Featured Application
This work will greatly improve the application of luciferase in biological imaging, protein labeling, and drug screening.
Abstract
The first luciferase from Antarctic krill (LAK) was cloned and successfully expressed in Escherichia coli BL21(DE3). LAK exhibits the unique ability to emit bright violet fluorescence at an emission wavelength of 350 nm, which represents the lowest reported bioluminescence wavelength for luciferases. However, its low thermal stability poses a limitation to its broader application. In this study, we employed a rational design approach to introduce three pairs of artificial disulfide bonds into LAK. Circular dichroism (CD) analysis revealed that the introduction of artificial disulfide bonds resulted in a significant increase in the secondary structural content of α-helices and β-sheets compared to the wild-type (WT) enzyme. However, these modifications did not influence the emission spectrum. Among the resultant mutant strains, two exhibited markedly enhanced thermal stability. Notably, Mut2 demonstrated a 6.18-fold increase in half-life at 50 °C. Molecular docking studies indicated that D-fluorescein can form additional hydrogen bonds with surrounding amino acid residues (A323, T347, and K534). The docking energies between the enzyme and substrate for WT and Mut2 were −19.5 kcal/mol and −23.4 kcal/mol, respectively, thereby establishing strong interactions within the catalytic pocket region. These interactions likely contribute to a 2.92-fold improvement in substrate affinity, as evidenced by a reduced Michaelis–Menten constant (Km). Our thermal stability and catalytic activity analyses revealed that the linker region between the N- and C-domains plays a crucial role in the overall stability of the enzyme. Furthermore, the C-terminus of LAK does not participate in substrate-binding and catalysis; its local excessive rigidity was found to restrict the release of the AMP product, thereby negatively impacting catalytic activity. These findings offer new insights into the mutagenesis of luciferases and pave the way for the further optimization of LAK for various biotechnological applications.
1. Introduction
Luciferase (EC1.13.12.5) is a natural enzyme that catalyzes the oxidation of luciferin in the presence of oxygen, ATP, and magnesium ions (Mg2+), resulting in the formation of oxyluciferin and the emission of light [1,2]. The intensity of this bioluminescence is directly proportional to the concentration of ATP within a specific range, rendering luciferase an invaluable tool as a reporter gene in various biological systems due to the ubiquitous and stable presence of ATP in living cells [3,4,5]. In the field of bacterial detection, the luminescence generated by the luciferase-catalyzed reaction is employed to quantify ATP released from bacterial cell lysates, thereby enabling the rapid and indirect enumeration of bacterial cells. For instance, Thore et al. successfully utilized luciferase to detect contamination by Listeria monocytogenesin food samples [6]. Similarly, Alison et al. demonstrated the rapid and accurate detection of industrial pollutants in water environments using luminescent bacteria [7]. These studies highlight the versatility of luminescence-based methods in various detection applications. The development of genetically engineered bacteria has further expanded the potential of luminescence-based detection. By introducing luciferase genes into recipient bacterial strains through genetic engineering, the modified bacterial cells acquire the capacity to catalyze luminescence reactions [8]. In practical applications, these genetically engineered bacteria are introduced into the test system. Upon stimulation by specific target microorganisms, the engineered bacteria undergo bioluminescence reactions, producing detectable luminescence. The intensity of this luminescence is directly proportional to the concentration of the target microorganisms, thereby allowing for quantitative and directional safety testing. This approach offers a highly sensitive and specific method for detecting and quantifying specific microorganisms in complex environmental or biological samples. Moreover, the high sensitivity of luciferase-based imaging allows for the direct monitoring and tracking of tumor cell metastasis and growth by detecting the bioluminescent signals from cells labeled with the luciferase gene [9,10,11]. A notable example is the work of Koutsoudakis in 2006, who successfully monitored the interaction between the hepatitis C virus and host cells using a dual cis–trans luciferase virus reporter genome [12]. This innovative approach not only provided an intuitive means of observing the impact of drug factors on the virus but also served as a platform for evaluating the efficacy of viral inhibitors [13].
Luciferases are broadly categorized into bacterial and firefly luciferases [14,15,16]. A common limitation of most naturally occurring luciferases is their poor thermal stability. For instance, the North American firefly luciferase, a well-known enzyme involved in bioluminescence, becomes inactive after being stored at 37 °C for just 3 min in vitro [16,17,18]. This thermal instability poses a significant challenge for the application of luciferases in various experimental and clinical settings.
