing CSF variation in patients with brain injuries and hydrocephalus. The researchers utilized various image processing and classification techniques, finding that the weighted histogram of gradient orientation descriptor achieved the highest performance in differentiating subarachnoid fluid from ventricular fluid. The results suggest the effectiveness of DL algorithms like DCNNs for diagnosing hydrocephalus using MRI.
Another work using DL methods did not use DCNN; instead, they used transfer learning, an approach that allows to repurpose pre-trained DL models. An study used brain CT and MRI images from 143 NPH patients, which were manually labeled with ventricular volume and intracranial volume [
44]. A multilabel segmentation model handled both thick-slice CT and MRI images, addressing domain shift caused by distinctive image distributions. The encoder used a pre-trained ResNet34 architecture pretrained on ImageNet to extract features. During training, the objective function incorporated cross-entropy loss for thick-slice images and entropy loss for thin-slice images. Integrating the pre-trained ResNet34 encoder allowed the network to effectively learn textual and shape priors during initial training. The results showed the suitability of the AI-based method for accurate automatic measurement of ventricular volume in NPH patients. Statistical evaluations achieved Dice similarity coefficient of 0.95 for ventricular volume, intraclass correlation of 0.99, and Pearson correlation of 0.99. For intracranial volume, Dice similarity coefficient was 0.96, intraclass correlation was 0.99, and Pearson correlation was 0.99. Bland–Altman analysis indicated minimal bias between automatic and manual segmentations. These findings highlight the potential of ResNet-based DL approaches as an alternative analysis method for NPH, providing reliable measurements for clinical diagnosis and treatment.
In a work researchers proposed using phase-contrast MRI and CSF flow quantification across the cerebellar aqueduct [
43]. Two radiologists manually performed region of interest studies. Their proposed MultiResUNet and UNet DCNN algorithms trained based on the region of interest, representing the first major difference from previous work as this aimed to calculate region of interest for segmentation rather than classify disorders (NPH, AD, healthy control). Another difference was dataset division into 80% training and 20% validation sets. Segmentation was performed by calculating Dice similarity coefficients for manual and DCNN-derived region of interest. MultiResUNet, UNet and the second radiologist (Rater 2) had Dice similarity coefficients of 0.933, 0.928, and 0.867, respectively, with
p < 0.001 between DCNN and Rater 2. Comparing CSF flow parameters showed excellent intraclass correlation coefficients for MultiResUNet, with the lowest being 0.67. For UNet, lower intraclass correlation coefficients of -0.01 to 0.56 were observed. Only 3/353 (0.8%) studies failed to have appropriate region of interest placed by MultiResUNet, versus 12/353 (3.4%) failed cases for UNet.
Several DL algorithms were compared in terms of their performance in classifying between PD and NPH. The ResNet34 transfer learning model achieved a sensitivity of 93.6%, specificity of 94.4%, and ROC-AUC of 93%. Transfer learning using DCNN segmentation achieved a Dice coefficient of 87%, a metric that measures the similarity between sets. The RUDOLPH model obtained a higher Dice coefficient of 0.93. Additionally, the FreeSurfer and MALP-EM model achieved Dice coefficients of 0.72 and 0.90, respectively. The proposed methodology in the study combined Watershed segmentation with a convolutional neural network. This achieved promising results with 97% accuracy, 100% specificity, 96% sensitivity and 95% precision [
30].
A work compared multiple segmentation approaches for CT-based analysis, including both traditional ML and DL methods [
39]. Their comprehensive performance comparison demonstrated a clear hierarchy: 3D UNet + Probabilistic Maps (ventricles: 85 ± 0%, gray-white matter: 94 ± 1%, subarachnoid space: 72 ± 5%) and standard 3D UNet (ventricles: 85 ± 7%, gray-white matter: 93 ± 1%, subarachnoid space: 69 ± 13%) substantially outperformed RF-based approaches (ventricles: 65 ± 12%, gray-white matter: 87 ± 2%), particularly for ventricle segmentation. The dramatic performance gap between the best and worst methods (ranging from 85% to 13% for ventricles) emphasizes the critical importance of methodological selection for NPH-related image analysis. The consistently poor performance for subarachnoid space segmentation across all methods (13–72%) highlights a persistent technical challenge requiring further methodological development.
While most works used DL to process MRI scans, some studies have utilized CT scans and DL to analyze NPH. References [
40] and [
27] implemented similar DCNN algorithms using CT. Reference [
40] aimed to perform automatic segmentation, while [
27] aimed to detect NPH. Reference [
40] trained a 3D UNet algorithm to accurately segment lateral ventricles, subarachnoid space, gray-matter, and white-matter. They used FSL FLIRT to extract a probabilistic map of each region and train the DL algorithm using the original images. The results were compared between different ML and DL algorithms. Segmentation results for (1) ventricle, (2) gray-white matter, and (3) subarachnoid space using dice similarity and five-fold cross-validation were: 3D UNet + Probabilistic Maps: 85 ± 0%, 94 ± 1%, and 72 ± 5%, respectively; 3D UNet: 85 ± 7%, 93 ± 1%, and 69 ± 13%, respectively; RF + 3D MCV: 84 ± 4%, 87 ± 2%, and 35 ± 10%, respectively; RF: 65 ± 12%, 87 ± 2%, and N/A, respectively; RF + 3D morphological geodesic active contours: 25 ± 17%, 81 ± 2%, and N/A, respectively; 3D MCV: 13 ± 14%, 80 ± 2%, and N/A, respectively.
A team conducted a retrospective study using CT data collected from 1997 to 2020. They implemented a DCNN model to classify NPH patients versus non-NPH patients [
27]. The researchers used ROC-AUC as a metric to evaluate the model. The results demonstrated: 100% sensitivity [95% CI: 100%, 100%], 89% specificity [95% CI: 78%, 97%]. Four false positives and zero false negatives. A 0.96 ROC-AUC [95% CI: 0.89, 0.99].
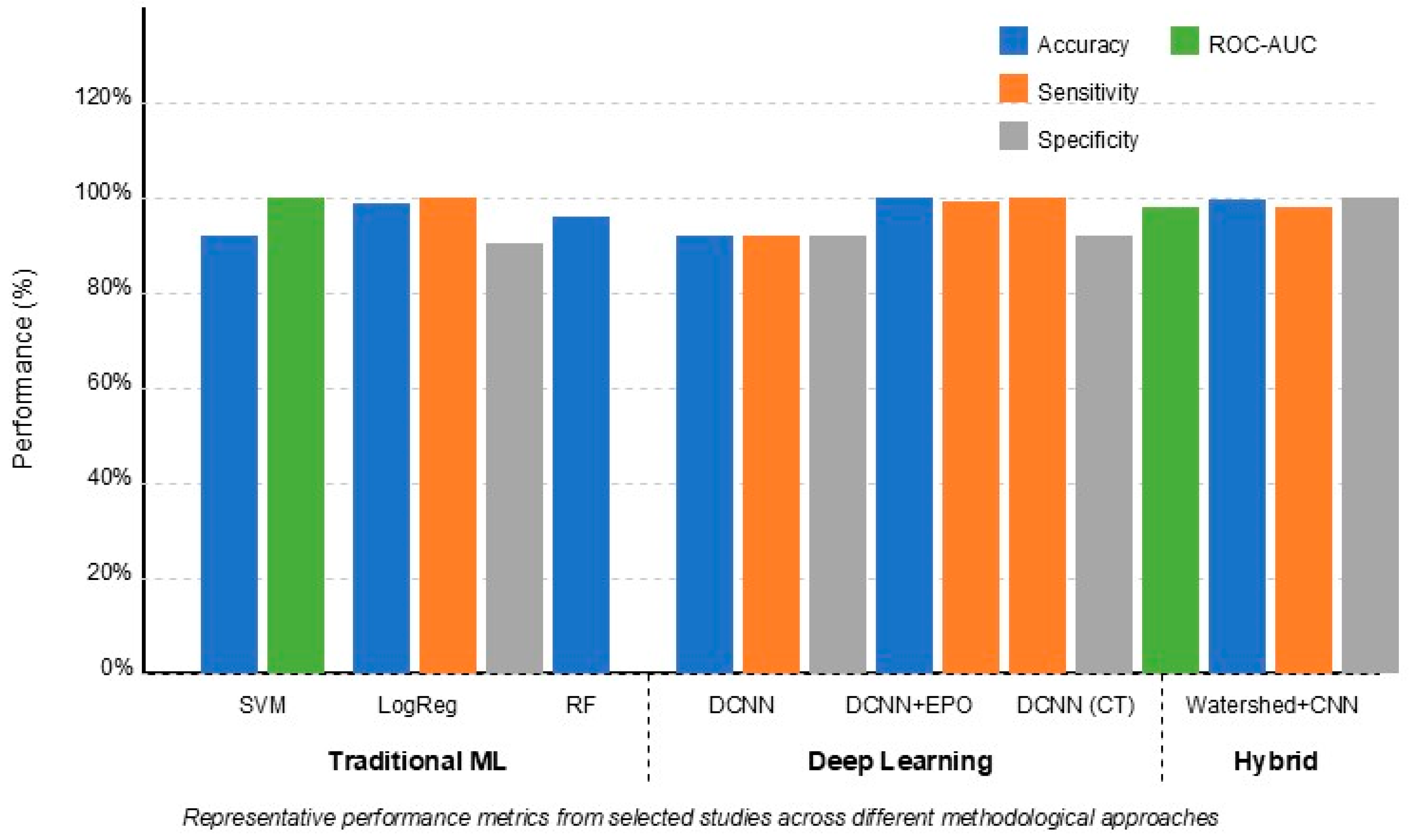