Antarctic krill, a keystone species in the Antarctic ecosystem, is found at densities of 10,000–30,000 individuals per cubic meter and represents one of the most successful animal species on Earth in terms of biomass energy, with an estimated total of approximately 500 million tons. These organisms possess specialized biological fluorescent organs capable of producing fluorescence. These organs are strategically distributed across different parts of the krill’s body: one pair in the eye stalks; another pair along the second to seventh thoracic and foot positions; and one in the abdomen. When threatened, Antarctic krill release an enzyme (luciferase) and a small molecular substrate (luciferin), which react in the presence of oxygen to emit bright violet fluorescence. This bioluminescent response not only masks the krill’s shadow, rendering it “invisible” to predators, but also plays a crucial role in mating and nighttime aggregation behaviors. Our research team has made significant strides in this field by completing the whole-genome sequencing of Antarctic krill [19]. Building on this foundation, we successfully cloned and expressed the first luciferase from Antarctic krill (LAK). LAK emits bright violet fluorescence with a weak tissue penetration ability and is less likely to be absorbed by skin tissue, making it particularly suitable for shallow tissue imaging. At 25 °C, LAK exhibits high catalytic activity and sensitivity, with Km values of 121 μmol/L for ATP and 53.6 μmol/L for the substrate luciferin, respectively. The minimum detectable level for ATP is as low as 1 fmol. However, the practical application of LAK is currently limited by its low thermal stability; after being heated at 50 °C for 10 min, the enzymatic activity of LAK is almost completely lost. Therefore, our objective was to engineer a LAK variant with enhanced thermostability and improved activity, characterized by a lower Km and a higher kcat value.
Enhancing enzyme thermostability through protein engineering to withstand extremely harsh conditions is a significant objective in contemporary research. Enzyme thermostability is influenced by several key factors, including hydrogen bonds, strong covalent disulfide bonds, π–π interactions, and salt bridges. These structural features collectively contribute to the overall stability of proteins under high-temperature conditions. To achieve this goal, a variety of protein engineering techniques have been developed and refined. These techniques include random mutagenesis, which introduces random mutations to explore beneficial changes in protein structure; DNA shuffling, which mimics natural recombination processes to generate diverse variants; truncation, which involves the removal of specific regions of the protein to enhance stability; and circularization, which can stabilize proteins by altering their structural topology [18,20,21,22,23]. Each of these methods has demonstrated potential in improving the thermostability of enzymes, thereby expanding their application scope in industrial and biotechnological processes. The introduction of novel disulfide bonds into proteins has been a widely employed strategy to improve protein stability [20,24,25,26]. However, not all engineered disulfide bonds result in increased stability, as there are instances where they have been reported to destabilize proteins. This highlights the need for more targeted approaches to reduce the screening effort required to identify enzymes with desired properties. Rational design based on protein structure has emerged as an efficient method to address this challenge [20,27]. The structural diversity among enzymes complicates the relationship between enzyme structure and function, necessitating a detailed study of the characteristics of individual enzymes. LAK, in particular, differs significantly from traditional enzyme molecules in that it possesses multiple catalytic domains, including binding sites for the substrate (luciferin), ATP/AMP, and Mg2+. Selecting appropriate mutation sites in such a complex enzyme is a formidable task. In this study, we introduced artificial disulfide bonds into LAK in the N-terminal and C-terminal connection region (linker peptide), the flexible surface loop region, and the C-terminal unstructured catalytic domain. We then investigated the effects of these mutations on the thermal stability and catalytic activity of LAK.
2. Materials and Methods
2.1. Homology Modeling and Selection of Mutation Sites
The tertiary structure of LAK was modeled using the Robetta online service “https://robetta.bakerlab.org” (accessed on 10 July 2024) with default parameters. The modeled structure was analyzed and visualized using PyMol 3.1.4 to identify potential mutation sites. Molecular docking was performed using AutoDock Vina 4.2 to further assess the interactions at these sites. The program Disulfide by Design 2.0 (Dombkowski, Wayne State University, Detroit, MI, USA) was employed to select sites for the insertion of disulfide bridges, focusing on cysteine pairs that are properly oriented to form stable disulfide bonds.
2.2. Plasmid Construction, Expression, and Purification of Luciferase Mutants
The plasmid map and DNA sequence of LAK are provided in the Supplementary Materials. The LAK gene and its mutants were cloned into the pCold II vector using standard molecular biology techniques. The recombinant luciferase was expressed in Escherichia coli BL21 (DE3) and purified using an Ni-affinity column (GE Healthcare, Chicago, IL, USA) according to the manufacturer’s protocol. The purity and molecular weight of the purified luciferase were confirmed by SDS-PAGE using a 15% polyacrylamide gel on a two-dimensional electrophoresis system (Bio-Rad, Hercules, CA, USA). The purified luciferase was stored at −80 °C for further analysis.