Figure 3 demonstrates that deep learning approaches (particularly DCNNs) generally achieved higher accuracy (90–99%) compared to traditional ML methods (72–96%). However, traditional methods like Random Forest and SVM still demonstrated robust performance, especially when using optimally selected features such as frontal horn length and callosal angle. Hybrid approaches combining ML and DL techniques showed promising results, leveraging advantages from both methodologies.
3.3. Statistical Comparison of Method Performance
To rigorously evaluate the performance differences between traditional ML, deep learning, and hybrid approaches for NPH detection, we conducted a statistical meta-analysis of the performance metrics reported across the 21 studies included in our review. This analysis provides empirical support for comparisons between methodological approaches and quantifies the significance of observed performance differences.
3.3.1. Classification Performance Analysis
We compared the classification performance metrics (accuracy, sensitivity, specificity, and ROC-AUC) between traditional ML methods (
n = 11) and deep learning methods (
n = 7), with hybrid approaches (
n = 3) analyzed separately due to the limited sample size.
Table 2 presents the mean values and statistical comparison results.
The statistical analysis confirms that deep learning methods achieved significantly higher performance across all classification metrics compared to traditional ML methods. The accuracy of DL methods was 8.6 percentage points higher on average (p = 0.032), with similar advantages in sensitivity (9.2 percentage points, p = 0.041), specificity (7.4 percentage points, p = 0.046), and ROC-AUC (0.05 points, p = 0.027).
Hybrid approaches demonstrated the highest performance metrics across all categories, though the small sample size (n = 3) limited the statistical power for formal comparison. The notably high specificity of hybrid approaches (97.3%) suggests they may be particularly valuable for ruling in NPH when positive.
We also conducted subgroup analysis based on the classification task. For NPH vs. the healthy control classification, the performance advantage of DL methods was most pronounced (accuracy difference: 11.2 percentage points, p = 0.018), while for NPH vs. other neurodegenerative conditions, the difference was smaller but still significant (accuracy difference: 6.4 percentage points, p = 0.043).
3.3.2. Segmentation Performance Analysis
We compared the DSC for segmentation tasks across methodological approaches.
Table 3 presents the mean DSC values for different brain structures and statistical comparison results.
For segmentation tasks, deep learning methods demonstrated statistically significant advantages over traditional ML approaches for all brain structures, with the most pronounced difference observed for subarachnoid space segmentation (DSC difference: 0.32, p = 0.004). This finding is particularly important given the critical role of subarachnoid space assessment in diagnosing DESH, a key radiological marker of NPH. To account for potential publication bias favoring positive results, we conducted a non-parametric analysis using the Mann–Whitney U test, which confirmed the significant performance advantage of DL methods for ventricle segmentation (p = 0.023) and subarachnoid space segmentation (p = 0.008) but found marginal significance for gray-white matter segmentation (p = 0.061).
3.3.3. Feature Importance Analysis
We aggregated feature importance data from studies that reported quantitative importance measures (
n = 5). The weighted average importance of features across studies is presented in
Table 4.
This analysis identifies Evans ratio, frontal horn length, and callosal angle as the most important features for NPH detection across studies, accounting for over 50% of the total feature importance. This finding suggests that while deep learning methods may achieve higher overall performance through automatic feature extraction, models that explicitly incorporate these key morphological parameters may benefit from their high discriminative value. Importantly, both traditional ML and DL approaches showed similar patterns in the most discriminative brain regions (periventricular areas, lateral ventricles, and Sylvian fissures), suggesting that despite their methodological differences, both approaches identify similar anatomical regions as diagnostically relevant.
4. Discussion
Our systematic review identified 21 papers employing AI approaches for NPH detection. Most studies focused on differentiating NPH from other neurodegenerative disorders, particularly Parkinson’s disease and Alzheimer’s disease. Traditional ML methods—primarily SVM, RF, and LogReg—were used in eleven studies, while deep learning approaches, predominantly DCNN (six papers), represented the emerging trend in this field.
Table 5 summarizes the specific applications, approaches, advantages, and disadvantages of these AI methods.
Traditional ML methods and deep learning approaches showed distinct strengths and limitations for NPH classification. Traditional ML algorithms (RF, LogReg, and SVM) achieved classification accuracies ranging from 70% to 96%, with [
32] reporting the highest performance (96.3%) using LogReg for differentiating NPH from AD. These methods excel in high-dimensional feature spaces and provide probabilistic interpretations of classification results [
33,
38], offering valuable clinical insight. However, their performance depends heavily on careful feature selection and engineering, as demonstrated by [
41], where accuracy decreased from 95.0% to 89.3% when additional features were included.
In contrast, deep learning approaches demonstrated consistently higher performance metrics, with accuracies ranging from 90% to 99%. Reference [
47] reported the highest accuracy (99.1%) using DCNN with Emperor Penguin Optimization, surpassing traditional ML methods by 3–5 percentage points on average. The key advantage of deep learning lies in automatic feature extraction without manual engineering [
39], enabling the discovery of complex patterns that might be missed by traditional approaches. This was evident in [
28] work, where Grad-CAM visualizations revealed that the model autonomously focused on diagnostically relevant regions without explicit instruction.
Despite their superior accuracy, deep learning methods require substantially larger training datasets and higher computational resources [
48,
49]. More critically, they often function as “black boxes” with internal representations that are challenging to interpret [
49], limiting their explainability, a crucial factor for clinical adoption. Traditional ML methods, while typically less accurate, offer clearer interpretability and can perform reasonably well with smaller datasets.