2.3. Determination of Luciferase Activity
Luciferase activity was measured at 25 °C using a Tecan Infinite M1000 PRO microplate reader equipped with an autoinjector module (Tecan Group, Männedorf, Switzerland). The reaction mixture consisted of 25 μL of incubated enzyme solution (0.5 mM DTT, 0.1 mg/mL BSA, 10.0 mM MgSO4, 25 mM HEPES) mixed with 25 μL of 0.3 mM D-luciferin stock solution in 96-well microplate wells. The luciferase reaction was initiated by injecting 50 μL of 1 mM ATP stock solution; the luminescence intensity was recorded at 10 s post injection.
2.4. Thermostability Analysis
The thermostability of the luciferase was assessed by preincubating the enzyme at temperatures ranging from 25 to 60 °C for 10 min using a thermal cycler (T-100, Bio-Rad, Hercules, CA, USA). The residual activity was immediately measured as described above, with three replicates performed per analysis. The deactivation constant (kd) was calculated from the semi-logarithmic plot of residual activity versus time using Equation (1), where Et is the residual enzyme activity after heat treatment for time t, and E0 is the initial enzyme activity before heat treatment. The half-lives of the WT and mutants were calculated using Equation (2), as follows:
ln[Et/E0] = −kdt
t1/2 = ln2/kd
2.5. Michaelis–Menten Kinetics Determination
The kinetic parameters were determined by monitoring the initial reaction velocities with increasing concentrations of luciferin (0.1 mM to 0.3 mM) at 25 °C. The affinity constant (Km) and the turnover number (kcat) were calculated using the nonlinear least-square fitting procedure for the Michaelis–Menten standard protocol in Prism software (version 5.0 for Windows, San Diego, CA, USA).
2.6. Circular Dichroism (CD) Spectroscopy Analysis
The purified luciferase solution (0.5 mg/mL) was dialyzed against water to remove any residual salts and buffer components. Subsequently, the secondary structure of the enzyme was characterized using circular dichroism (CD) spectroscopy. The CD spectra were recorded over a wavelength range of 180–260 nm, with a step resolution of 1 nm and a scan rate of 30 nm/min. Each spectrum was obtained as the average of three independent scans to ensure reproducibility and accuracy. The CD data were expressed in terms of mean residue ellipticity [θ].
3. Results and Discussions
3.1. Homology Modeling
The amino acid sequence of LAK was used as a reference to model its tertiary structure, leveraging the crystal structure of Lampyridae (PDB ID: 6K4C) to ascertain the relative positioning of LAK residues. Following molecular dynamics (MD) energy minimization, a validation check was performed. A Ramachandran plot analysis revealed that 93.28% of the residues were situated in the most favored regions, with an additional 5.9% in allowed regions, indicating the high quality of the modeled structures. The tertiary structure of the LAK protein comprises a larger N-terminal domain (residues 1–447), a flexible linker peptide (residues 448–454), and a smaller C-terminal domain (residues 455–551) positioned opposite the N-terminal region (Figure 1a). The opposing surfaces of the N- and C-terminal domains form a cleft containing numerous conserved residues, which is presumed to be the active site. Upon substrate-binding, the N- and C-terminal regions converge to clamp the substrate, forming a sandwich-like structure. The primary N-terminal domain features a compact architecture with a distorted antiparallel β-barrel and two β-sheets flanked by α-helices, creating a five-layered αβαβα structure. The C-terminal domain adopts an α+β conformation. The conserved catalytic region of LAK exhibits characteristics typical of the adenylate-forming domain superfamily, commonly involved in binding CoA, Mg2+, AMP, and ATP. A conserved tri-peptide sequence, serine–lysine–leucine (SKL), is located at the C-terminus, playing a role in peroxidase localization and AMP release (Figure 1b).
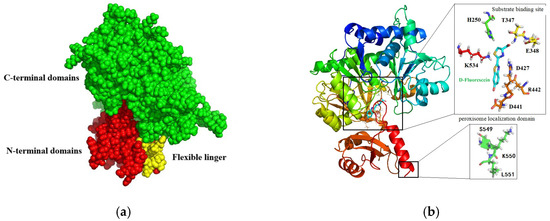
Figure 1.
Models of the simulated spatial structure of LAK: (a) the structure shown on the surface. N-terminal, C-terminal, and the flexible linker are colored in green, red and yellow, respectively; and (b) the complex of LAK and D-fluorescein., D-fluorescein, the binding sites, and the conserved tri-peptide are shown in rainbow sticks.
3.2. Construction of Mutants
Strong covalent disulfide bonds are pivotal in protein stabilization, primarily through entropic effects that reduce conformational chain entropy and increase the free energy of the denatured state. The wild-type (WT) LAK, a single-chain molecule devoid of disulfide bonds, may inherently lack thermal stability. Utilizing Disulfide by Design, we identified 49 residue pairs geometrically suited for disulfide bridge formation upon cysteine mutation. To refine our selection, we adhered to the following two criteria: (1) a relatively low bond energy with a Chi3 (×3 torsion angle) value between ±90 and ±110 and a disulfide bond energy less than 8.2 kcal/mol−1; and (2) a distance of 25–75 amino acids between the cysteine residues forming the disulfide bond, ensuring proper LAK secondary structure folding. Consequently, three disulfide bridges were designed in silico (Table 1), located in the flexible loop region of the N-terminal domain (Mut1), the flexible linker region between the N- and C-domains (Mut2), and the C-terminal domain (Mut3) (Figure 2).

Table 1.
Disulfide bond screening results of LAK using Disulfide by Design.
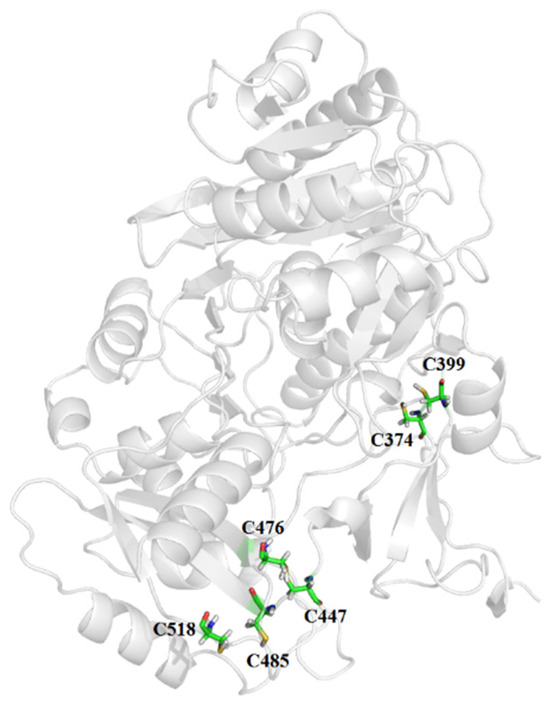
Figure 2.
The predicted mutant sites of LAK. The whole structure is shown in the gray cartoon and the mutant sites are shown in rainbow sticks.
3.3. Plasmid Construction and Expression of Luciferase Mutants
The genes encoding the WT and mutant luciferases were cloned into the pCold II vector and transformed into Escherichia coli BL21 (DE3) competent cells. Expression of the recombinant luciferase was induced with isopropyl-β-D-thiogalactopyranoside (IPTG) at 16 °C. The expressed proteins were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) using a 15% polyacrylamide gel. The results showed that the mutant luciferases exhibited a specific band with a molecular mass of approximately 61 kDa, while no recombinant protein was detected in the control E. coli BL21 (DE3)/pCold II cells (Figure 3a).
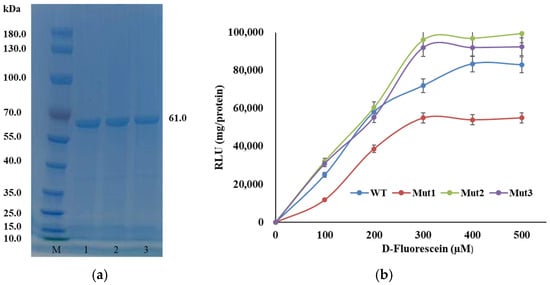
Figure 3.
The purification and the activity of LAK and mutants: (a) SDS–PAGE (15%) analysis of purified luciferase. The lanes are as follows: M, marker proteins with relative molecular masses. Lane 1–3 Mut1–Mut3; and (b) Luciferase activity was measured at 25 °C. Data are given as the means ± SD, n = 3.
In this study, luminescence intensity was employed as a proxy for luciferase activity. All mutants demonstrated detectable luciferase activity, with luminescence intensity increasing progressively as the concentration of luciferin was raised. Notably, the luminescence intensity of Mut2 reached 9.6 × 104 relative light units (RLU) per mg of protein (Figure 3b). It is important to highlight that, since luminescence intensity was measured rather than the actual concentration of the product, the calculated kinetic parameters (Km and kcat) are considered apparent or relative values. These values are derived from the luminescence measurements and should be interpreted with this caveat in mind.
3.4. Enzymatic Thermostability
The thermal stability of the WT and mutant enzymes was evaluated as described in the Section 2. As illustrated in Figure 4a, the activities of both WT and mutant luciferases decreased with increasing temperatures. After incubation at 40 °C for 10 min, the enzymatic activities of WT and Mut1 luciferases were almost completely lost. In contrast, Mut2 and Mut3 showed significantly enhanced thermal tolerance, retaining approximately 76% and 62% of their activity, respectively. The inactivation half-lives of the WT and mutant luciferases at 50 °C were also monitored (Figure 4b). The inactivation curves revealed that Mut2 and Mut3 had the slowest inactivation rates. After 30 min at 50 °C, Mut2 and Mut3 still maintained residual activities of 64% and 38%, respectively, compared to only about 7% for WT luciferase, while Mut1 almost completely lost its activity.