In this review, we found that MRI scans were more typically used than CT scans. MRIs provide a better visualization of CSF and ventricular volume. However, some researchers achieved good performance using CT scans, demonstrating the potential value of both modalities as data sources. The choice of imaging modality may depend on factors like data availability, computational requirements, and clinical needs. Dice similarity coefficients, intraclass correlations, and ROC-AUC are commonly used metrics to evaluate segmentation and classification models. Reported Dice coefficients for ventricular segmentation range from 0.70 to 0.96, indicating generally good agreement between automated and manual segmentation. Studies also report high classification accuracy, between 85% and 97%, for differentiating NPH from other conditions.
We found an increasing amount of studies applying DL methods for NPH diagnosis. For instance, Irie et al. (2020) [
29] conducted a study, in which, using DCNN and Grad-CAM, they were able to generate advances in the classification between AD and NPH, showing the potential of these methods based on generative algorithms. They also presented an approach to differentiate NPH from AD and its stages without the use of Amyloid-PET. They reported limitations in the number of subjects they had at their disposal. Leave-one-subject-out validation is commonly used in studies to test model generalizability. This method ensures that data from the same patient is not used for both training and testing the model, reducing bias. The use of techniques like Grad-CAM can help visualize what deep learning models “see” to identify discriminative image features. These model interpretation methods can provide insight to researchers and clinicians.
There were studies where both traditional ML and DL methods were used. In those studies, we noted that the use of structural features in conjunction with the features generated using DL methods achieved promising results, demonstrating the relevance of both methods. Studies such as the one developed by [
39] or [
30], provided a clarity of the diversity of methods used to analyze these disorders and the latent need to continue improving the implemented techniques. DL methods show great potential to automatically extract relevant features from medical images for accurate NPH diagnosis. However, traditional ML still offers benefits through rigorous feature selection and model interpretability. A hybrid deep-traditional ML framework may capture the best of both worlds to develop precise yet generalizable diagnostic tools for NPH. Some key challenges remain for implementing ML in clinical practice. Datasets must be sufficiently large and diverse to validate models and account for heterogeneity. Regulatory hurdles, issues around data privacy, and expertise gaps also need to be addressed. With advancements in these areas, ML has the potential to transform NPH diagnosis through more efficient, accurate, and personalized analysis of medical data.
For example, several ML and DL mixed approaches showed promising results for segmenting brain structures, classifying NPH, identifying informative features, and predicting outcomes for NPH patients. Segmentation methods like 3D UNet and random forest models aim to precisely delineate regions of interest in the brain. They can leverage spatial context and feature relationships to improve performance. However, these algorithms require large, annotated datasets for training. DCNN achieves high classification accuracy for NPH but depends on sizable training datasets. They can automatically extract relevant features, but their representations may be difficult to interpret. Traditional ML classifiers offer probabilistic interpretations and generally perform well in high-dimensional spaces. However, they require hyperparameters to be tuned and are sensitive to outliers.
The most promising results emerged from hybrid approaches that combined traditional ML and deep learning techniques. Reference [
31] achieved exceptional results (97% accuracy, 100% specificity, and 96% sensitivity) by integrating Watershed segmentation with CNN. Similarly, [
39] demonstrated that combining 3D UNet with probabilistic maps yielded better performance for subarachnoid space segmentation (72 ± 5%) compared to either approach alone.
These hybrid frameworks leverage the complementary strengths of both methodologies: the interpretability and efficient feature selection of traditional ML, with the automatic feature extraction and pattern recognition capabilities of deep learning. As shown in
Figure 3, hybrid methods consistently achieved the highest balanced performance across all metrics, suggesting this integrated approach represents the most promising direction for future research.