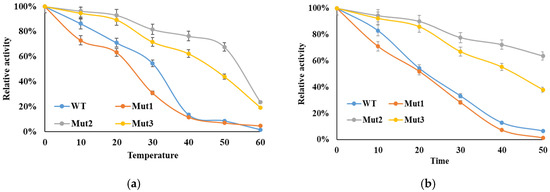
Figure 4.
Thermostability and inactivation curves of WT luciferase and its mutants. (a) The thermal stability of the WT and mutant enzymes. (b) The thermal inactivation of the enzyme at 50 °C was determined from 0 to 50 min, with residual activities measured immediately after heat treatment. The activity of the pre-incubated enzyme at 4 °C was taken as 100%. Data are given as the means ± SD, n = 3.
Enzymes from cold-adapted organisms typically exhibit greater flexibility, a characteristic feature of psychrophilic proteins. As the temperature increases, the flexible regions of these enzymes begin to fold to accommodate the changes in entropy, which is often a contributing factor to their poor stability. Luciferases catalyze two essential enzymatic steps for bioluminescence: (i) adenylation of D-luciferin; and (ii) oxygenation of adenyl-luciferin. In the second step, luciferase functions as an oxygenase, catalyzing the oxygenation of adenyl-luciferin to form an energy-rich dioxetanone intermediate. Throughout this process, luciferase undergoes significant conformational changes. Although Mut1 (F374C-G399C) is located in a highly flexible loop region and is more likely to unfold at an early stage; it is also in close proximity to the active pocket region. The excessive local rigidity caused by steric contacts reduces the enzyme’s ability to fold at high temperatures, negatively impacting its activity. This suggests that increasing local rigidity can enhance thermal stability; it may also interfere with the necessary conformational changes required for enzyme activity, highlighting the delicate balance needed in protein engineering for improved thermal stability without sacrificing function.
3.5. Thermal Deactivation Analysis
The deactivation constants (kd) for the first-order thermal deactivation were calculated using Equation (1). The kd values for Mut2 and Mut3 at 50 °C were 8.9 × 10−3 min−1 and 1.88 × 10−2 min−1, respectively, compared to 5.5 × 10−2 min−1 for the WT luciferase (Figure 5). The half-lives of Mut3 and WT luciferase were determined using Equation (2). Mut2 exhibited a half-life of 77.9 min, significantly longer than that of the WT luciferase (12.6 min). Compared to the WT, the half-lives of mutants Mut2 and Mut3 were increased by 6.18 and 2.92 times, respectively. The introduction of artificial disulfide bonds thus yielded two thermally stable mutant strains.
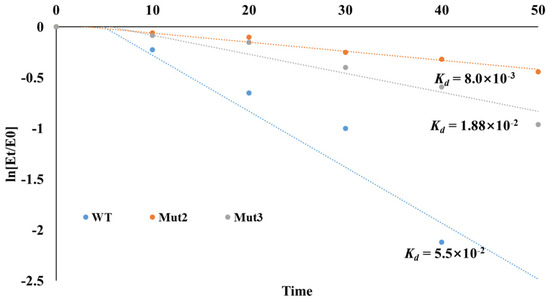
Figure 5.
Thermal deactivation curves of WT luciferase and its mutants.
3.6. Kinetic Analysis
To assess the impact on enzyme activity, the kinetic constants Km and kcat of the WT and mutants were measured. Luciferase activity was detected at 25 °C with gradually increasing substrate concentrations. The kinetic constants were determined using the Lineweaver–Burk graphical method (Table 2). Although the thermal stability of Mut3 improved, its affinity for the substrate luciferin decreased significantly (high Km). Mut3 (A485C-T518C) is located near the C-terminus, close to the conserved domain Ser-Lys-Leu (SKL), which is involved in the release of the product AMP during enzyme catalysis. The oxidation of luciferin is facilitated by the activating effect of AMP, which is crucial for the formation of the dioxetanone ring. The introduction of disulfide bonds increased the rigidity of this region, hindering the release of AMP and becoming the rate-limiting step in the enzyme-catalyzed reaction. Consequently, Mut3 exhibited lower affinity compared to the WT.

Table 2.
Kinetic constants of WT and mutants’ enzyme.
The thermal stability of Mut2 was significantly enhanced; its substrate affinity increased by 1.97 times. Generally, an increase in the number of hydrogen bonds between side chains of residues improves intermolecular interaction forces [28]. The enzyme–substrate interactions of Mut2 were investigated. As the substrate luciferin approaches the active site, the region gradually closes to encapsulate the substrate. The WT luciferase had two residues, T347 and D441, which interacted with the substrate through two hydrogen bonds (Figure 6a). For Mut2, the introduction of artificial disulfide bonds increased the intermolecular force in this region. In this study, molecular docking analysis revealed that D-fluorescein can form additional hydrogen bonds with the surrounding amino acid residues A323, T347, and K534, thereby establishing strong interactions with the substrate within the catalytic pocket region (Figure 6b). These interactions likely contribute to enhanced substrate-binding stability. Furthermore, while the WT enzyme exhibited no π–alkyl interactions (Figure 6c), Mut2 displayed π–alkyl interactions between D-fluorescein and residues D427 and R442. Additionally, a π–sulfur interaction was observed between D-fluorescein and residue H250 in Mut2 (Figure 6d). The formation of these π–alkyl and π–sulfur interactions significantly strengthened the binding affinity of D-fluorescein to Mut2, resulting in higher substrate affinity compared to the WT enzyme. The stability of a molecular docking conformation is inversely related to its binding energy; lower binding energy indicates a more stable conformation and a higher likelihood of interaction between the enzyme and substrate. In this study, the docking energies for WT, Mut2 were −19.5 kcal/mol and −23.4 kcal/mol, respectively. The more negative docking energies of Mut2 suggest stronger binding interactions with D-fluorescein compared to the WT enzyme. This finding may partly explain the decreased Km values observed for Mut2 relative to the WT enzyme, indicating enhanced substrate affinity due to the optimized interactions within the catalytic pocket.
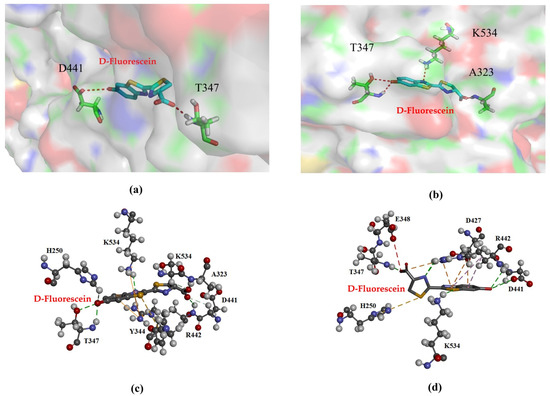
Figure 6.
Hydrogen bonds and non-bond interactions of WT and Mut2. (a) Hydrogen bonds between WT and substrate. (b) Hydrogen bonds between Mut2 and substrate. (c) Non-bond interactions between WT and substrate. (d) Non-bond interactions between Mut2 and substrate. Red: hydrogen bonds. Orange: π–sulfur interactions. Green: conventional interactions. Pink: π–alkyl interactions.
3.7. CD and Luminescence Spectra Analysis
Circular dichroism (CD) spectroscopy is a powerful tool for characterizing the secondary structural changes in proteins, including enzymes. In this study, CD analysis was employed to examine the secondary structure composition of the WT enzyme and its mutants. The results revealed that the WT enzyme contained 29.95% α-helix, 19.06% β-sheet, and 43.92% random coil (Figure 7a). In contrast, Mut2 exhibited a slightly higher proportion of α-helix (31.4%) and β-sheet (19.96%), with 48.64% random coil. These findings indicate that the introduction of artificial disulfide bonds in the mutants led to an increase in the content of α-helix and β-sheet compared to the WT enzyme. This structural change suggests enhanced stability due to the additional disulfide cross-linking, which may contribute to the improved thermostability of the mutants. Similar observations have been reported in studies on 1,4-α-glucan branching enzyme (GBE), where the introduction of disulfide bonds also resulted in increased secondary structural content and stability [29].
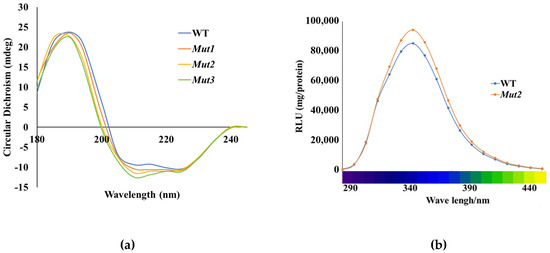
Figure 7.
CD spectroscopy and normalized luminescence spectra of WT and Muts. (a) CD spectroscopy of WT and Muts. (b) Normalized luminescence spectra of WT and Mut2. The luminescence spectra were measured at 25 °C.
Numerous studies have confirmed that mutations in certain amino acids within luciferase can affect the luminescence spectrum, leading to “red shift” and “violet shift” phenomena. To investigate the impact of mutations, we used a spectrophotometer to scan the WT luciferase and determine if the mutations affected the luminescence spectrum. The luminescence spectra of Mut2 luciferase are shown in Figure 7b. The bioluminescence spectra of WT and Mut2 were very similar, with a maximum peak at a wavelength of 350 nm. This indicates that the artificial disulfide bonds had no effect on the emission spectrum of luciferase.