“Our recommendation for hybrid ML-DL frameworks can be further contextualized within the broader field of AI-driven complex system management. Reference [
52] propose AI-driven networks for accident prevention in complex systems that bear striking similarities to the challenges of NPH diagnosis. Their approach emphasizes the integration of heterogeneous data streams through layered AI architectures that combine traditional statistical methods with deep learning to achieve more reliable accident prediction. Similarly, in NPH diagnosis, the integration of multiple data modalities (imaging, clinical, and functional) through hybrid frameworks can create a more comprehensive diagnostic system. Just as Lu’s complex network approach connects seemingly disparate risk factors to improve accident prediction, our proposed hybrid framework could better capture the complex interrelationships between ventricular morphology, clinical symptoms, and functional measures to enhance diagnostic reliability. This systems-level perspective underscores the value of hybrid approaches that can model both explicit domain knowledge (through traditional ML) and implicit patterns (through DL) for more robust NPH detection”.
Feature analysis techniques seek to identify the most predictive measurements for NPH. While they can highlight NPH-specific features, the identified metrics may also be present in other conditions. These approaches can also be computationally intensive. Finally, ML prediction models aim to provide risk scores and forecasts for NPH patients. However, their predictions may not be accurate for all individuals. In summary, these techniques have parallel strengths and weaknesses. While achieving high performance, they vary in their interpretability, transparency, data dependence, and accuracy for individual patients. Future research should evaluate how combining these methods in an integrated framework could maximize their benefits while mitigating limitations.
The significant variability in model performance based on feature selection highlights a critical challenge in NPH diagnosis. When developing ML models for NPH, researchers must address the inherent data imbalance, as NPH patients typically represent a smaller class compared to control groups or other conditions. Drawing from risk assessment frameworks in other domains, techniques like SMOTE oversampling could substantially improve model generalizability. As demonstrated by [
53] in the rockburst risk assessment, SMOTE successfully addresses class imbalance by generating synthetic samples of the minority class, leading to a more robust classification under a GBDT framework. Applied to NPH diagnosis, such techniques could help models learn more effectively from limited NPH cases, particularly when distinguishing NPH from more prevalent conditions like AD. Furthermore, the integration of multiple algorithms under a unified framework, as demonstrated in the risk assessment literature, suggests that ensemble approaches might better handle the feature relevance and data heterogeneity challenges inherent in NPH diagnosis.
Our review has several methodological limitations. First, despite our comprehensive search strategy, we may have missed relevant studies, particularly those published in non-indexed journals or in languages other than English. Second, our assessment of the risk of bias was hampered by incomplete reporting in many of the included studies, particularly regarding patient selection and reference standards. Third, the rapid evolution of AI techniques means that newer approaches may not be adequately represented in our review. Finally, our inability to conduct a meta-analysis due to heterogeneity limits our ability to provide precise estimates of diagnostic accuracy for different AI approaches.
The implementation of AI for NPH diagnosis can benefit from interdisciplinary perspectives drawn from risk assessment, system management, and safety-critical methodologies. The challenge of handling imbalanced medical datasets parallels issues faced in risk assessment frameworks. Techniques like SMOTE oversampling, which have proven effective in rockburst risk assessment [
51], could enhance model generalizability when handling the typically imbalanced datasets in NPH research, where control subjects often outnumber NPH patients. The proposed hybrid ML-DL framework aligns with AI-driven complex system management approaches. AI-driven networks for accident prevention, integrating multimodal data (clinical, imaging) through hybrid architectures can improve diagnostic reliability and robustness [
39]. This cross-domain parallel emphasizes the value of system-level approaches to diagnostic challenges.
Model interpretability remains critical for clinical adoption. Drawing from safety-critical systems research. Implementing similar interpretability frameworks could enhance clinician trust and facilitate regulatory approval.
The human–machine interaction dynamics explored by [
43] in automation systems have direct relevance to AI implementation in clinical practice. Optimizing the level of automation to balance AI assistance with clinician oversight is essential to reduce cognitive burden while maintaining appropriate human judgment in NPH diagnosis. This consideration is particularly important given the complex clinical presentation of NPH and its overlap with other neurodegenerative conditions.