4. Conclusions
In this study, we explored strategies to enhance the stability of Antarctic krill luciferase (LAK) while preserving its catalytic activity. LAK holds significant promise for applications in medical tissue imaging, primarily due to its unique capability to emit violet fluorescence. However, the inherent structural complexity of LAK, characterized by multiple substrate-binding sites and a multifaceted catalytic mechanism, presents challenges for molecular modification. Our findings indicate that the introduction of artificial disulfide bonds can increase the content of α-helix and β-sheet, subtly alter the secondary structure of LAK, and reduce the overall molecular free energy. These changes collectively contribute to improved protein thermostability. Additionally, the increase in hydrogen bonds within the protein’s interior may enhance conformational stability without compromising catalytic performance. The formation of π–alkyl and π–sulfur interactions between the substrate and the catalytic region further strengthens the substrate-binding affinity. Notably, Mut2 exhibited a 6.18-fold increase in half-life at 50 °C and a 2.92-fold enhancement in substrate affinity (Km), findings that align with those reported by Vogt et al. [30]. Importantly, our results demonstrate that the introduction of disulfide bonds need not be restricted to surface flexible regions. For cold-adapted enzymes, the linker region is also a hotspot for mutation. These regions exhibit greater spatial conformational changes upon temperature increases; stiffening them can significantly improve thermal stability. While the C-terminal domain is crucial for maintaining the enzyme’s overall stability, local steric constraints within this region can impede product release, thereby reducing catalytic efficiency. In conclusion, the work presented here indicates that the introduction of artificial disulfide bonds is an effective method for protein engineering.
Despite the effectiveness of disulfide bond engineering in improving protein stability, several factors limit its widespread application. First, detailed three-dimensional (3D) structural information of the proteins is essential for designing disulfide bridges using appropriate computational strategies. For enzymes with low sequence homology or those whose structures are difficult to resolve, such as aminopeptidases, luciferases, and dehalogenases, selecting optimal mutation sites poses a significant challenge. Second, enzymes that act on complex biomolecular substrates, such as chitooligosaccharides or carrageen, possess multiple functional regions, including catalytic active centers, substrate-binding sites, coenzyme-binding sites, and product-release sites. Their intricate catalytic mechanisms mean that single-point mutations often result in detrimental effects or even destabilization. This underscores the need for a deeper understanding of the fine spatial structure of enzyme molecules and a clear elucidation of their catalytic mechanisms.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/app15073563/s1, Figure S1: The plasmid map of pCold II, Table S1: DNA sequence of LAK.
Author Contributions
Y.M., writing, idea proposal, experimental data validation; Y.Z., methodology, conceptualization, formal analysis; X.J., software development, submission, and preparation; J.S., review, resources, and editing. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Key Research and Development Program of China (Grant No. 2022YFC2807505).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors on request.
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| CD | Circular dichroism |
| LAK | Antarctic krill luciferase |
| IPTG | isopropyl-β-D-thiogalactopyranoside |
| WT | wild-type |
References
- Stefan, S.; Al-Handawi, M.B. Mechanically Assisted Bioluminescence with Natural Luciferase. Angew. Chem. Int. Ed. Engl. 2020, 59, 16485–16489. [Google Scholar]
- Kaskova, Z.M.; Tsarkova, A.S. 1001 lights: Luciferins, luciferases, their mechanisms of action and applications in chemical analysis, biology and medicine. Chem. Soc. Rev. 2016, 45, 6048–6077. [Google Scholar]
- Viviani, V.R. The origin, diversity, and structure function relationships of insect luciferases. CMLS Cell. Mol. Life Sci. 2002, 59, 1833–1850. [Google Scholar]
- Contag, C.H.; Spilman, S.D. Visualizing bioluminescent gene expression in 1iving mammals using reporter. Photochem. Photobiol. 1997, 66, 523–531. [Google Scholar] [PubMed]
- Niu, G.; Chen, X. Molecular imaging with activatable reporter systems. Theranostics 2012, 2, 413–423. [Google Scholar] [PubMed]
- Thore, A.; Ansehn, S. Detection of bacteriuria by luciferase assay of adenosine triphosphate. J. Clin. Microbiol. 1975, 1, 1–8. [Google Scholar] [PubMed]
- Alison, M.; Horsburgh, D.P. On-line microbial biosensing and fingerprinting of water pollutants. Biosens. Bioelectron. 2002, 17, 495–501. [Google Scholar]
- Yan, S.L.; Miao, S.N. ATP bioluminescence rapid detection of total viable count in soy sauce. Luminescence 2012, 27, 34–38. [Google Scholar]
- Deb, D.K.; Srivastava, K.K. Bioluminescent rapid vitro and high in Mycobacterium throughput aurum expressing screening of firefly luciferase antimycobacterial infected drugs for in macrophages. Biochem. Biophys. Res. Commun. 2000, 279, 457–461. [Google Scholar]
- Andrey, A.K.; Robert, A.D. Rapid and Sensitive Detection of Retrovirus Entry by Using a Novel Luciferase-Based Content-Mixing Assay. J. Virol. 2004, 78, 5124–5132. [Google Scholar]
- Kocher, B.; Piwnicaworms, D. Illuminating Cancer Systems with Genetically-Engineered Mouse Models and Coupled Luciferase Reporters In Vivo. Cancer Discov. 2013, 3, 616–628. [Google Scholar] [CrossRef]
- Koutsoudakis, G.; Kaul, A. Characterization of the early steps of hepatitis C virus infection by using luciferase reporter viruses. J. Virol. 2006, 80, 5308–5320. [Google Scholar] [CrossRef] [PubMed]
- Pietschmann, T.; Kaul, A. Construction and characterization of infectious intragenotypic and intergenotypic hepatitis C virus chimeras. Proc. Natl. Acad. Sci. USA 2006, 103, 7408–7413. [Google Scholar] [CrossRef] [PubMed]
- Wood, K.V. The chemical mechanism and evolutionary development of beetle bioluminescence. Photochem. Photobiol. 1995, 62, 662–673. [Google Scholar] [CrossRef]
- Gui-Chun, L.; Ru, Z. Cloning and Characterization of Luciferase from the Chinese Firefly Lamprigera yunnana. Photochem. Photobiol. 2019, 95, 1186–1194. [Google Scholar]
- Tania, P.; Farhima, A. Firefly Luciferase Mutant with Enhanced Activity and Thermostability. ACS Omega 2018, 3, 2628–2633. [Google Scholar]
- Mitani, Y.; Futahashi, R. Tibetan Firefly Luciferase with Low Temperature Adaptation. Photochem. Photobiol. 2017, 93, 466–472. [Google Scholar] [CrossRef]
- Law, G.H.; Gandelman, O.A. Mutagenesis of solvent-exposed amino acids in Photinus pyralis luciferase improves thermostability and pH-tolerance. Biochem. J. 2006, 397, 305–312. [Google Scholar] [CrossRef] [PubMed]
- Changwei, S.; Shuai, S. The enormous repetitive Antarctic krill genome reveals environmental adaptations and population insights. Cell 2023, 186, 1279–1294.e19. [Google Scholar]
- Imani, M.; Hosseinkhani, S. Design and introduction of a disulfide bridge in firefly luciferase: Increase of thermostability and decrease of pH sensitivity. Photochem. Photobiol. Sci. 2010, 9, 1167–1177. [Google Scholar] [CrossRef]
- Eijsink, V.G.H.; Bjørk, A. Rational engineering of enzyme stability. J. Biotechnol. 2004, 113, 105–120. [Google Scholar] [CrossRef]
- Tokurik, N.; Tawfik, D.S. Stability effects of mutations and protein evolvability. Curr. Opin. Struct. Biol. 2009, 19, 596–604. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Ma, X. Protein cyclization enhanced thermostability and exopeptidase resistanceof green fluorescent protein. J. Microbiol. Biotechnol. 2010, 20, 460–466. [Google Scholar] [PubMed]
- Gilbert, H.F. Protein Disulfide Isomerase and Assisted Protein Folding. J. Biol. Chem. 1997, 272, 29399–29402. [Google Scholar] [CrossRef]
- Mamathambika, B.S.; Bardwell, J.C. Disulfide-linked protein folding pathways. Annu. Rev. Cell Dev. Biol. 2008, 24, 211–235. [Google Scholar] [CrossRef]
- Kaushik, M.; Sinha, P. Protein engineering and de novo designing of a biocatalyst. J. Mol. Recognit. 2016, 29, 499–503. [Google Scholar] [CrossRef] [PubMed]
- Le, Q.A.T.; Joo, J.C. Development of thermostable Candida antarctica lipase b through novel in silico design of disulfide bridge. Biotechnol. Bioeng. 2012, 109, 867–876. [Google Scholar] [CrossRef]
- Hou, Q.Q.; Li, N. Design and regulation of the surface and interfacial behavior of protein molecules. Chin. J. Chem. Eng. 2020, 28, 2837–2847. [Google Scholar] [CrossRef]
- Li, C.; Ban, X. Rational design of disulfide bonds for enhancing the thermostability of the 1,4 α-glucan branching enzyme from Geobacillus thermoglucosidans STB02. J. Agric. Food Chem. 2020, 68, 13791–13797. [Google Scholar] [CrossRef]
- Vogt, G.; Woell, S. Protein thermal stability, hydrogen bonds, and ion pairs. J. Mol. Biol. 1997, 269, 631–643. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).